β-Alanine Metabolism Leads to Increased Extracellular pH during the Heterotrophic Ammonia Oxidation of Pseudomonas putida Y-9
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganism and Culture Media
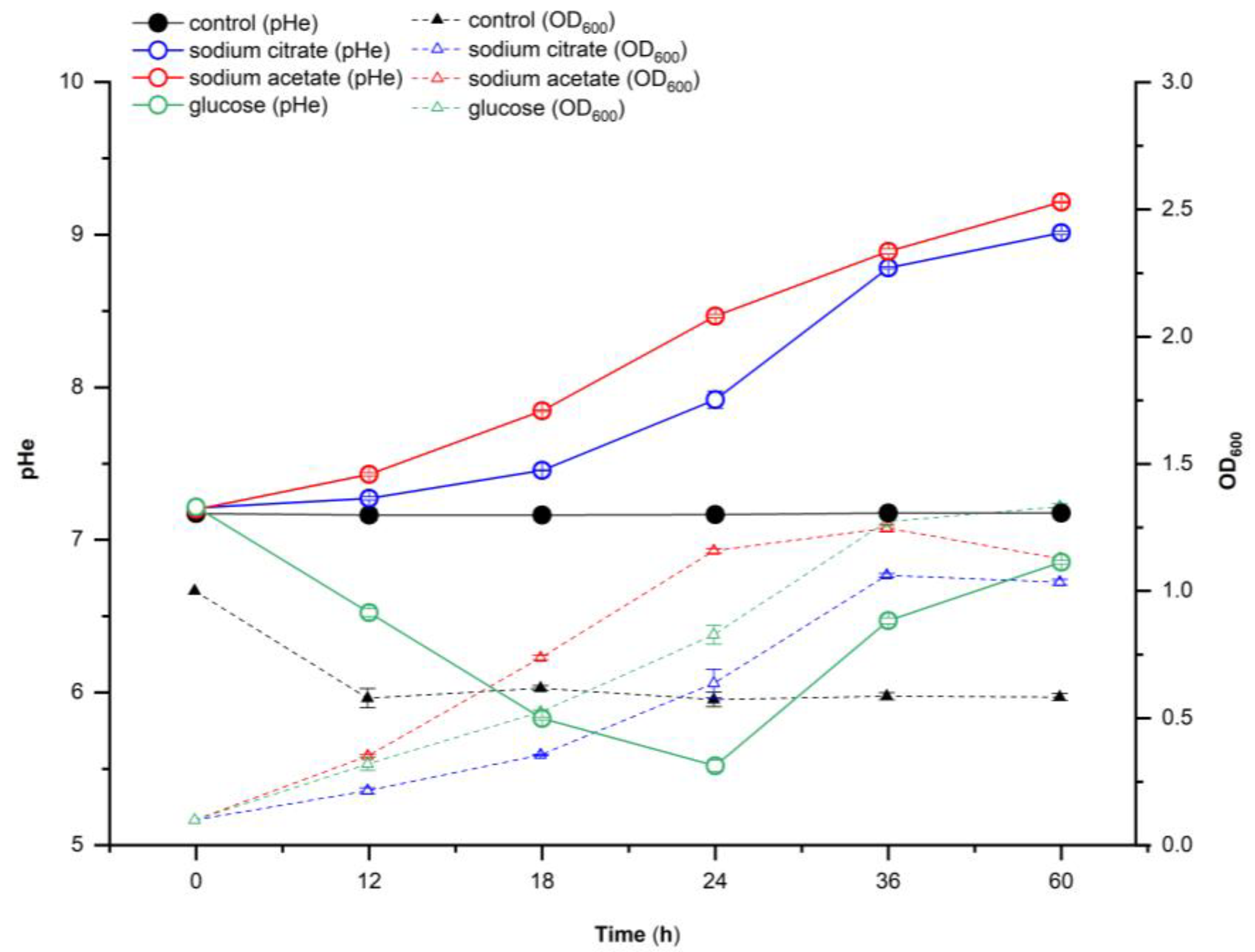
2.2. Effect of Different Carbon Sources on Extracellular pH and Growth of Strain Y-9
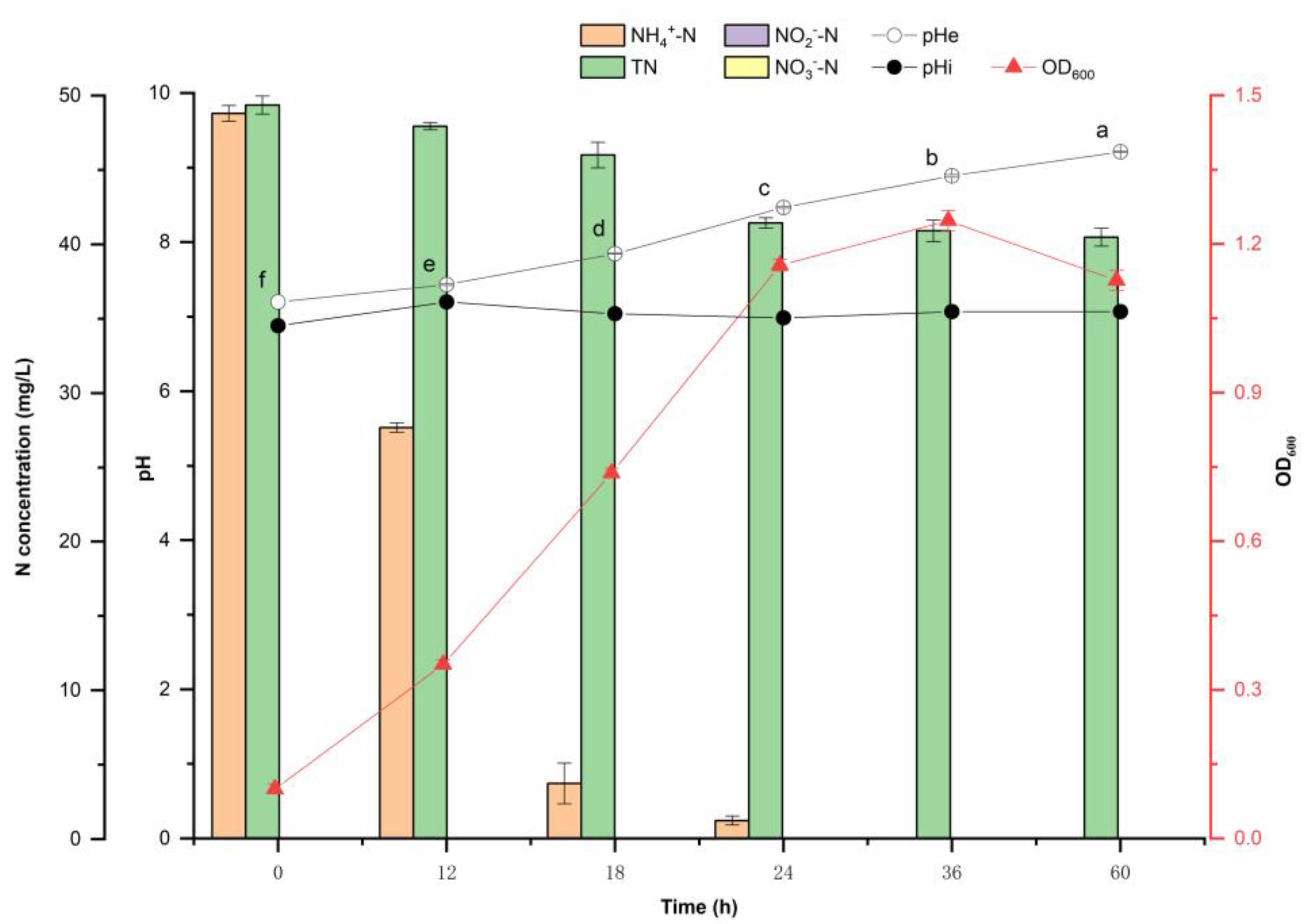
2.3. Ammonium Oxidation Capacity of Y-9
2.4. Detection of Extracellular Metabolites Produced by P. putida Y-9
2.5. Real-Time Quantitative PCR
2.6. Analytical Methods
2.7. Statistical Analysis and Graphical Work
3. Results
3.1. Effect of Different Carbon Sources on the Extracellular pH of P. putida Y-9
3.2. P. putida Y-9 Upregulates External pH during the Heterotrophic Ammonia Oxidation
3.3. Analysis of Significantly Different Metabolites of Y-9 Strains at Different Culture Times
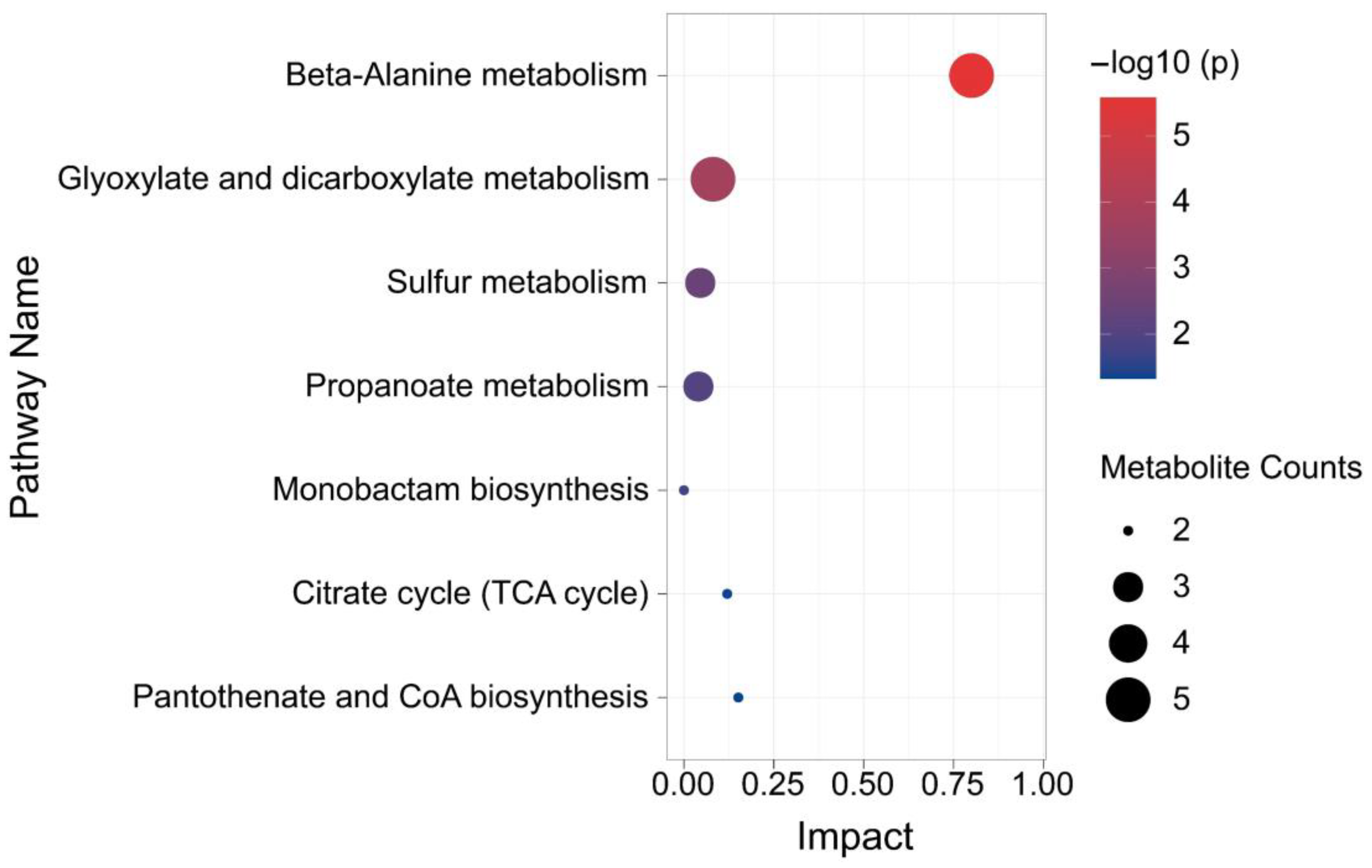
3.4. Analysis of Metabolic Pathway Enrichment of Y-9 Differential Metabolites
3.5. Correlation Analysis between FC of Metabolite and FC of the Medium’s pH
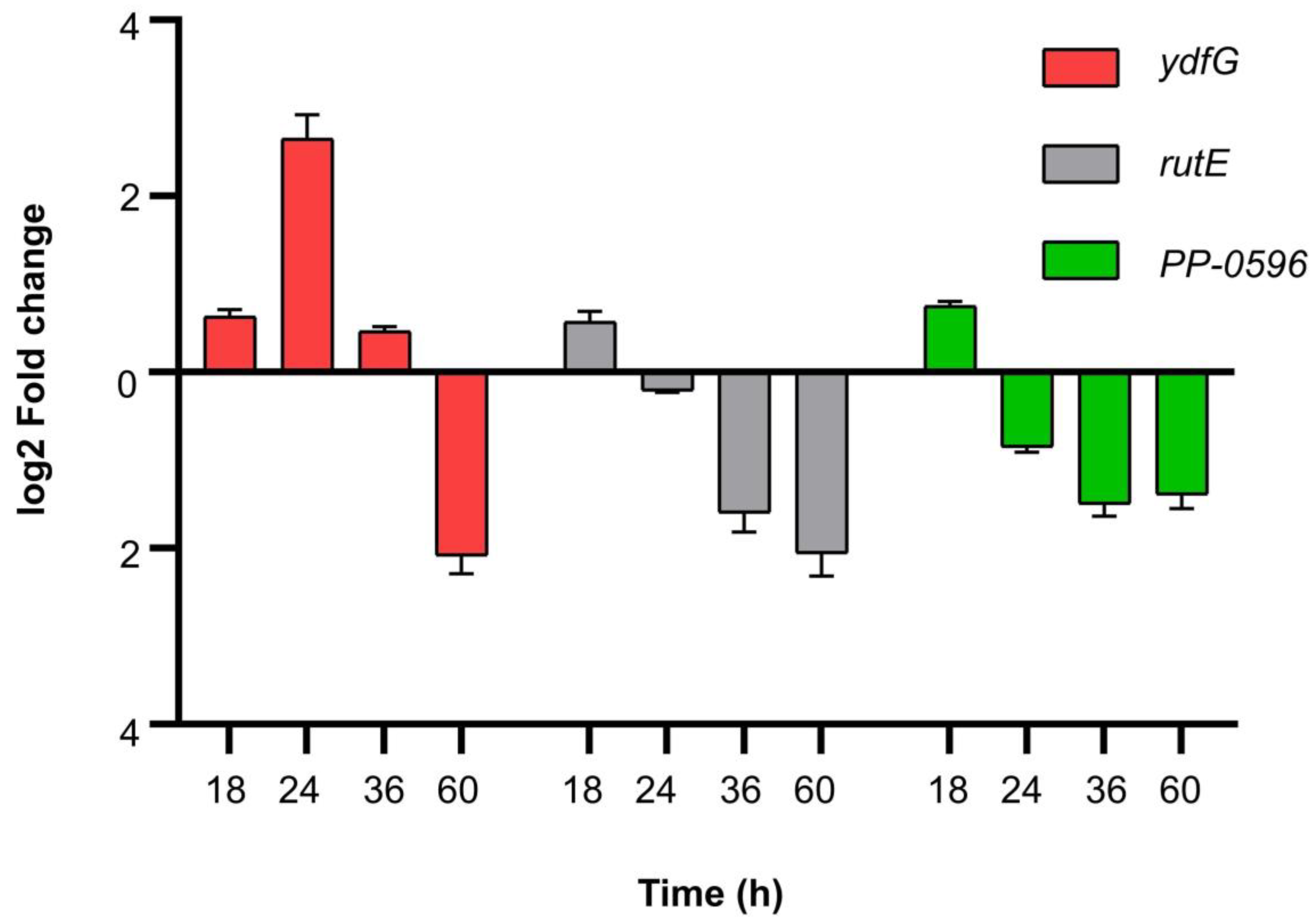
3.6. RT-qPCR Validation of Related Genes in β-alanine Metabolism
4. Discussion
4.1. Response of Extracellular pH of Y-9 to Different Carbon Sources
4.2. Nitrogen Metabolism Is Not the Reason Why Y-9 Causes the Increase in Extracellular pH
4.3. Analysis of the Reasons for the Increase in Extracellular pH Using Metabolites
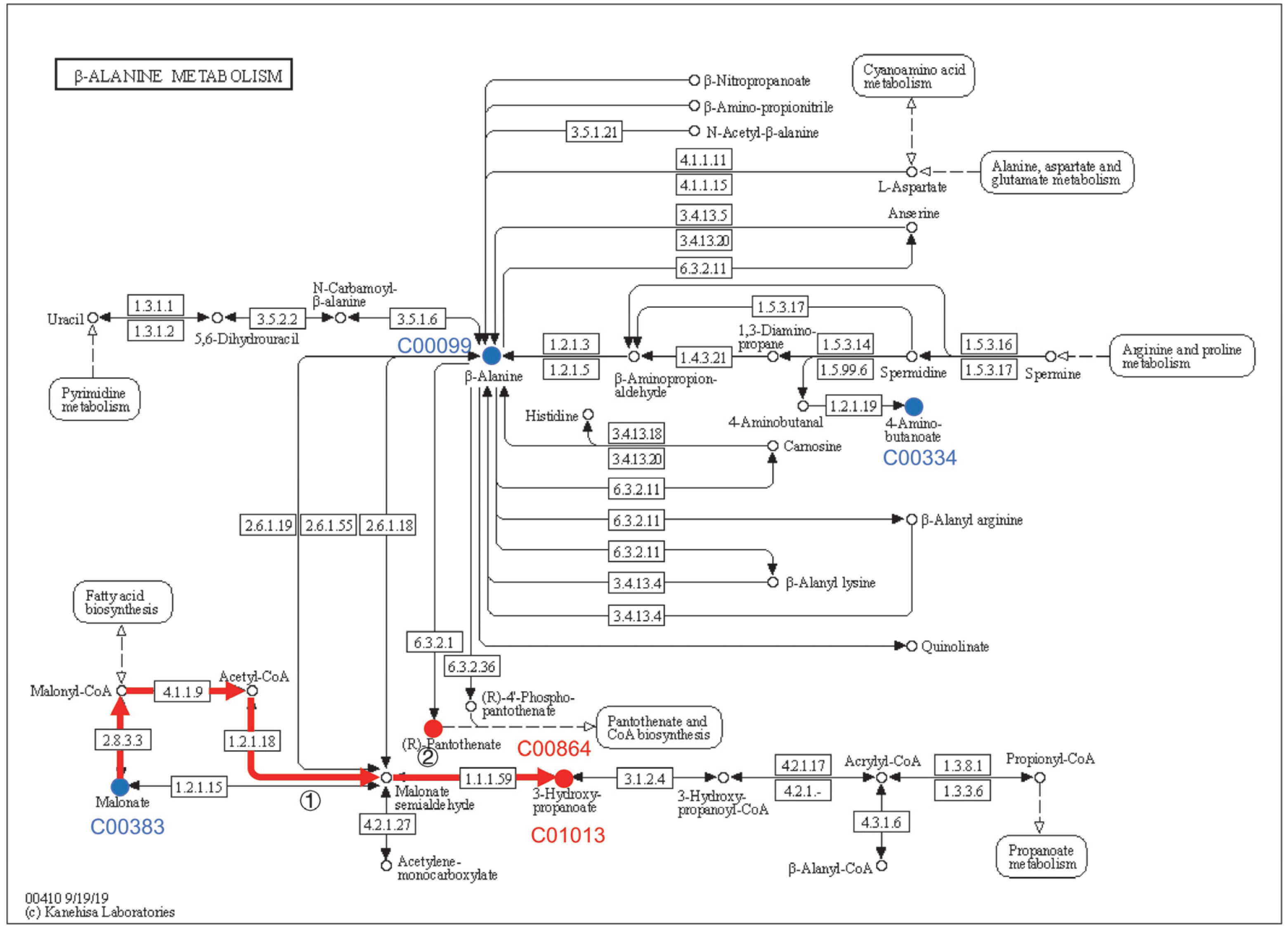
4.4. Effect of β-alanine Metabolism and Glyoxylate and Dicarboxylate Metabolism on the Rise of pH in the Culture Medium
4.5. Correlation Analysis and RT-qPCR Verification Support That the Rise in Extracellular pH Is Caused by β-alanine Metabolism
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lehtovirta-Morley, L.E. Ammonia oxidation: Ecology, physiology, biochemistry and why they must all come together. FEMS Microbiol. Lett. 2018, 365, fny058. [Google Scholar] [CrossRef] [Green Version]
- Kuypers, M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef]
- Ju, L.; Nallagatla, P.B. Monitoring and kinetic study of ammonia oxidation using dissolved oxygen electrode and NAD (P) H fluorometer. Water Res. 2003, 37, 188–196. [Google Scholar] [CrossRef]
- Song, T.; Zhang, X.; Li, J.; Wu, X.; Feng, H.; Dong, W. A review of research progress of heterotrophic nitrification and aerobic denitrification microorganisms (HNADMs). Sci. Total Environ. 2021, 801, 149319. [Google Scholar] [CrossRef]
- Xie, F.; Thiri, M.; Wang, H. Simultaneous heterotrophic nitrification and aerobic denitrification by a novel isolated Pseudomonas mendocina X49. Bioresour. Technol. 2021, 319, 124198. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, S.; Xue, W.; Guo, H.; Li, X.; Zou, G.; Zhao, T.; Dong, H. The characteristics of carbon, nitrogen and sulfur transformation during cattle manure composting—Based on different aeration strategies. Int. J. Environ. Res. Public Health 2019, 16, 3930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stancik, L.M.; Stancik, D.M.; Schmidt, B.; Barnhart, D.M.; Yoncheva, Y.N.; Slonczewski, J.L. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 2002, 184, 4246–4258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padan, E.; Bibi, E.; Ito, M.; Krulwich, T.A. Alkaline pH homeostasis in bacteria: New insights. Biochim. Biophys. Acta BBA-Biomembr. 2005, 1717, 67–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Clemente, R.; Guijo, M.I.; Nogales, J.; Blasco, R. Carbon source influence on extracellular pH changes along bacterial cell-growth. Genes 2020, 11, 1292. [Google Scholar] [CrossRef]
- Huang, X.; Xu, Y.; He, T.; Jia, H.; Feng, M.; Xiang, S.; Wang, S.; Ni, J.; Xie, D.; Li, Z. Ammonium transformed into nitrous oxide via nitric oxide by Pseudomonas putida Y-9 under aerobic conditions without hydroxylamine as intermediate. Bioresour. Technol. 2019, 277, 87–93. [Google Scholar] [CrossRef]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, Z.; Wang, L.; Liu, R.; Song, L. The suppressor of cytokine signaling 2 (SOCS2) modulating the neurotransmitters release in Eriocheir sinensis. Fish Shellfish Immun. 2013, 35, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kim, K.; Pelton, J.G.; Inwood, W.B.; Andersen, U.; Kustu, S.; Wemmer, D.E. The Rut pathway for pyrimidine degradation: Novel chemistry and toxicity problems. J. Bacteriol. 2010, 192, 4089–4102. [Google Scholar] [CrossRef] [Green Version]
- Danhof, H.A.; Vylkova, S.; Vesely, E.M.; Ford, A.E.; Gonzalez-Garay, M.; Lorenz, M.C. Robust extracellular pH modulation by Candida albicans during growth in carboxylic acids. mBio 2016, 7, e01646-16. [Google Scholar] [CrossRef] [Green Version]
- Lund, P.; Tramonti, A.; De Biase, D. Coping with low pH: Molecular strategies in neutralophilic bacteria. FEMS Microbiol. Rev. 2014, 38, 1091–1125. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Xue, Y.; Zhang, J.; Hu, X. Removal of lead from acidic wastewater by bio-mineralized bacteria with pH self-regulation. Chemosphere 2020, 241, 125041. [Google Scholar] [CrossRef]
- Jiang, H.; Liu, G.H.; Ma, Y.; Xu, X.; Chen, J.; Yang, Y.; Liu, X.; Wang, H. A pilot-scale study on start-up and stable operation of mainstream partial nitrification-anammox biofilter process based on online pH-DO linkage control. Chem. Eng. J. 2018, 350, 1035–1042. [Google Scholar] [CrossRef]
- He, T.; Xie, D.; Li, Z.; Ni, J.; Sun, Q. Ammonium stimulates nitrate reduction during simultaneous nitrification and denitrification process by Arthrobacter arilaitensis Y-10. Bioresour. Technol. 2017, 239, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, Y.; Chen, H.; Lyu, Y. Ammonium removal characteristics of an acid-resistant bacterium Acinetobacter sp. JR1 from pharmaceutical wastewater capable of heterotrophic nitrification-aerobic denitrification. Bioresour. Technol. 2019, 274, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Toribio, V.; García-Martín, A.B.; Martínez, M.J.; Martínez, Á.T.; Guillén, F. Induction of extracellular hydroxyl radical pro duction by white-rot fungi through quinone redox cycling. Appl. Environ. Microbiol. 2009, 75, 3944–3953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Fouchécour, F.; Sánchez-Castañeda, A.; Saulou-Berion, C.; Spinnler, H.E. Process engineering for microbial production of 3-hydroxypropionic acid. Biotechnol. Adv. 2018, 36, 1207–1222. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Sun, G.; Wang, Z.; Xiao, J.; Yang, J. Production of 3-hydroxypropionate using a novel malonyl-CoA-mediated biosynthetic pathway in genetically engineered E. coli strain. Green Chem. 2019, 21, 6103–6115. [Google Scholar] [CrossRef]
- Zhou, S.; Lama, S.; Jiang, J.; Sankaranarayanan, M.; Park, S. Use of acetate for the production of 3-hydroxypropionic acid by metabolically-engineered Pseudomonas denitrificans. Bioresour. Technol. 2020, 307, 123194. [Google Scholar] [CrossRef]
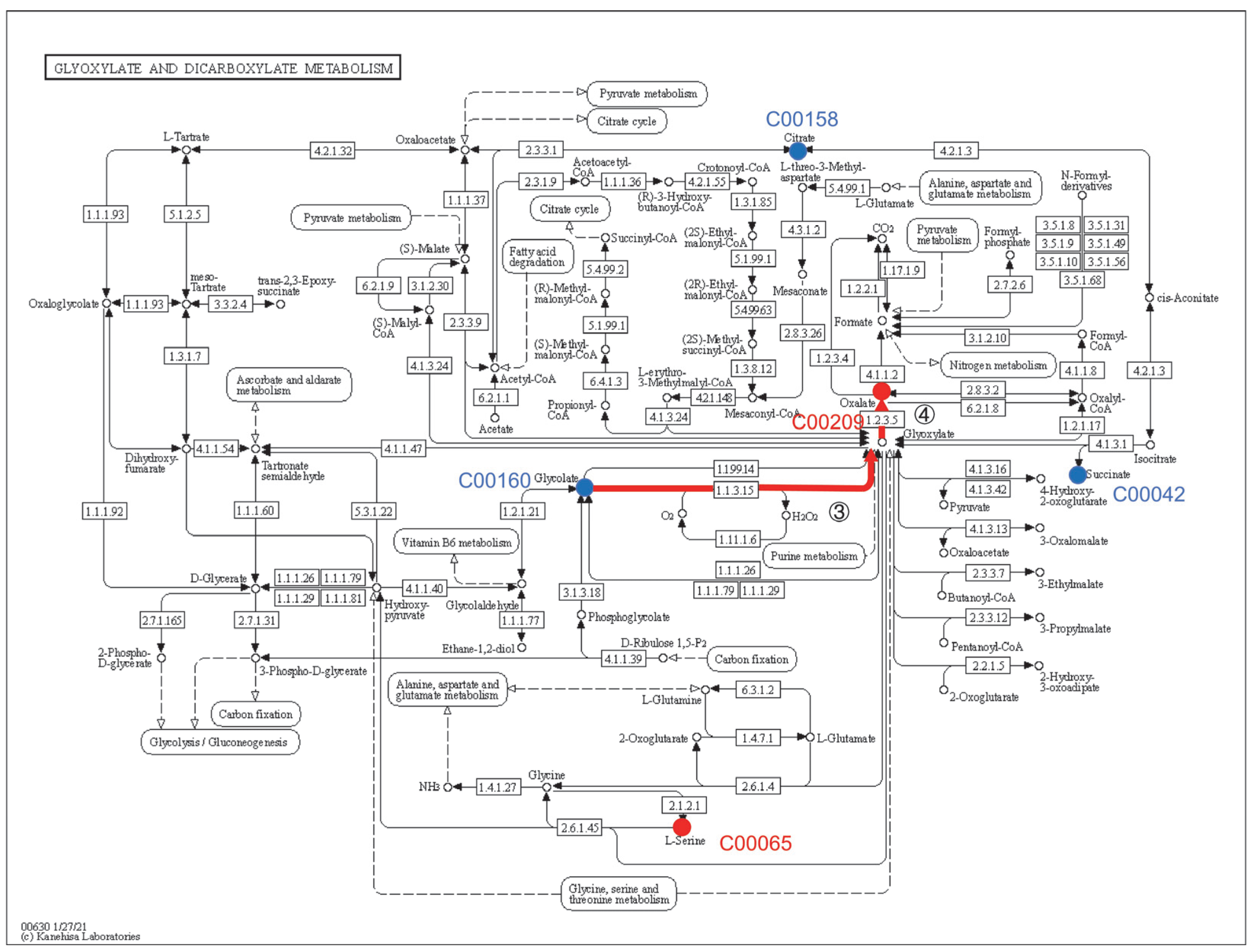


| Metabolite | KEGG ID | Class | Fold Change | |||
|---|---|---|---|---|---|---|
| S1 (18 h VS. 12 h) | S2 (24 h VS. 12 h) | S3 (36 h VS. 12 h) | S4 (60 h VS. 12 h) | |||
| Myo-inositol | C00137 | Alcohols and polyols | 417,387.89 | 1,550,810.60 | 944,079.06 | 917,126.45 |
| Syclohexylsulfamic acid 1 | C02824 | Sulfamic acid derivatives | 111,839.17 | 1,661,137.85 | 2,703,052.95 | 2,550,359.01 |
| Serine 1 | C00065 | Carboxylic acids and derivatives | 2.57 | 18.58 | 30.65 | 30.59 |
| 3-Hydroxypropionic acid 1 | C01013 | Hydroxy acids and derivatives | 2.18 | 11.01 | 15.71 | 14.65 |
| Alanine 1 | C01401 | - | 1.98 | 6.87 | 10.16 | 10.43 |
| Oxalic acid | C00209 | Carboxylic acids and derivatives | 1.74 | 5.56 | 4.75 | 5.06 |
| Pantothenic acid | C00864 | Carboxylic acids and derivatives | 0.83 | 498,303.28 | 634,225.94 | 628,839.31 |
| Myristic acid | C06424 | Fatty acyls | - | 8.73 | 24.21 | 22.64 |
| Sulfuric acid | C00059 | - | - | 1.51 | 2.36 | 2.36 |
| Phenylphosphoric acid | C02734 | - | - | 1.43 | - | - |
| Adenine | C00147 | Imidazopyrimidines | - | - | - | 1.52 |
| 2-Ketobutyric acid 1 | C00109 | Keto acids and derivatives | 0.88 | - | - | - |
| 4-Aminobutyric acid 3 | C00334 | Carboxylic acids and derivatives | 0.83 | - | - | - |
| Beta-alanine 2 | C00099 | Carboxylic acids and derivatives | 0.83 | - | - | - |
| Malonic acid 1 | C00383 | Carboxylic acids and derivatives | 0.78 | 0.53 | 0.38 | 0.41 |
| Maleamate 4 | C01596 | - | 0.68 | 0.10 | 0.00 | 0.04 |
| Succinic acid | C00042 | Carboxylic acids and derivatives | - | 0.69 | 0.45 | 0.76 |
| Glycolic acid | C00160 | Hydroxy acids and derivatives | - | - | 0.68 | 0.81 |
| Citric acid | C00158 | Carboxylic acids and derivatives | - | - | 0.00 | 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nie, M.; Li, K.; Li, Z. β-Alanine Metabolism Leads to Increased Extracellular pH during the Heterotrophic Ammonia Oxidation of Pseudomonas putida Y-9. Microorganisms 2023, 11, 356. https://doi.org/10.3390/microorganisms11020356
Nie M, Li K, Li Z. β-Alanine Metabolism Leads to Increased Extracellular pH during the Heterotrophic Ammonia Oxidation of Pseudomonas putida Y-9. Microorganisms. 2023; 11(2):356. https://doi.org/10.3390/microorganisms11020356
Chicago/Turabian StyleNie, Ming, Kaili Li, and Zhenlun Li. 2023. "β-Alanine Metabolism Leads to Increased Extracellular pH during the Heterotrophic Ammonia Oxidation of Pseudomonas putida Y-9" Microorganisms 11, no. 2: 356. https://doi.org/10.3390/microorganisms11020356
APA StyleNie, M., Li, K., & Li, Z. (2023). β-Alanine Metabolism Leads to Increased Extracellular pH during the Heterotrophic Ammonia Oxidation of Pseudomonas putida Y-9. Microorganisms, 11(2), 356. https://doi.org/10.3390/microorganisms11020356






