Supplementation of Sulfide or Acetate and 2-Mercaptoethane Sulfonate Restores Growth of the Methanosarcina acetivorans ΔhdrABC Deletion Mutant during Methylotrophic Methanogenesis
Abstract
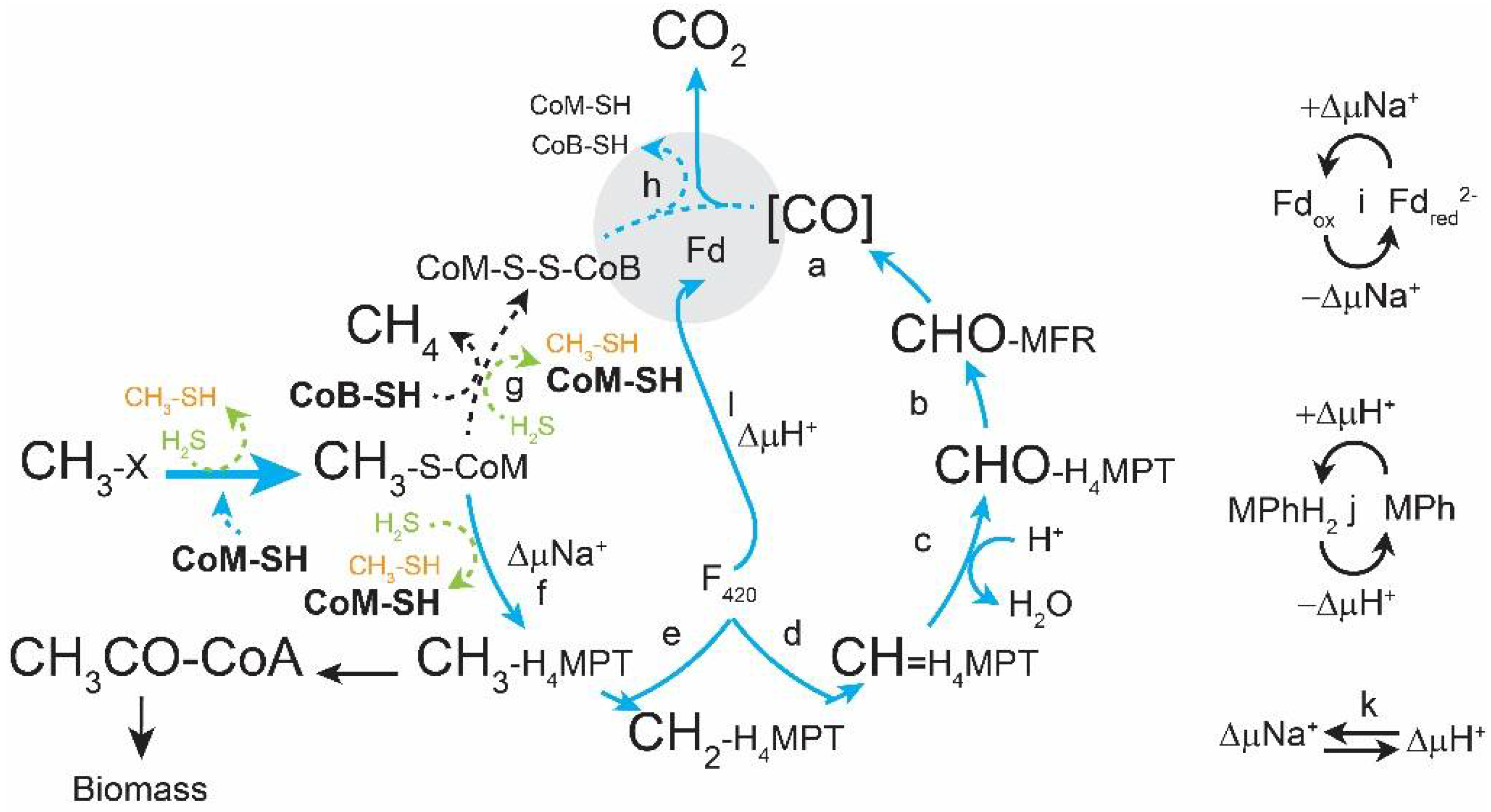
:1. Introduction
2. Materials and Methods
2.1. Culture Conditions
2.2. Strain Validation
2.3. Genome-Scale Metabolic Model of Methanosarcina acetivorans
3. Results
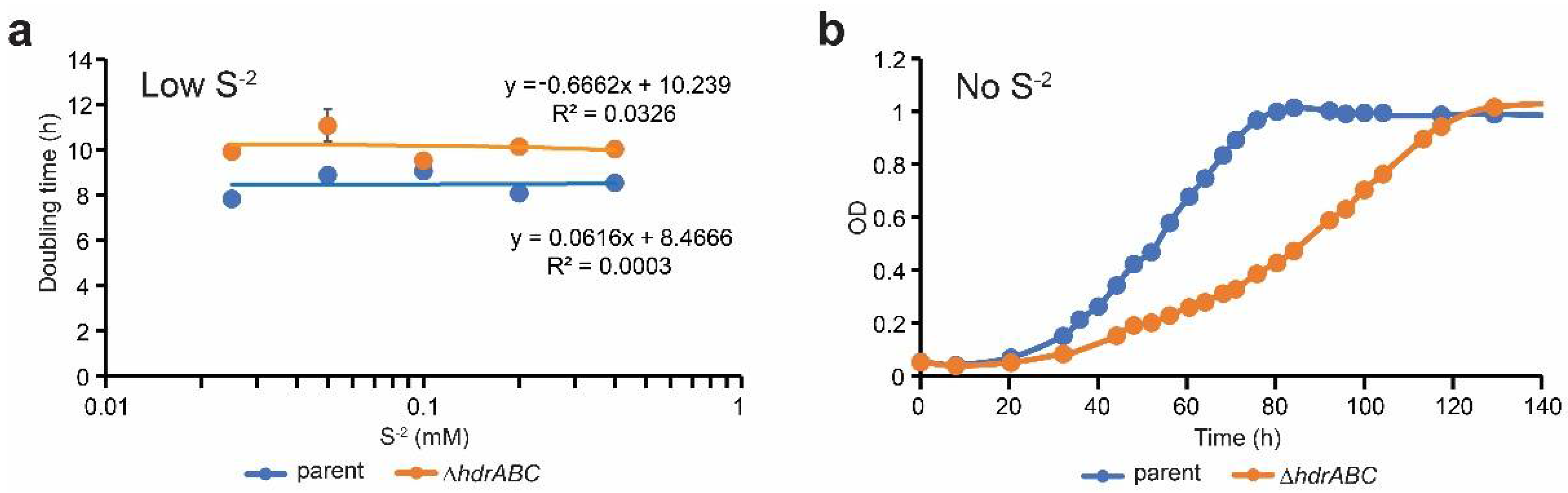
3.1. Sulfide Partially Rescues the ΔhdrABC Mutant Phenotype on Methanol
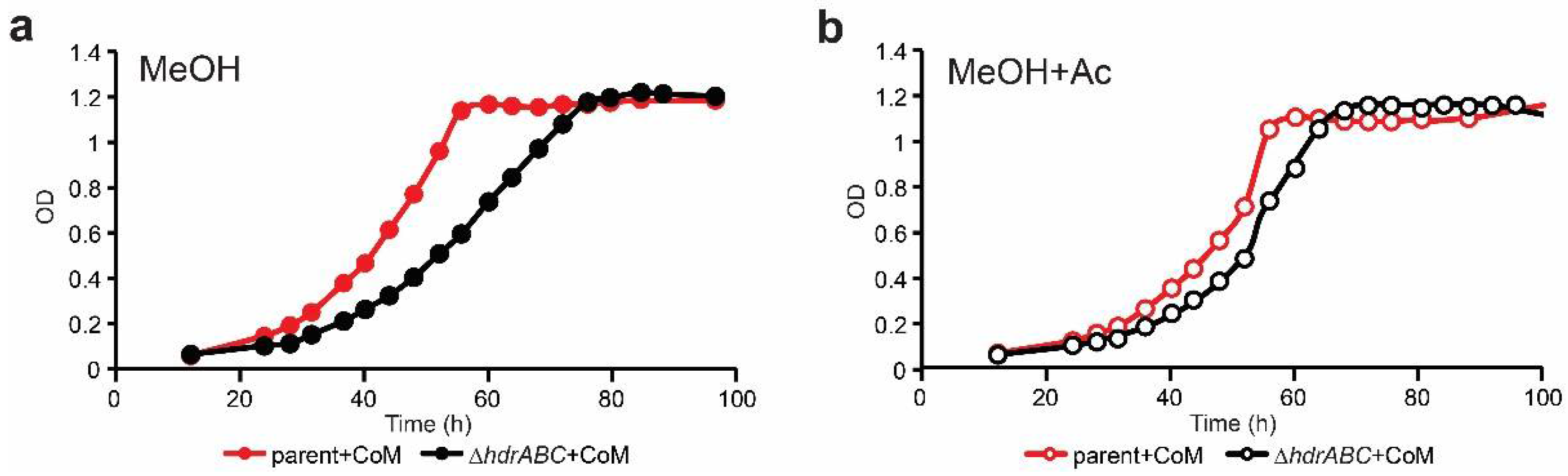
3.2. Mixotrophic Growth on Methanol + Acetate Does Not Rescue the DhdrABC Mutant Phenotype
3.3. CoM Supplementation Alone Does Not Improve Methylotrophic Growth
3.4. CoM-SH and Acetate Supplementation Phenotypically Compensate for Lack of HdrABC
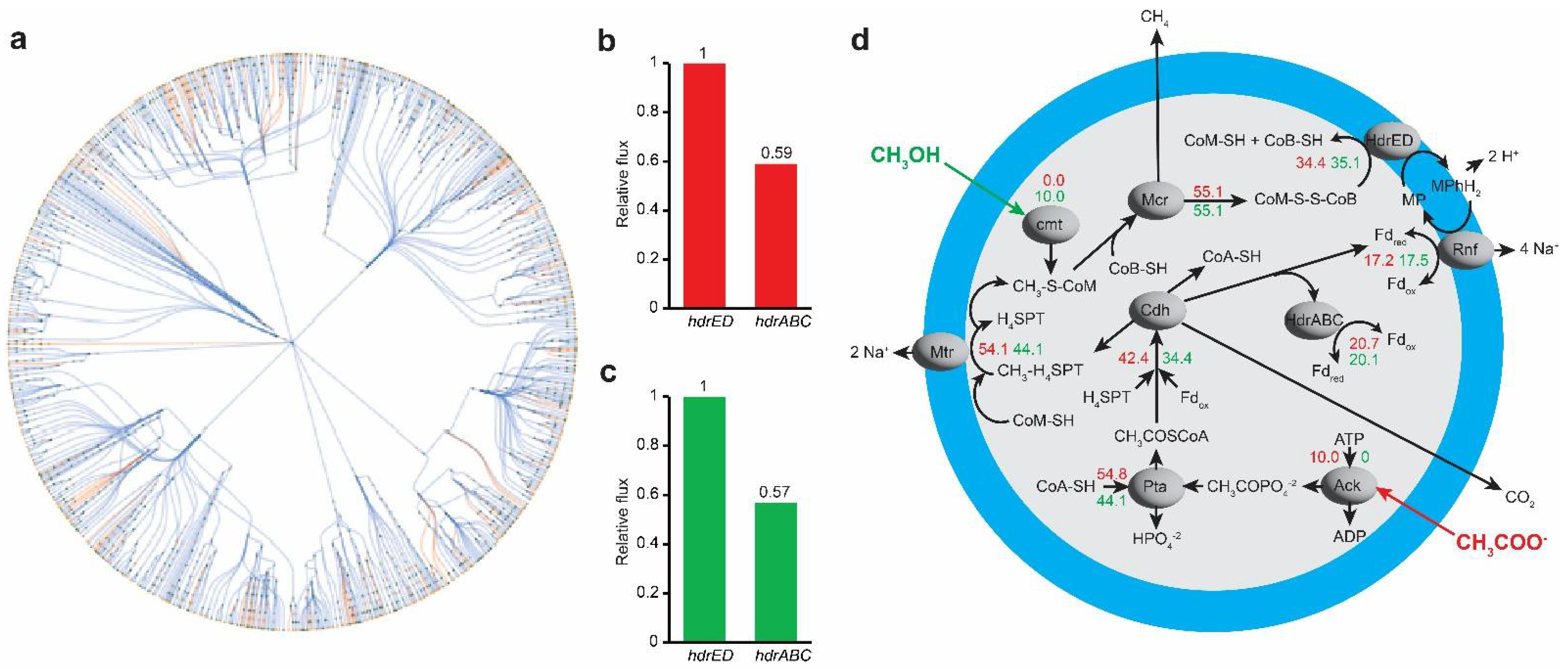
3.5. In Silico Analysis for Metabolic Bypass of HdrABC Activity by Acetate and Sulfide
4. Discussion
5. Summary and Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Buan, N.R.; Metcalf, W.W. Methanogenesis by Methanosarcina acetivorans involves two structurally and functionally distinct classes of heterodisulfide reductase. Mol. Microbiol. 2010, 75, 843–853. [Google Scholar] [CrossRef]
- Catlett, J.L.; Ortiz, A.M.; Buan, N.R. Rerouting Cellular Electron Flux To Increase the Rate of Biological Methane Production. Appl. Environ. Microbiol. 2015, 81, 6528–6537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buan, N.R. Methanogens: Pushing the boundaries of biology. Emerg. Top. Life Sci. 2018, 2, 629–646. [Google Scholar] [CrossRef] [Green Version]
- Sowers, K.R.; Boone, J.E.; Gunsalus, R.P. Disaggregation of Methanosarcina spp. and Growth as Single Cells at Elevated Osmolarity. Appl. Environ. Microbiol. 1993, 59, 3832–3839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horne, A.J.; Lessner, D.J. Assessment of the oxidant tolerance of Methanosarcina acetivorans. FEMS Microbiol. Lett. 2013, 343, 13–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jennings, M.E.; Schaff, C.W.; Horne, A.J.; Lessner, F.H.; Lessner, D.J. Expression of a bacterial catalase in a strictly anaerobic methanogen significantly increases tolerance to hydrogen peroxide but not oxygen. Microbiology 2014, 160, 270–278. [Google Scholar] [CrossRef] [Green Version]
- Guss, A.M.; Rother, M.; Zhang, J.K.; Kulkarni, G.; Metcalf, W.W. New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea 2008, 2, 193–203. [Google Scholar] [CrossRef] [Green Version]
- Peterson, J.R.; Thor, S.; Kohler, L.; Kohler, P.R.A.; Metcalf, W.W.; Luthey-Schulten, Z. Genome-wide gene expression and RNA half-life measurements allow predictions of regulation and metabolic behavior in Methanosarcina acetivorans. BMC Genom. 2016, 17, 924. [Google Scholar] [CrossRef] [Green Version]
- Lewis, N.E.; Hixson, K.K.; Conrad, T.M.; Lerman, J.A.; Charusanti, P.; Polpitiya, A.D.; Adkins, J.N.; Schramm, G.; Purvine, S.O.; Lopez-Ferrer, D.; et al. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol. Syst. Biol. 2010, 6, 390. [Google Scholar] [CrossRef]
- Rauch, B.J.; Perona, J.J. Efficient Sulfide Assimilation in Methanosarcina acetivorans Is Mediated by the MA1715 Protein. J. Bacteriol. 2016, 198, 1974–1983. [Google Scholar] [CrossRef]
- Hari, A.; Lobo, D. Fluxer: A web application to compute, analyze and visualize genome-scale metabolic flux networks. Nucleic Acids Res. 2020, 48, W427–W435. [Google Scholar] [CrossRef]
- Li, Q.; Li, L.; Rejtar, T.; Karger, B.L.; Ferry, J.G. Proteome of Methanosarcina acetivorans Part II: Comparison of protein levels in acetate- and methanol-grown cells. J. Proteome. Res. 2005, 4, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Galagan, J.E.; Nusbaum, C.; Roy, A.; Endrizzi, M.G.; Macdonald, P.; FitzHugh, W.; Calvo, S.; Engels, R.; Smirnov, S.; Atnoor, D.; et al. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 2002, 12, 532–542. [Google Scholar] [CrossRef] [Green Version]
- Catlett, J.L.; Carr, S.; Cashman, M.; Smith, M.D.; Walter, M.; Sakkaff, Z.; Kelley, C.; Pierobon, M.; Cohen, M.B.; Buan, N.R. Metabolic Synergy between Human Symbionts Bacteroides and Methanobrevibacter. Microbiol. Spectr. 2022, 10, e0106722. [Google Scholar] [CrossRef] [PubMed]
- Catlett, J.L.; Catazaro, J.; Cashman, M.; Carr, S.; Powers, R.; Cohen, M.B.; Buan, N.R. Metabolic Feedback Inhibition Influences Metabolite Secretion by the Human Gut Symbiont Bacteroides thetaiotaomicron. mSystems 2020, 5, e00252-20. [Google Scholar] [CrossRef]
- Chandrasekaran, S.; Price, N.D. Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2010, 107, 17845–17850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Rai, N.; Zorraquino, V.; Tagkopoulos, I. Multi-omics integration accurately predicts cellular state in unexplored conditions for Escherichia coli. Nat. Commun. 2016, 7, 13090. [Google Scholar] [CrossRef] [Green Version]
- Jasso-Chavez, R.; Apolinario, E.E.; Sowers, K.R.; Ferry, J.G. MrpA Functions in Energy Conversion during Acetate-Dependent Growth of Methanosarcina acetivorans. J. Bacteriol. 2013, 195, 3987–3994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vepachedu, V.R.; Ferry, J.G. Role of the fused corrinoid/methyl transfer protein CmtA during CO-dependent growth of Methanosarcina acetivorans. J. Bacteriol. 2012, 194, 4161–4168. [Google Scholar] [CrossRef] [Green Version]
- Rohlin, L.; Gunsalus, R.P. Carbon-dependent control of electron transfer and central carbon pathway genes for methane biosynthesis in the Archaean, Methanosarcina acetivorans strain C2A. BMC Microbiol. 2010, 10, 62. [Google Scholar] [CrossRef]
- Bose, A.; Kulkarni, G.; Metcalf, W.W. Regulation of putative methyl-sulphide methyltransferases in Methanosarcina acetivorans C2A. Mol. Microbiol. 2009, 74, 227–238. [Google Scholar] [CrossRef]
- Bose, A.; Pritchett, M.A.; Metcalf, W.W. Genetic analysis of the methanol- and methylamine-specific methyltransferase 2 genes of Methanosarcina acetivorans C2A. J. Bacteriol. 2008, 190, 4017–4026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.Y.; Li, Q.B.; Rohlin, L.; Kim, U.; Salmon, K.; Rejtar, T.; Gunsalus, R.P.; Karger, B.L.; Ferry, J.G. Quantitative proteomic and microarray analysis of the archaeon Methanosarcina acetivorans grown with acetate versus methanol. J. Proteome Res. 2007, 6, 759–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sprenger, W.W.; Hackstein, J.H.; Keltjens, J.T. The competitive success of Methanomicrococcus blatticola, a dominant methylotrophic methanogen in the cockroach hindgut, is supported by high substrate affinities and favorable thermodynamics. FEMS Microbiol. Ecol. 2007, 60, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Samuel, B.S.; Hansen, E.E.; Manchester, J.K.; Coutinho, P.M.; Henrissat, B.; Fulton, R.; Latreille, P.; Kim, K.; Wilson, R.K.; Gordon, J.I. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc. Natl. Acad. Sci. USA 2007, 104, 10643–10648. [Google Scholar] [CrossRef] [Green Version]
- Roden, E.E.; Wetzel, R.G. Competition between Fe(III)-reducing and methanogenic bacteria for acetate in iron-rich freshwater sediments. Microb. Ecol. 2003, 45, 252–258. [Google Scholar] [CrossRef]


| Primers | |||
|---|---|---|---|
| Name | Sequence | Purpose | Source |
| oNB121 | GCACCCAGGCACATTGTTC | hdrA 301 rev | [2] |
| oNB122 | TACTGGGGTTTCTGGGAGAC | hdrA 1024 rev | [2] |
| oNB123 | ATGCCCTCTCCGTAAATGAG | hdrA 1880 fwd | [2] |
| oNB124 | GATTCAAGCACACTGCGATC | hdrC 2616 rev | [2] |
| Methanosarcina acetivorans C2A strains | |||
| 34 | parent | Δhpt::φC31 int attP | [7] |
| 36 | ΔhdrABC | Δhpt::φC31 int attP ΔhdrA1B1C1 | [1] |
| Strain | S−2 (mM) | Doubling Time (h) a | Std Dev a | P vs. Parent b | P vs. DhdrABC b |
|---|---|---|---|---|---|
| parent | 0 c | 15.3 | 1.95 | 0.003 | 0.014 |
| 0.025 | 7.8 | 0.27 | 0.003 | 0.001 | |
| 0.05 | 8.9 | 0.29 | 0.478 | 0.026 | |
| 0.1d | 9.1 | 0.33 | 1 | 0.126 | |
| 0.2 | 8.1 | 0.32 | 0.010 | 0.002 | |
| 0.4 | 8.5 | 0.21 | 0.038 | 0.001 | |
| DhdrABC | 0 c | 27.8 | 3.62 | 0.001 | 0.006 |
| 0.025 | 9.9 | 0.24 | 0.023 | 0.152 | |
| 0.05 | 11.1 | 0.73 | 0.005 | 0.031 | |
| 0.1 d | 9.5 | 0.21 | 0.126 | 1 | |
| 0.2 | 10.1 | 0.20 | 0.009 | 0.042 | |
| 0.4 | 10.0 | 0.09 | 0.343 | 0.034 |
| Strain | Energy Source a | CoM-SH b | Doubling Time (h) c | Std Dev c | P vs. Parent d,e | P vs. ΔhdrABC e,f |
|---|---|---|---|---|---|---|
| parent | MeOH | − | 9.2 | 0.35 | 1 | |
| + | 10.1 | 0.15 | 0.01 | |||
| MeOH+Ac | − | 10.8 | 0.14 | 0.00 | ||
| + | 10.3 | 0.21 | 0.00 | |||
| DhdrABC | MeOH | − | 12.6 | 1.01 | 0.00 | 1 |
| + | 11.5 | 0.22 | 0.00 | 0.20 | ||
| MeOH+Ac | − | 11.8 | 0.26 | 0.00 | 0.18 | |
| + | 10.1 | 0.29 | 0.68 | 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salvi, A.M.; Chowdhury, N.B.; Saha, R.; Buan, N.R. Supplementation of Sulfide or Acetate and 2-Mercaptoethane Sulfonate Restores Growth of the Methanosarcina acetivorans ΔhdrABC Deletion Mutant during Methylotrophic Methanogenesis. Microorganisms 2023, 11, 327. https://doi.org/10.3390/microorganisms11020327
Salvi AM, Chowdhury NB, Saha R, Buan NR. Supplementation of Sulfide or Acetate and 2-Mercaptoethane Sulfonate Restores Growth of the Methanosarcina acetivorans ΔhdrABC Deletion Mutant during Methylotrophic Methanogenesis. Microorganisms. 2023; 11(2):327. https://doi.org/10.3390/microorganisms11020327
Chicago/Turabian StyleSalvi, Alicia M., Niaz Bahar Chowdhury, Rajib Saha, and Nicole R. Buan. 2023. "Supplementation of Sulfide or Acetate and 2-Mercaptoethane Sulfonate Restores Growth of the Methanosarcina acetivorans ΔhdrABC Deletion Mutant during Methylotrophic Methanogenesis" Microorganisms 11, no. 2: 327. https://doi.org/10.3390/microorganisms11020327
APA StyleSalvi, A. M., Chowdhury, N. B., Saha, R., & Buan, N. R. (2023). Supplementation of Sulfide or Acetate and 2-Mercaptoethane Sulfonate Restores Growth of the Methanosarcina acetivorans ΔhdrABC Deletion Mutant during Methylotrophic Methanogenesis. Microorganisms, 11(2), 327. https://doi.org/10.3390/microorganisms11020327






