Association between the Presence of Resistance Genes and Sanitiser Resistance of Listeria monocytogenes Isolates Recovered from Different Food-Processing Facilities
Abstract
:1. Introduction
2. Materials and Methods
2.1. L. monocytogenes Isolates and Culture Conditions
2.2. Occurrence of Resistant QAC Genes in Selected L. monocytogenes Isolates
2.3. Minimum Inhibitory Concentration (MIC) Assays
2.4. Genetic Analysis
2.5. Statistical Analysis
3. Results and Discussion
3.1. Occurrence of Genes Conferring Resistance to QACs in the L. monocytogenes Isolates
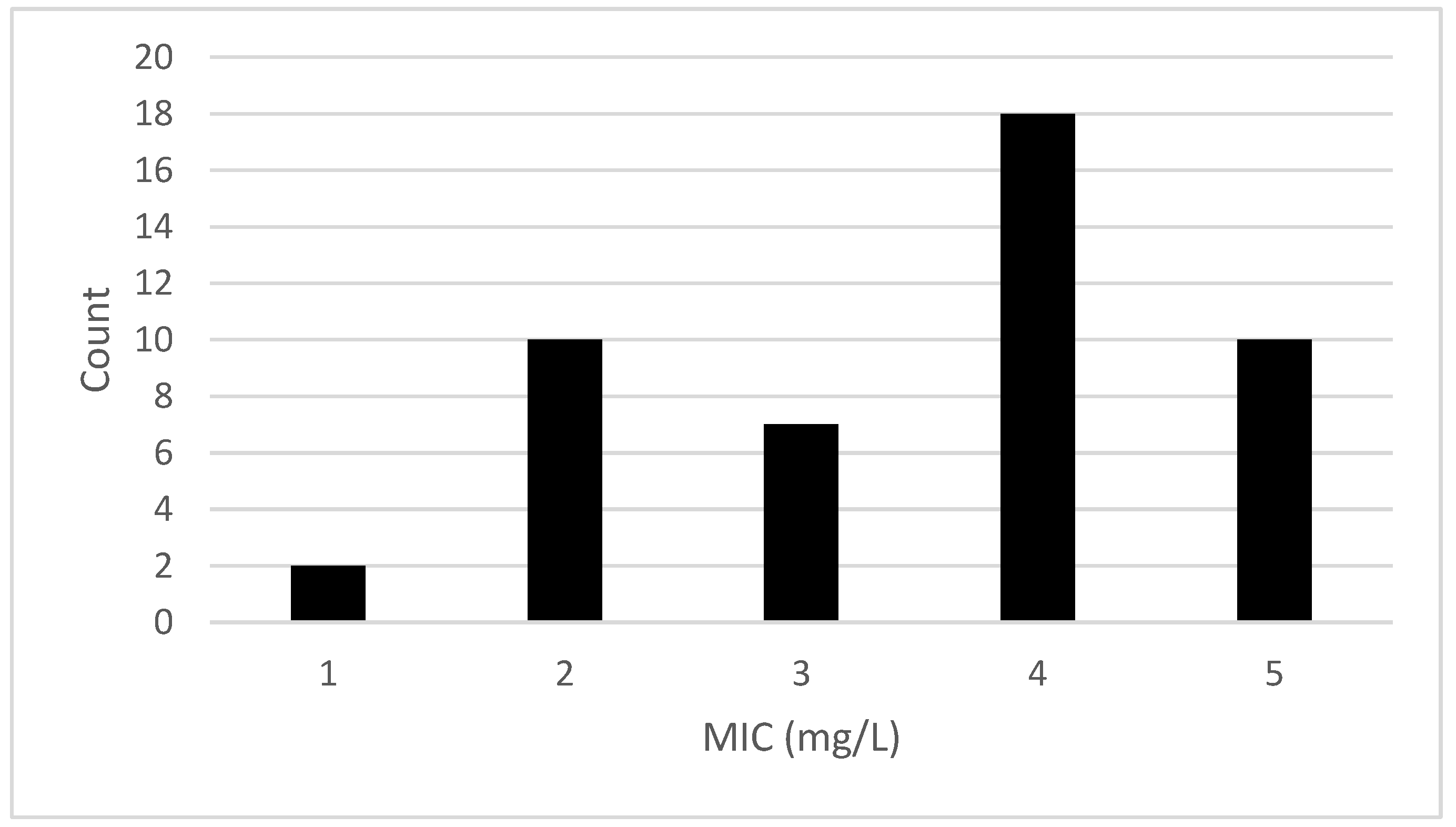
3.2. MIC Analysis
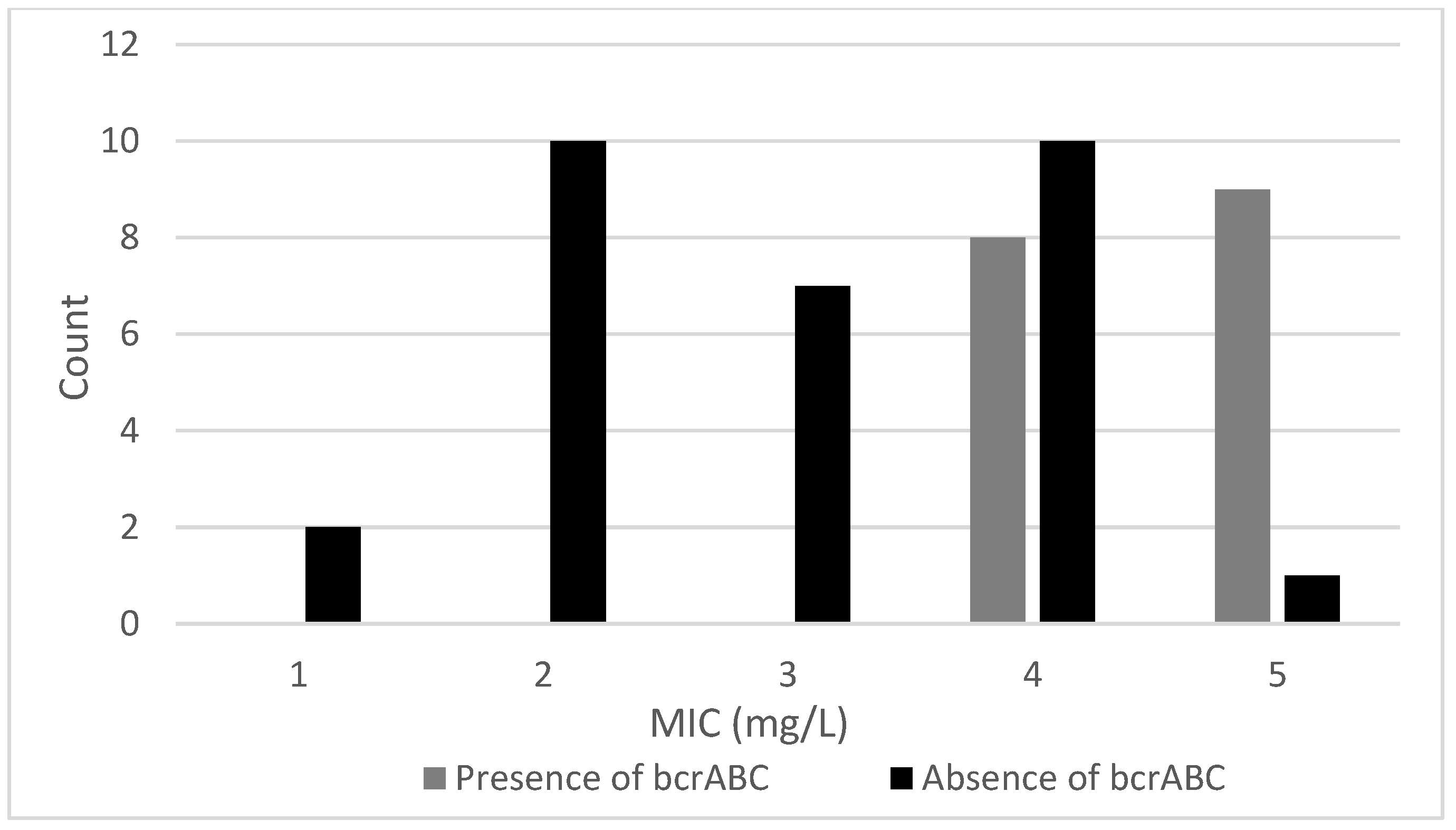
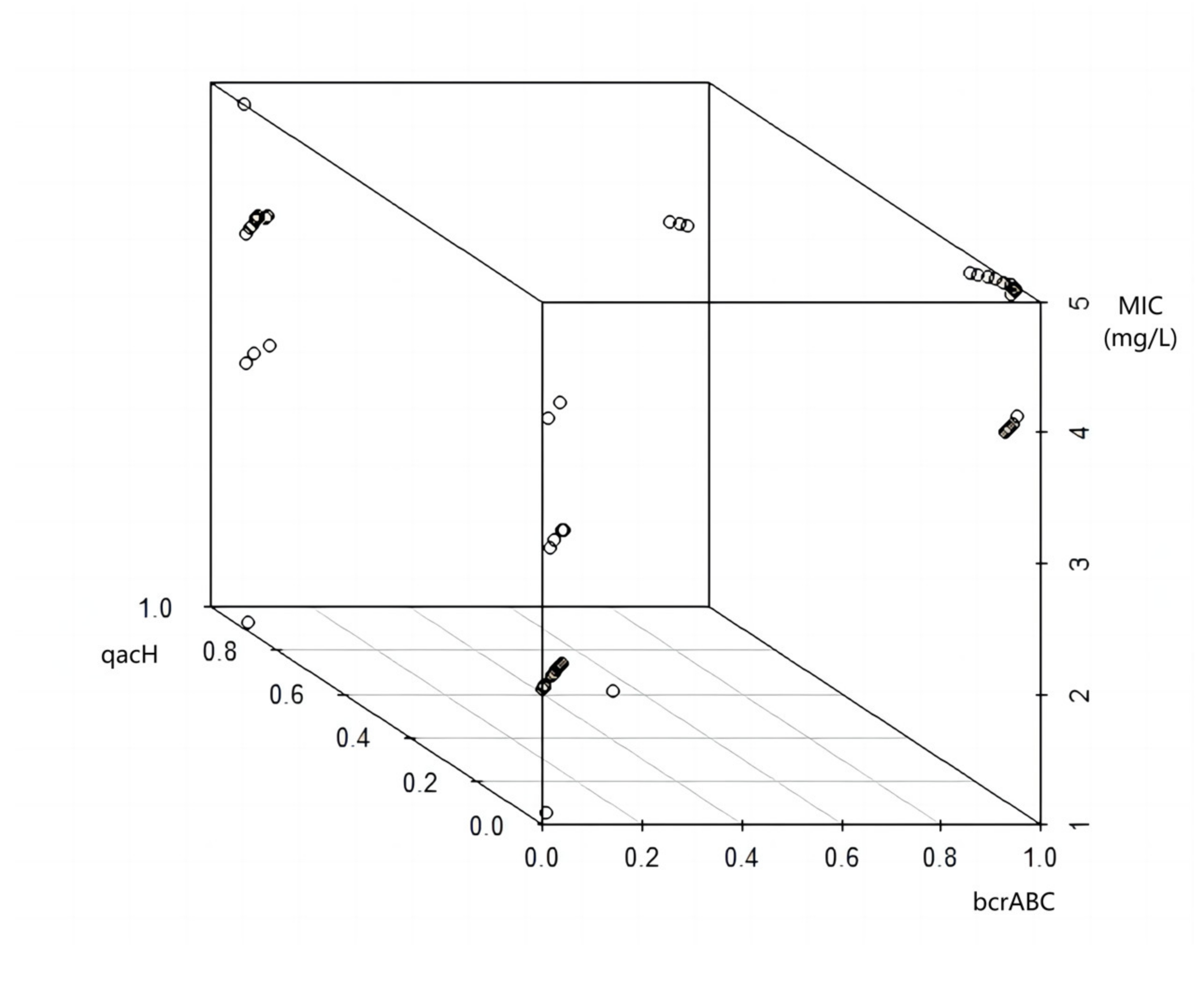
3.2.1. Effects of bcrABC and qacH Genes on BAC Resistance of L. monocytogenes
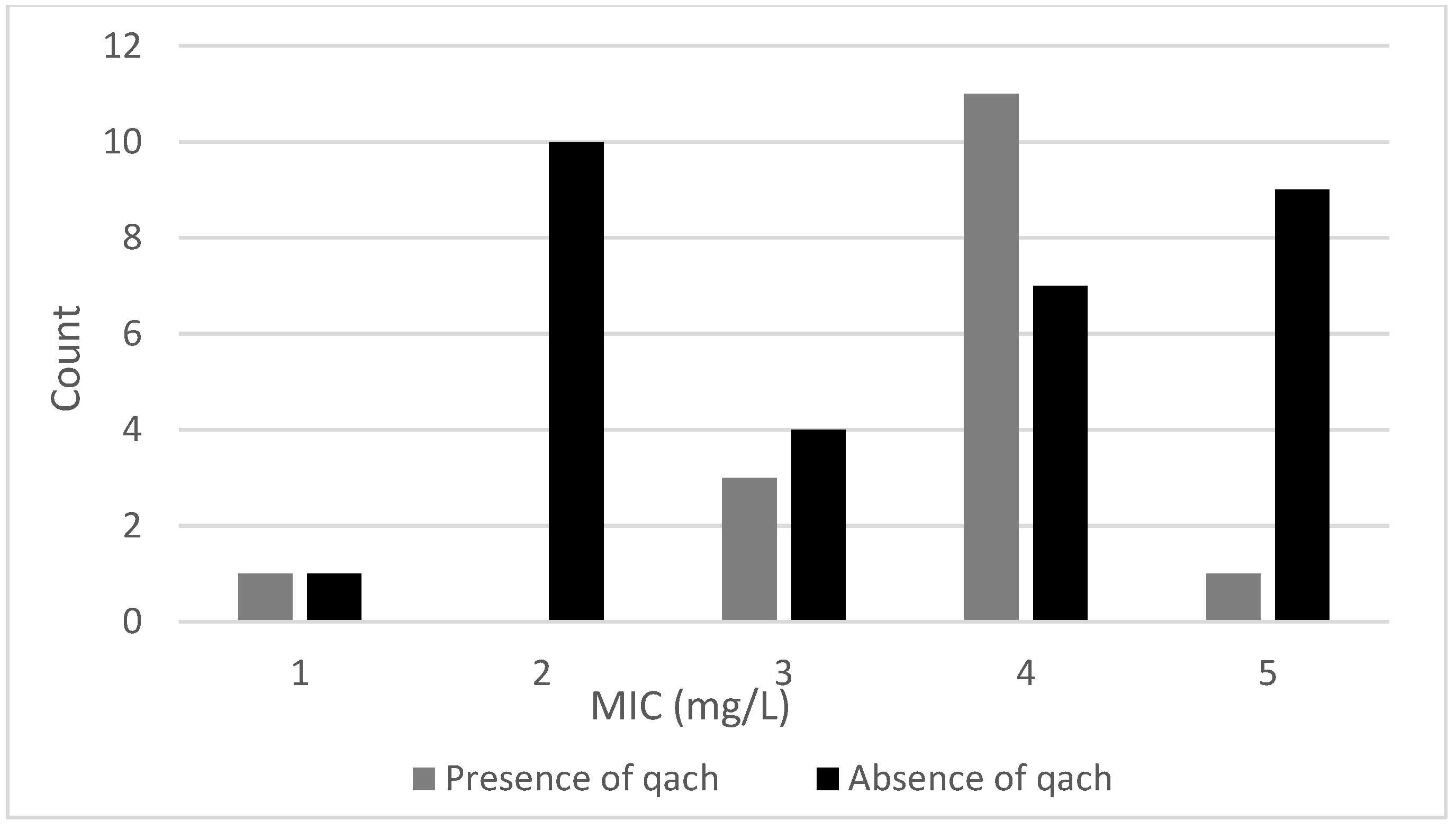
3.2.2. Differences in Sensitivity to BAC between Groups of qacH-Positive Isolates
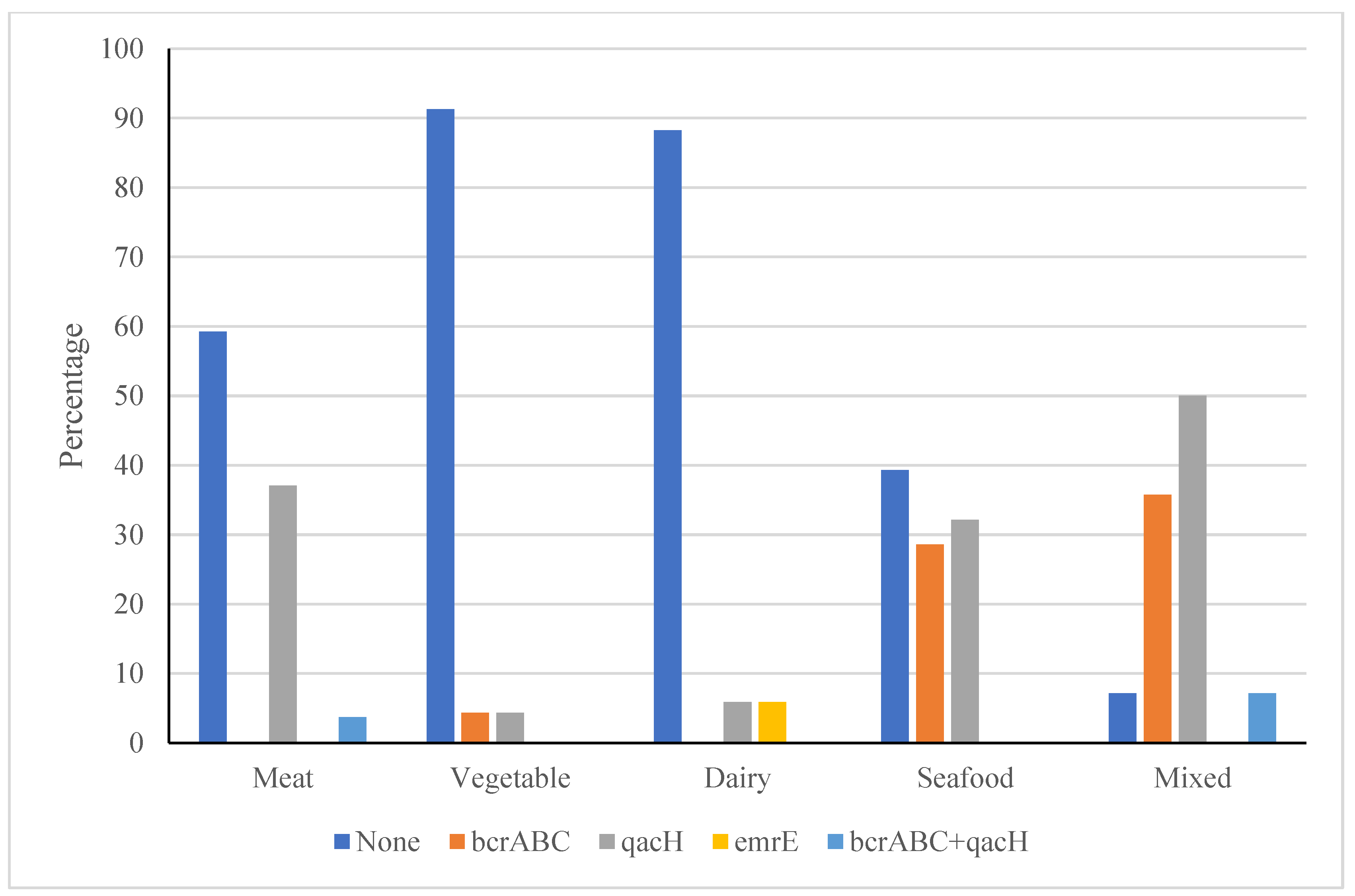
3.2.3. Effect of Process Environment on BAC Resistance of L. monocytogenes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Olaimat, A.N.; Al-Holy, M.A.; Shahbaz, H.M.; Al-Nabulsi, A.A.; Abu Ghoush, M.H.; Osaili, T.M.; Ayyash, M.M.; Holley, R.A. Emergence of Antibiotic Resistance in Listeria monocytogenes Isolated from Food Products: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1277–1292. [Google Scholar] [CrossRef]
- Volokhov, D.; Rasooly, A.; Chumakov, K.; Chizhikov, V. Identification of Listeria Species by Microarray-Based Assay. J. Clin. Microbiol. 2002, 40, 4720–4728. [Google Scholar] [CrossRef]
- Allam, M.; Tau, N.; Smouse Shannon, L.; Mtshali Phillip, S.; Mnyameni, F.; Khumalo Zamantungwa, T.H.; Ismail, A.; Govender, N.; Thomas, J.; Smith Anthony, M. Whole-Genome Sequences of Listeria monocytogenes Sequence Type 6 Isolates Associated with a Large Foodborne Outbreak in South Africa, 2017 to 2018. Genome Announc. 2018, 6, e00538-18. [Google Scholar] [CrossRef]
- Dufour, C. Application of EC regulation no. 2073/2005 regarding Listeria monocytogenes in ready-to-eat foods in retail and catering sectors in Europe. Food Control 2011, 22, 1491–1494. [Google Scholar] [CrossRef]
- Matereke, L.T.; Okoh, A.I. Listeria monocytogenes Virulence, Antimicrobial Resistance and Environmental Persistence: A Review. Pathogens 2020, 9, 528. [Google Scholar] [CrossRef]
- Desai, A.N.; Anyoha, A.; Madoff, L.C.; Lassmann, B. Changing epidemiology of Listeria monocytogenes outbreaks, sporadic cases, and recalls globally: A review of ProMED reports from 1996 to 2018. Int. J. Infect. Dis. 2019, 84, 48–53. [Google Scholar] [CrossRef]
- Mashak, Z.; Banisharif, F.; Banisharif, G.; Reza Pourian, M.; Eskandari, S.; Seif, A.; Safarpoor Dehkordi, F.; Alavi, I. Prevalence of Listeria species and serotyping of Listeria monocytogenes bacteria isolated from seafood samples. Egypt. J. Vet. Sci. 2021, 52, 1–9. [Google Scholar] [CrossRef]
- Matle, I.; Mbatha Khanyisile, R.; Madoroba, E. A review of Listeria monocytogenes from meat and meat products: Epidemiology, virulence factors, antimicrobial resistance and diagnosis. Onderstepoort J. Vet. Res. 2020, 87, 1–20. [Google Scholar]
- Melero, B.; Stessl, B.; Manso, B.; Wagner, M.; Esteban-Carbonero, Ó.J.; Hernández, M.; Rovira, J.; Rodriguez-Lázaro, D. Listeria monocytogenes colonization in a newly established dairy processing facility. Int. J. Food Microbiol. 2019, 289, 64–71. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, S.; Chen, X.; Qu, C. Review controlling Listeria monocytogenes in ready-to-eat meat and poultry products: An overview of outbreaks, current legislations, challenges, and future prospects. Trends Food Sci. Technol. 2021, 116, 24–35. [Google Scholar] [CrossRef]
- Listeria Outbreaks. Available online: https://www.cdc.gov/listeria/outbreaks/index.html (accessed on 1 November 2023).
- Møretrø, T.; Schirmer, B.C.T.; Heir, E.; Fagerlund, A.; Hjemli, P.; Langsrud, S. Tolerance to quaternary ammonium compound disinfectants may enhance growth of Listeria monocytogenes in the food industry. Int. J. Food Microbiol. 2017, 241, 215–224. [Google Scholar] [CrossRef]
- Gerba Charles, P.; Müller, V. Quaternary Ammonium Biocides: Efficacy in Application. Appl. Environ. Microbiol. 2015, 81, 464–469. [Google Scholar] [CrossRef]
- Minarovičová, J.; Véghová, A.; Mikulášová, M.; Chovanová, R.; Šoltýs, K.; Drahovská, H.; Kaclíková, E. Benzalkonium chloride tolerance of Listeria monocytogenes strains isolated from a meat processing facility is related to presence of plasmid-borne bcrABC cassette. Antonie Van Leeuwenhoek 2018, 111, 1913–1923. [Google Scholar] [CrossRef]
- Kim, M.; Weigand Michael, R.; Oh, S.; Hatt Janet, K.; Krishnan, R.; Tezel, U.; Pavlostathis Spyros, G.; Konstantinidis Konstantinos, T.; Dozois Charles, M. Widely Used Benzalkonium Chloride Disinfectants Can Promote Antibiotic Resistance. Appl. Environ. Microbiol. 2018, 84, e01201-18. [Google Scholar] [CrossRef]
- Duze, S.T.; Marimani, M.; Patel, M. Tolerance of Listeria monocytogenes to biocides used in food processing environments. Food Microbiol. 2021, 97, 103758. [Google Scholar] [CrossRef]
- Tong, C.; Hu, H.; Chen, G.; Li, Z.; Li, A.; Zhang, J. Disinfectant resistance in bacteria: Mechanisms, spread, and resolution strategies. Environ. Res. 2021, 195, 110897. [Google Scholar]
- Romanova, N.A.; Wolffs, P.F.G.; Brovko, L.Y.; Griffiths, M.W. Role of Efflux Pumps in Adaptation and Resistance of Listeria monocytogenes to Benzalkonium Chloride. Appl. Environ. Microbiol. 2006, 72, 3498. [Google Scholar]
- Mereghetti, L.; Quentin, R.; Marquet-Van Der Mee, N.; Audurier, A. Low Sensitivity of Listeria monocytogenes to Quaternary Ammonium Compounds. Appl. Environ. Microbiol. 2000, 66, 5083. [Google Scholar]
- Haubert, L.; Zehetmeyr, M.L.; da Silva, W.P. Resistance to benzalkonium chloride and cadmium chloride in Listeria monocytogenes isolates from food and food-processing environments in southern Brazil. Can. J. Microbiol. 2019, 65, 429–435. [Google Scholar]
- Tamburro, M.; Ripabelli, G.; Vitullo, M.; Dallman, T.J.; Pontello, M.; Amar, C.F.L.; Sammarco, M.L. Gene expression in Listeria monocytogenes exposed to sublethal concentration of benzalkonium chloride. Comp. Immunol. Microbiol. Infect. Dis. 2015, 40, 31–39. [Google Scholar] [CrossRef]
- Dutta, V.; Elhanafi, D.; Kathariou, S. Conservation and distribution of the benzalkonium chloride resistance cassette bcrABC in Listeria monocytogenes. Appl. Environ. Microbiol. 2013, 79, 6067–6074. [Google Scholar]
- Müller, A.; Rychli, K.; Muhterem-Uyar, M.; Zaiser, A.; Stessl, B.; Guinane, C.M.; Cotter, P.D.; Wagner, M.; Schmitz-Esser, S. Tn6188—A novel transposon in Listeria monocytogenes responsible for tolerance to benzalkonium chloride. PLoS ONE 2013, 8, e76835. [Google Scholar]
- Kovacevic, J.; Ziegler, J.; Wałecka-Zacharska, E.; Reimer, A.; Kitts, D.D.; Gilmour, M.W. Tolerance of Listeria monocytogenes to quaternary ammonium sanitizers is mediated by a novel efflux pump encoded by emrE. Appl. Environ. Microbiol. 2016, 82, 939–953. [Google Scholar]
- Roedel, A.; Dieckmann, R.; Brendebach, H.; Hammerl, J.A.; Kleta, S.; Noll, M.; Al Dahouk, S.; Vincze, S. Biocide-tolerant Listeria monocytogenes isolates from German food production plants do not show cross-resistance to clinically relevant antibiotics. Appl. Environ. Microbiol. 2019, 85, e01253-19. [Google Scholar]
- Gilmour, M.W.; Graham, M.; Van Domselaar, G.; Tyler, S.; Kent, H.; Trout-Yakel, K.M.; Larios, O.; Allen, V.; Lee, B.; Nadon, C. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genom. 2010, 11, 120. [Google Scholar]
- Yamamoto, T.; Hara, H.; Tsuchiya, K.; Sakai, S.; Fang, R.; Matsuura, M.; Nomura, T.; Sato, F.; Mitsuyama, M.; Kawamura, I.; et al. Listeria monocytogenes strain-specific impairment of the TetR regulator underlies the drastic increase in cyclic di-AMP secretion and beta interferon-inducing ability. Infect. Immun. 2012, 80, 2323–2332. [Google Scholar]
- Schug, A.R.; Bartel, A.; Scholtzek, A.D.; Meurer, M.; Brombach, J.; Hensel, V.; Fanning, S.; Schwarz, S.; Feßler, A.T. Biocide susceptibility testing of bacteria: Development of a broth microdilution method. Vet. Microbiol. 2020, 248, 108791. [Google Scholar]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 1–9. [Google Scholar]
- Seemann T, Mlst. Github. Available online: https://github.com/tseemann/mlst (accessed on 11 January 2023).
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 1–15. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar]
- Kelly, B.G.; Vespermann, A.; Bolton, D.J. The role of horizontal gene transfer in the evolution of selected foodborne bacterial pathogens. Food Chem. Toxicol. 2009, 47, 951–968. [Google Scholar]
- Nordholt, N.; Kanaris, O.; Schmidt, S.B.I.; Schreiber, F. Persistence against benzalkonium chloride promotes rapid evolution of tolerance during periodic disinfection. Nat. Commun. 2021, 12, 6792. [Google Scholar]
- Cloete, T.E. Resistance mechanisms of bacteria to antimicrobial compounds. Int. Biodeterior. Biodegrad. 2003, 51, 277–282. [Google Scholar]
- Yu, T.; Jiang, X.; Zhang, Y.; Ji, S.; Gao, W.; Shi, L. Effect of Benzalkonium Chloride Adaptation on Sensitivity to Antimicrobial Agents and Tolerance to Environmental Stresses in Listeria monocytogenes. Front. Microbiol. 2018, 9, 2906. [Google Scholar]
- Zhu, Z.; Shan, L.; Zhang, X.; Hu, F.; Zhong, D.; Yuan, Y.; Zhang, J. Effects of bacterial community composition and structure in drinking water distribution systems on biofilm formation and chlorine resistance. Chemosphere 2021, 264, 128410. [Google Scholar]
- Cruz, C.D.; Fletcher, G.C. Assessing manufacturers’ recommended concentrations of commercial sanitizers on inactivation of Listeria monocytogenes. Food Control 2012, 26, 194–199. [Google Scholar]
- Martínez-Suárez, J.V.; Ortiz, S.; López-Alonso, V. Potential Impact of the Resistance to Quaternary Ammonium Disinfectants on the Persistence of Listeria monocytogenes in Food Processing Environments. Front. Microbiol. 2016, 7, 638. [Google Scholar]
- Van Acker, H.; Van Dijck, P.; Coenye, T. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 2014, 22, 326–333. [Google Scholar]
- Romanova, N.A.; Gawande, P.V.; Brovko, L.Y.; Griffiths, M.W. Rapid methods to assess sanitizing efficacy of benzalkonium chloride to Listeria monocytogenes biofilms. J. Microbiol. Methods 2007, 71, 231–237. [Google Scholar]


| Resistance Gene | GenBank Number | Size | Isolation Source | Reference |
|---|---|---|---|---|
| bcrABC | JX023284.1 | 1406 bp | Food-processing plant | [22] |
| qacH | MK944277.1 | 378 bp | Food-processing environment | [25] |
| emrE | CP001602.2 | 324 bp | RTE food | [26] |
| mdrL | AB671769.1 | 1194 bp | Yoshihiro Asano Ehime University, Ehime, Japan | [27] |
| Resistance Gene | mdrL | bcrABC | qacH | emrE |
|---|---|---|---|---|
| Number of isolates | 150 | 23 | 40 | 1 |
| Isolate No. | CC | ST | Resistance Genes | Source | MIC (mg/L) | |||
|---|---|---|---|---|---|---|---|---|
| mdrL | bcrABC | qacH | emrE | |||||
| 1564 | 2 | 2 | + | + | + | − | Meat | 4 |
| 1387 | 2 | 2 | + | − | − | − | Seafood | 3 |
| 2183 | 2 | 2 | + | − | − | − | Vegetables | 2 |
| 1413 | 2 | 2 | + | − | − | − | Vegetables | 3 |
| 1374 | 3 | 3 | + | − | − | − | Meat | 3 |
| 1382 | 4 | 4 | + | − | − | − | Dairy | 2 |
| F1524_16 | 5 | 5 | + | + | − | − | Mixed food | 4 |
| 1386 | 5 | 5 | + | + | − | − | Seafood | 5 |
| F347_16 | 5 | 5 | + | + | − | − | Mixed food | 5 |
| F742_17 | 5 | 5 | + | + | − | − | Mixed food | 5 |
| F991_16 | 5 | 5 | + | + | − | − | Mixed food | 5 |
| F1857_15 | 5 | 5 | + | + | − | − | Mixed food | 5 |
| F2166_17 | 5 | 5 | + | + | − | − | Mixed food | 5 |
| F2299_15 | 5 | 5 | + | + | − | − | Mixed food | 5 |
| 1392 | 6 | 6 | + | − | − | − | Seafood | 2 |
| 1445 | 7 | 7 | + | − | − | − | Meat | 2 |
| 1880 | 8 | 8 | + | − | − | − | Vegetables | 2 |
| 958 | 9 | 9 | + | + | − | − | Vegetables | 4 |
| 1021 | 9 | 9 | + | − | − | + | Dairy | 4 |
| 1370 | 9 | 9 | + | − | − | − | Meat | 3 |
| 1379 | 14 | 14 | + | − | − | − | Dairy | 2 |
| 1389 | 31 | 31 | + | + | − | − | Seafood | 4 |
| 1095 | 37 | 37 | + | − | − | − | Mixed food | 2 |
| 1006 | 37 | 37 | + | − | − | − | Meat | 1 |
| 2256 | 121 | 121 | + | − | + | − | Vegetables | 5 |
| 1123 | 121 | 121 | + | − | + | − | Seafood | 4 |
| 1373 | 121 | 121 | + | − | + | − | Meat | 3 |
| 1513 | 121 | 121 | + | − | + | − | Seafood | 1 |
| 1989 | 121 | 121 | + | − | + | − | Seafood | 4 |
| 6179 | 121 | 121 | + | − | + | − | Dairy | 4 |
| F112_17 | 121 | 121 | + | − | + | − | Mixed food | 3 |
| F347_17 | 121 | 121 | + | − | + | − | Mixed food | 4 |
| F1230_16 | 121 | 121 | + | − | + | − | Meat | 4 |
| F1646_17 | 121 | 121 | + | − | + | − | Mixed food | 4 |
| F2151_17 | 121 | 121 | + | − | + | − | Meat | 4 |
| F2152_17 | 121 | 121 | + | − | + | − | Meat | 4 |
| F2165_17 | 121 | 121 | + | − | + | − | Mixed food | 3 |
| F113_17 | 121 | 132 | + | + | + | − | Mixed food | 4 |
| F1099_17 | 121 | 132 | + | + | + | − | Mixed food | 4 |
| 1385 | 155 | 155 | + | − | − | − | Seafood | 2 |
| 1679 | 204 | 204 | + | + | − | − | Seafood | 4 |
| 1309 | 204 | 204 | + | − | − | − | Seafood | 4 |
| 2226 | 220 | 220 | + | − | − | − | Vegetables | 2 |
| 1268 | 224 | 224 | + | − | − | − | Dairy | 2 |
| 1306 | NOVEL | 836 | + | + | − | − | Seafood | 4 |
| 1427 | NOVEL | 836 | + | + | − | − | Seafood | 5 |
| 1428 | NOVEL | 836 | + | + | − | − | Seafood | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, Y.; Mousavi, Z.E.; Pennone, V.; Hurley, D.; Butler, F. Association between the Presence of Resistance Genes and Sanitiser Resistance of Listeria monocytogenes Isolates Recovered from Different Food-Processing Facilities. Microorganisms 2023, 11, 2989. https://doi.org/10.3390/microorganisms11122989
Cheng Y, Mousavi ZE, Pennone V, Hurley D, Butler F. Association between the Presence of Resistance Genes and Sanitiser Resistance of Listeria monocytogenes Isolates Recovered from Different Food-Processing Facilities. Microorganisms. 2023; 11(12):2989. https://doi.org/10.3390/microorganisms11122989
Chicago/Turabian StyleCheng, Yue, Zeinabossadat Ebrahimzadeh Mousavi, Vincenzo Pennone, Daniel Hurley, and Francis Butler. 2023. "Association between the Presence of Resistance Genes and Sanitiser Resistance of Listeria monocytogenes Isolates Recovered from Different Food-Processing Facilities" Microorganisms 11, no. 12: 2989. https://doi.org/10.3390/microorganisms11122989
APA StyleCheng, Y., Mousavi, Z. E., Pennone, V., Hurley, D., & Butler, F. (2023). Association between the Presence of Resistance Genes and Sanitiser Resistance of Listeria monocytogenes Isolates Recovered from Different Food-Processing Facilities. Microorganisms, 11(12), 2989. https://doi.org/10.3390/microorganisms11122989







