Epsilon-Caprolactam- and Nylon Oligomer-Degrading Bacterium Brevibacterium epidermidis BS3: Characterization and Potential Use in Bioremediation
Abstract
:1. Introduction
2. Material and Methods
2.1. Chemicals
2.2. Bacterial Strain and Growth Conditions
2.3. Microscopy
2.4. Characterization of the Physiological and Biochemical Properties
2.5. Identification of the Strain BS3
2.5.1. 16. S rRNA Gene Sequencing and Phylogenetic Analysis
2.5.2. G + C Analysis and DNA-DNA Hybridization
2.6. Caprolactam Degradation Experiment
2.7. Caprolactam Tolerance Test
2.8. Gas Chromatography Analysis
2.9. Statistical Data Processing
3. Results
3.1. Morphology and Ultrastructure of Cells
3.2. Physiological and Biochemical Properties
3.3. Identification of the Strain BS3
3.4. Caprolactam Tolerance
3.5. Caprolactam Degradation
3.6. Growth of Strain BS3 on Caprolactam Intermediates and Nylon Oligomers
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Negoro, S. Biodegradation of nylon and other synthetic polyamides. Biopolymers 2002, 9, 395–415. [Google Scholar] [CrossRef]
- Baxi, N.N. Feasibility studies of in situ bioremediation of nylon-6 oligomer waste contaminated soil. Curr. Res. Microbiol. Biotechnol. 2014, 3, 378–383. [Google Scholar]
- Sheldon, T. Chromosomal damage induced by caprolactam in human lymphocytes. Mutat. Res. 1989, 224, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, R.S.; Kanekar, P.P. Bioremediation of ε-caprolactam from nylon-6 waste water by use of Pseudomonas aeruginosa MCM B-407. Curr. Microbiol. 1998, 37, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Baxi, N.N.; Shah, A.K. Biological treatment of the components of solid oligomeric waste from a nylon-6 production plant. World J. Microbiol. Biotechnol. 2000, 16, 835–840. [Google Scholar] [CrossRef]
- Baxi, N.N. Influence of ε-caprolactam on growth and physiology of environmental bacteria. Ann. Microbiol. 2013, 63, 1471–1476. [Google Scholar] [CrossRef]
- Sanuth, H.A.; Yadav, A.; Fagade, O.E.; Shouche, Y. ε-Caprolactam utilization by Proteus sp. and Bordetella sp. isolated from solid waste dumpsites in Lagos State, Nigeria, first report. Indian J. Microbiol. 2013, 53, 221–226. [Google Scholar] [CrossRef]
- Rajoo, S.; Ahn, J.O.; Lee, H.W.; Jung, J.K. Isolation and characterization of a novel ε-caprolactam-degrading microbe, Acinetobacter calcoaceticus, from industrial wastewater by chemostat-enrichment. Biotechnol. Lett. 2013, 35, 2069–2072. [Google Scholar] [CrossRef]
- Mehta, S.K.; Panchal, P.A.; Butala, B.N.; Sane, S.A. Bacillus cereus mediated ε-caprolactam degradation: An initiative for waste water treatment of nylon-6 production plant. J. Bioremediat. Biodegrad. 2014, 5, 5. [Google Scholar] [CrossRef]
- Shama, G.; Wase, D.A. The biodegradation of ε-caprolactam and some related compounds. Int. Biodeterior. Bull. 1981, 17, 1–7. [Google Scholar]
- Esikova, T.Z.; Ponamoreva, O.N.; Baskunov, B.P.; Taran, S.A.; Boronin, A.M. Transformation of low-molecular linear caprolactam oligomers by caprolactam-degrading bacteria. J. Chem. Technol. Biotechnol. 2012, 87, 1284–1290. [Google Scholar] [CrossRef]
- Kinoshita, S.; Kageyama, S.; Iba, K.; Yamada, Y.; Okada, H. Utilization of a cyclic dimer and linear oligomers of ε-aminocaproic acid by Achromobacter guttatus KI 72. Agric. Biol. Chem. 1975, 39, 1219–1223. [Google Scholar] [CrossRef]
- Esikova, T.Z.; Akatova, E.V.; Taran, S.A. Bacteria that degrade low molecular linear epsilon-caprolactam oligomers. Appl. Biochem. Microbiol. 2014, 5, 463–470. [Google Scholar] [CrossRef]
- Belova, S.E.; Suzina, N.E.; Rijpstra, W.I.C.; Damst, J.S.S.; Dedysh, S.N. Edaphobacter lichenicola sp. nov., a member of the family Acidobacteriaceae from lichen-dominated forested tundra. Int. J. Syst. Evol. Microbiol. 2018, 68, 1265–1270. [Google Scholar] [CrossRef]
- Chung, Y.; Kobayashi, T.; Kanai, H.; Akiba, T.; Kudo, T. Purification and properties of extracellular amylase from the hyperthermophilic archeon Thermococcus profundus DT5432. Appl. Environ. Microbiol. 1995, 61, 1502–1506. [Google Scholar] [CrossRef]
- Kovacs, N. Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature 1956, 178, 703–704. [Google Scholar] [CrossRef]
- Lane, D. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematic; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- BLAST Software Package. Available online: https://www.ncbi.nlm.nih.gov/blast (accessed on 1 November 2022).
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position’specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Owen, R.L.; Lapage, S.P. The terminal denaturation of partly purified bacterial deoxyribonucleic acid and its taxonomic application. J. Appl. Bacteriol. 1976, 41, 335–340. [Google Scholar] [CrossRef]
- He, L.; Li, W.; Huang, Y.; Wang, L.M.; Liu, Z.H.; Lanoot, B.J.; Bancanneyt, M.; Swings, J. Streptomyces jietaisiensis sp. nov., isolated from soil in northern China. Int. J. Syst. Evol. Microbiol. 2005, 55, 1939–1944. [Google Scholar] [CrossRef]
- Microsoft Excel 2007 Program. Available online: https://www.microsoft.com/ru-ru/microsoft-365/previous-versions/download-office-2007 (accessed on 1 November 2022).
- Gruner, E.; Pfyffer, G.E.; Graevenitz, A.V. Characterization of Brevibacterium spp. from clinical specimens. J. Clin. Microbiol. 1993, 31, 1408–1412. [Google Scholar] [CrossRef]
- Otzen, M.; Palacio, C.; Janssen, D.B. Characterization of the caprolactam degradation pathway in Pseudomonas jessenii using mass spectrometry-based proteomics. Appl. Microbiol. Biotechnol. 2018, 102, 6699–6711. [Google Scholar] [CrossRef] [PubMed]
- Palacio, C.M.; Roseboom, H.J.; Lanfranchi, E.; Meng, Q.; Otzen, M.; Janssen, D.B. Biochemical properties of a Pseudomonas aminotransferase involved in caprolactam metabolism. FEBS J. 2019, 286, 4086–4102. [Google Scholar] [CrossRef] [PubMed]
- Girard, L.; Lood, C.; Höfte, M.; Vandamme, P.; Rokni-Zadeh, H.; van Noort, V.; Lavigne, R.; de Mot, R. The ever-expanding Pseudomonas genus: Description of 43 new species and partition of the Pseudomonas putida group. Microorganisms 2021, 9, 1766. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, H.; Tanabe, I.; Fukumura, T.; Kato, K. Taxonomic study on the ε-caprolactam-utilizing bacteria. J. Appl. Microbiol. 1967, 13, 125–137. [Google Scholar] [CrossRef]
- Esikova, T.Z.; Grishchenkov, V.G.; Boronin, A.M. Plasmids that control ε-caprolactam biodegradation. Microbiology 1990, 59, 547–552. [Google Scholar]
- Baxi, N.N.; Patel, S.; Hansoti, D. An Arthrobacter citreus strain suitable for degrading ε-caprolactam in polyamide waste and accumulation of glutamic acid. AMB Express 2019, 9, 161. [Google Scholar] [CrossRef]
- Panov, A.V.; Volkova, O.V.; Puntus, I.F.; Esikova, T.Z.; Kosheleva, I.A.; Boronin, A.M. scpA, a new salicylate hydroxylase gene localized in salicylate/caprolactam degradation plasmids. Mol. Biol. 2013, 47, 116–123. [Google Scholar] [CrossRef]
- Collins, M.D.; Farrow, J.A.E.; Goodfellow, M.; Minnikin, D.E. Brevibacterium casei sp. nov. and Brevibacterium epidermidis sp. nov. Syst. Appl. Microbiol. 1983, 4, 388–395. [Google Scholar] [CrossRef]
- Wang, C.C.; Lee, C.M. Isolation of the ε-caprolactam denitrifying bacteria from a wastewater treatment system manufactured with acrylonitrile-butadiene-styrene resin. J. Hazard Mater. 2007, 145, 136–141. [Google Scholar] [CrossRef]
- Fukumura, T.; Takeuchi, M.; Bann, I. Stepwise loss of metabolism of ε-aminocaproic acid cyclic dimer in Alcaligenes species D-2. Eur. J. Appl. Microbiol. Biotechnol. 1982, 14, 120–126. [Google Scholar] [CrossRef]
- Yasura, K.; Uedo, Y.; Takeo, M.; Kato, D.; Negoro, S. Genetic organization of nylon-oligomer-degrading enzymes from alcalophilic bacterium, Agromyces sp. KY5R. J. Biosci. Bioeng. 2007, 104, 521–524. [Google Scholar] [CrossRef]
- Fukumura, T. Bacterial breakdown of ε-caprolactam and its cyclic oligomers. Plant Cell Physiol. 1966, 7, 93–104. [Google Scholar] [CrossRef]
- Kinoshita, S.; Terada, T.; Taniguchi, T.; Taken, Y.; Masuda, S.; Matsunaga, N.; Okada, H. Purification and characterization of 6-aminohexanoic-acid-oligomer hydrolase of Flavobacterium sp. KI72. Eur. J. Biochem. 1981, 116, 547–555. [Google Scholar] [CrossRef]
- Prijambada, I.; Negoro, S.; Yomo, T.; Urabe, I. Emergence of nylon oligomer degradation enzymes in Pseudomonas aeruginosa PAO through experimental evolution. Appl. Environ. Microbiol. 1995, 61, 2020–2022. [Google Scholar] [CrossRef]
- Rybkina, D.O.; Plotnikova, E.G.; Dorofeeva, L.V.; Mironenko, Y.; Demakov, V.A. A new aerobic gram-positive bacterium with a unique ability to degrade ortho- and para-clorinated biphenyls. Microbiology 2003, 72, 759–765. [Google Scholar] [CrossRef]
- Baxi, N.N.; Shah, A.K. ε-Caprolactam-degradation by Alcaligenes faecalis for bioremediation of wastewater of a nylon-6 production plant. Biotechnol. Lett. 2002, 24, 1177–1180. [Google Scholar] [CrossRef]
- Fortmann, L.; Rosenberg, A. Fate of ε-caprolactam in the aquatic environment. Chemosphere 1984, 13, 53–65. [Google Scholar] [CrossRef]
- Boronin, A.M.; Naumova, R.P.; Grishchenkov, V.G.; Ilijinskaya, O.N. Plasmids specifying ε-caprolactam degradation in Pseudomonas strains. FEMS Microbiol. Lett. 1984, 22, 167–170. [Google Scholar] [CrossRef]
- Kulkarnik, R.S.; Kanekar, P.P. Effect of some curing agents on phenotypic stability in Pseudomonas putida degrading ε-caprolactam. World J. Microbiol. Biotechnol. 1998, 14, 255–257. [Google Scholar] [CrossRef]
- Kinoshita, S.; Kobayashi, E.; Okada, H. Degradation of e-caprolactam by Achromobacter guttatus KF71. J. Ferment. Technol. 1973, 51, 719–725. [Google Scholar]
- Sokolov, A.B.; Pechatnikov, M.G.; Krizhanovskii, A.S.; Petrov, G.G. Kombinirovanie khimicheskikh i biologicheskikh sposobov ochistki kaprolaktamisoderzhashchikh stokov [Combining chemical and biological methods for cleaning caprolactam-containing wastes]. Ross. Khim. Zh. 2006, 50, 48–55. [Google Scholar]




| Characteristics | |
|---|---|
| Cell form/size | Round-ended short rods, 1.0–1.4 × 0.6–0.8 μm |
| Gram stain | Positive |
| Motility | Negative |
| Relation to oxygen | Aerobic |
| Oxidase | Negative |
| Catalase | Positive |
| Nitrate reduction | Positive |
| Urease | Negative |
| Esculin hydrolysis | Negative |
| Starch hydrolysis | Negative |
| Casein hydrolysis | Positive |
| Gelatin liquefaction | Positive |
| β-Galactosidase, arginine dihydrolase | Negative |
| Indole production | Negative |
| Hydrolysis of Tweens | Negative |
| Growth temperature (optimum), °C | 10–37 (25–28) |
| Growth pH range (optimum) | 6.0–10.5 (7.5–8.0) |
| NaCl tolerance, % (optimum) | 0–18.0 (0.2) |
| DNA G + C content, mol. % | 62.6 |
| Assimilation of substrates | |
| Fructose, cellobiose, galactose, glucose, ribose, sucrose | Positive |
| Xylose, arabinose, maltose, rhamnose, fucose, lactose | Negative |
| Dulcitol, inositol, adonitol, arobitol, sorbitol | Negative |
| Mannitol, glycerol | Positive |
| Malate, phenylacetate, sodium citrate, potassium gluconate, capric acid, succinate | Positive |
| Formation of acid from | |
| Inositol | Negative |
| Mannitol | Positive |
| Rhamnose | Negative |
| Salicylate | Negative |
| Sorbitol | Negative |
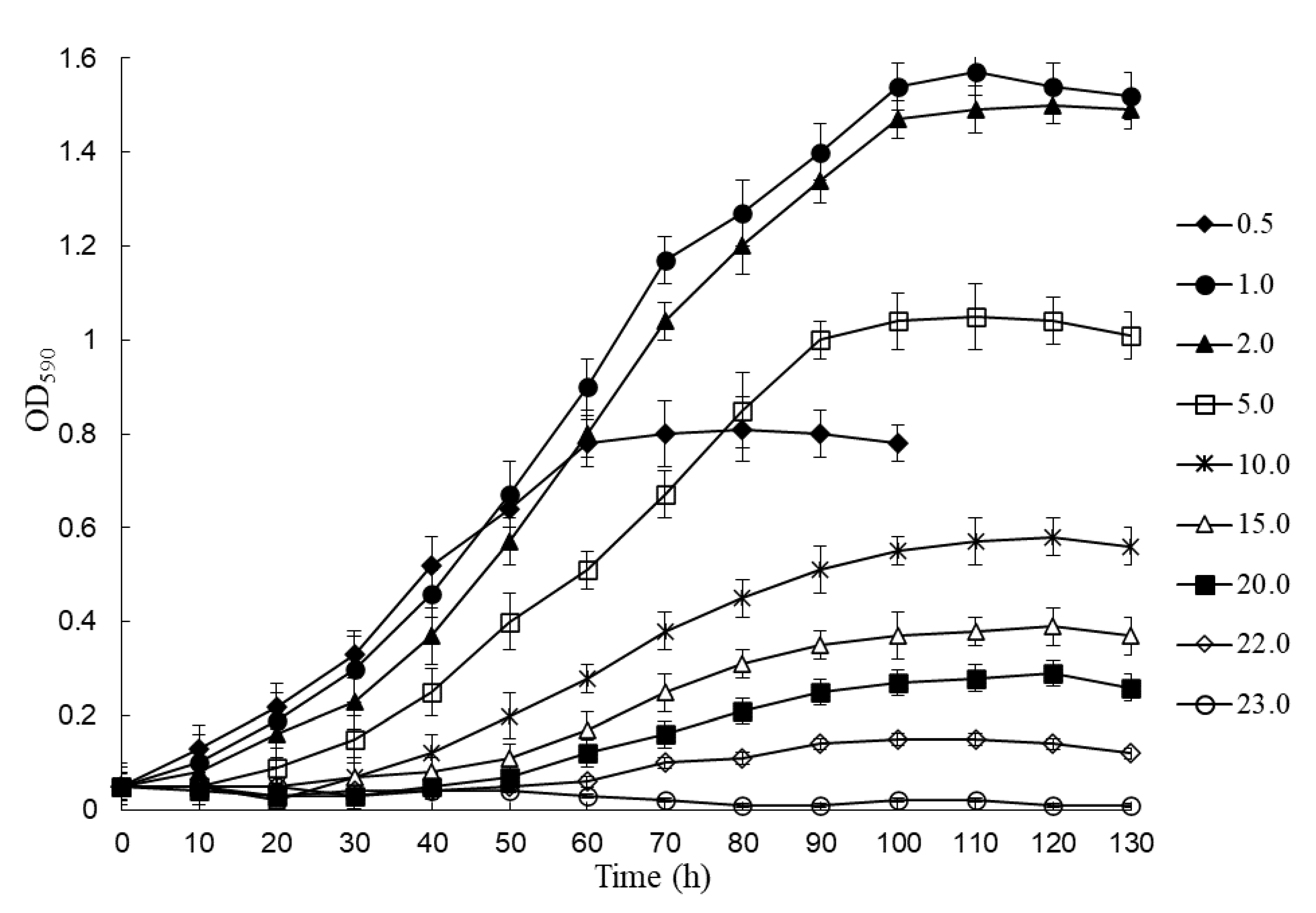
| CAP Concentration, g/L | Lag Phase, h | µmax, h−1 | OD590 | Dry Cell Weight, g/mL |
|---|---|---|---|---|
| 0.4 | 0 | 0 | 0.03 | 0 |
| 0.5 | 1.0 | 0.143 | 0.81 | 0.48 ± 0.03 |
| 1.0 | 6.0 | 0.236 | 1.57 | 0.98 ± 0.09 |
| 2.0 | 7.0 | 0.224 | 1.50 | 0.92± 0.07 |
| 5.0 | 15 | 0.146 | 1.05 | 0.64 ± 0.05 |
| 10.0 | 27 | 0.080 | 0.58 | 0.31 ± 0.02 |
| 15.0 | 40 | 0.0543 | 0.39 | 0.24 ± 0.02 |
| 20.0 | 50 | 0.041 | 0.29 | 0.176 ± 0.01 |
| 22.0 | 60 | 0.022 | 0.15 | 0.08 ± 0.001 |
| 23.0 | 0 | 0.03 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esikova, T.Z.; Akatova, E.V.; Solyanikova, I.P. Epsilon-Caprolactam- and Nylon Oligomer-Degrading Bacterium Brevibacterium epidermidis BS3: Characterization and Potential Use in Bioremediation. Microorganisms 2023, 11, 373. https://doi.org/10.3390/microorganisms11020373
Esikova TZ, Akatova EV, Solyanikova IP. Epsilon-Caprolactam- and Nylon Oligomer-Degrading Bacterium Brevibacterium epidermidis BS3: Characterization and Potential Use in Bioremediation. Microorganisms. 2023; 11(2):373. https://doi.org/10.3390/microorganisms11020373
Chicago/Turabian StyleEsikova, Tatiana Z., Ekaterina V. Akatova, and Inna P. Solyanikova. 2023. "Epsilon-Caprolactam- and Nylon Oligomer-Degrading Bacterium Brevibacterium epidermidis BS3: Characterization and Potential Use in Bioremediation" Microorganisms 11, no. 2: 373. https://doi.org/10.3390/microorganisms11020373
APA StyleEsikova, T. Z., Akatova, E. V., & Solyanikova, I. P. (2023). Epsilon-Caprolactam- and Nylon Oligomer-Degrading Bacterium Brevibacterium epidermidis BS3: Characterization and Potential Use in Bioremediation. Microorganisms, 11(2), 373. https://doi.org/10.3390/microorganisms11020373







