Characterization of Bacillus Strains from Natural Honeybee Products with High Keratinolytic Activity and Antimicrobial Potential
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microbial Growth Conditions and Growth Determination
2.2. Experimental Design, Sampling, DNA Extraction and 16S rRNA Sequencing
2.3. Genome Sequencing, Assembly and Annotation
2.4. Taxonomic Analysis
2.5. Keratinase Activity Detection Assay
2.6. Growth Inhibition Bioassay
2.7. Analysis of Gene Clusters with Antimicrobial Potential
3. Results
3.1. Isolation and Selection of Bacillus Strains from Raw Honeybee Products
3.2. Taxonomy of the Bacillus Isolates and General Features of Their Genomes
3.3. Antimicrobial Assays
3.4. Characterization of the Keratin-Degrading Activity of the Isolated Bacillus Strains
3.5. Gene Cluster Associated with the Biosynthesis of Putative Antimicrobial Metabolites
3.6. B. licheniformis CG1 Keratinase Activity Detected across a Wide Range of pH
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Converting Poultry Feathers Discarded Waste into Valuable Raw Materials. Available online: https://cordis.europa.eu/article/id/418077-converting-poultry-feathers-from-discarded-waste-into-valuable-raw-materials (accessed on 9 October 2022).
- Lyndall, M.P.; Kurtböke, D.I. Development of an Environmentally Friendly Biofertilizer with Keratin Degrading and Antiobiotic Producing Actinomycetes. Actinomycetologica 2004, 18, 34–42. [Google Scholar]
- Zouari-Fakhfakh, N.; Haddar, A.; Hmidet, N.; Frikha, F.; Nasri, M. Application of statistical experimental design for optimization of keratinases production by Bacillus pumilus A1 grown on chicken feather and some biochemical properties. Process Biochem. 2010, 45, 617–626. [Google Scholar] [CrossRef]
- Verma, A.; Singh, H.; Anwar, S.; Chattopadhay, A.; Tiwari, K.K.; Kaur, S.; Dhilo, G.S. Microbial keratinases: Industrial enzymes with waste management potential. Crit. Rev. Biotechnol. 2017, 37, 476–491. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.R.; Reddy, K.S.; Chouhan, R.; Bee, H.; Reddy, G. Effective feather degradation and keratinase production by Bacillus pumilus GRK for its application as bio-detergent additive. Bioresour. Tehcnol. 2017, 243, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Tamreihao, K.; Mukherjee, S.; Khunjamayum, R.; Devi, L.J.; Asem, R.S.; Ningthoujam, D.S. Feather degradation by keratinolytic bacteria and biofertilizing potential for sustainable agricultural production. J. Basic Microbiol. 2019, 59, 4–13. [Google Scholar] [CrossRef]
- Dariot, D.J.; Brandelli, A. A current assessment on the production of bacterial keratinases. Crit. Rev. Biotechnol. 2013, 34, 372–384. [Google Scholar] [CrossRef]
- Lin, X.; Lee, C.G.; Casale, E.S.; Shih, J.C. Purification and Characterization of a Keratinase from a Feather-Degrading Bacillus licheniformis Strain. Appl. Environ. Microbiol. 1992, 58, 3271–3275. [Google Scholar] [CrossRef]
- Bordel, S.; Martín-González, D.; Muñoz, R.; Santos-Beneit, F. Genome sequence analysis and characterization of Bacillus altitudinis B12, a polylactic acid- and keratin-degrading bacterium. Mol. Genet. Genom. 2022. [Google Scholar] [CrossRef]
- Bonifer, K.S.; Wen, X.; Hasim, S.; Phillips, E.K.; Dunlap, R.N.; Gann, E.R.; DeBruyn, J.M.; Reynolds, T.B. Bacillus pumilus B12 degrades polylactic acid and degradation is affected changing nutrient conditions. Front. Microbiol. 2019, 10, 2548. [Google Scholar] [CrossRef]
- Golden, C.E.; Rothrock, M.K.; Mishra, A. Comparison Between Random Forest and Gradient Boosting Machine Methods for Predicting Listeria spp. Prevalence in the Environment of Pastured Poultry Farms. Int. Food Res. J. 2019, 122, 47–55. [Google Scholar] [CrossRef]
- Velasquez, C.G.; Macklin, K.S.; Kumar, S.; Bailey, M.; Ebner, P.E.; Oliver, H.F.; Martin-Gonzalez, F.S.; Singh, M. Prevalence and antimicrobial resistance patterns of Salmonella isolated from poultry farms in southeastern United States. Poult. Sci. 2018, 97, 2144–2152. [Google Scholar] [CrossRef]
- Neoigi, S.B.; Islam, M.M.; Islam, S.K.S.; Akhter, A.H.M.T.; Sikder, M.M.H.; Yamasaki, S.; Kabir, S.M.L. Risk of multi-drug resistant Campylobacter spp. and residual antimicrobials at poultry farms and live bird markets in Bangladesh. BMC Infect. Dis. 2020, 20, 278. [Google Scholar] [CrossRef]
- Santos-Beneit, F.; Ceniceros, A.; Nikolaou, A.; Salas, J.A.; Gutierrez-Merino, J. Identification of Antimicrobial Compounds in Two Streptomyces sp. Strains Isolated From Beehives. Front. Microbiol. 2022, 13, 742168. [Google Scholar] [CrossRef]
- Gutierrez-Merino, J.; Isla, B.; Combes, T.; Martinez-Estrada, F.; Maluquer De Motes, C. Beneficial bacteria activate type-I interferon production via the intracellular cytosolic sensors STING and MAVS. Gut Microbes 2020, 11, 771–788. [Google Scholar] [CrossRef]
- Santos-Beneit, F.; Roberts, D.M.; Cantlay, S.; McCormick, J.R.; Errington, J. A mechanism for FtsZ-independent proliferation in Streptomyces. Nat. Commun. 2017, 8, 1378. [Google Scholar] [CrossRef]
- Fernández-Martínez, L.T.; Santos-Beneit, F.; Martín, J.F. Is PhoR-PhoP partner fidelity strict? PhoR is required for the activation of the pho regulon in Streptomyces coelicolor. Mol. Genet. Genom. 2012, 287, 565–573. [Google Scholar] [CrossRef]
- Santos-Beneit, F. Genome sequencing analysis of Streptomyces coelicolor mutants that overcome the phosphate-depending vancomycin lethal effect. BMC Genom. 2018, 19, 457. [Google Scholar] [CrossRef]
- Amin, F.A.Z.; Sabri, S.; Ismail, M.; Chan, K.W.; Isamail, N.; Esa, N.M.; Lila, M.A.M.; Zawawi, N. Probiotic Properties of Bacillus Strains Isolated from Stingless Bee (Heterotrigona itama) Honey Collected across Malaysia. Int. J. Environ. Res. Public Health 2020, 17, 278. [Google Scholar] [CrossRef]
- Santorelli, L.A.; Wilkinson, T.; Abdulmalik, R.; Rai, Y.; Creevey, C.J.; Huws, S.; Gutierrez-Merino, J. Beehives possess their own distinct microbiomes. Environ. Microbiome 2023, 18, 1. [Google Scholar] [CrossRef]
- Stedman, A.; Vilet, A.H.M.; Chambers, M.A.; Gutierrez-Merino, J. Gut commensal bacteria show beneficial properties as wildlife probiotics. Ann. N. Y. Acad. Sci. 2020, 1467, 112–132. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Teeling, H.; Meyerdierks, A.; Bauer, M.; Amann, R.; Glöckner, F.O. Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ. Microbiol. 2004, 6, 938–947. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Kembhavi, A.A.; Kulkarni, A.; Pant, A. Salt-tolerant and thermostable alkaline protease from Bacillus subtilis NCIM no. 64. Appl. Biochem. Biotechnol. 1993, 38, 83–92. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- van Heel, A.J.; de Jong, A.; Montalbán-López, M.; Kok, J.; Kuipers, O.P. BAGEL3: Automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 2013, 41, W448–W453. [Google Scholar] [CrossRef]
- Gudiña, E.J.; Teixeira, J.A. Bacillus licheniformis: The unexplored alternative for the anaerobic production of lipopeptide biosurfactants? Biotechnol. Adv. 2022, 60, 108013. [Google Scholar] [CrossRef]
- Chen, X.; Lu, Y.; Shan, M.; Zhao, H.; Lu, Z.; Lu, Y. A mini-review: Mechanism of antimicrobial action and application of surfactin. World J. Microbiol. Biotechnol. 2022, 38, 143. [Google Scholar] [CrossRef]
- Barbosa, J.C.; Silva, Í.C.; Caetano, T.; Mösker, E.; Seidel, M.; Lourenço, J.; Süssmuth, R.D.; Santos, N.C.; Gonçalves, S.; Mendo, S. Assessing the potential of the two-peptide lantibiotic lichenicidin as a new generation antimicrobial. World J. Microbiol. Biotechnol. 2022, 38, 18. [Google Scholar] [CrossRef]
- Dischinger, J.; Josten, M.; Szekat, C.; Sahl, H.G.; Bierbaum, G. Production of the Novel Two-Peptide Lantibiotic Lichenicidin by Bacillus licheniformis DSM 13. PLoS ONE 2009, 4, e6788. [Google Scholar] [CrossRef]
- Gupta, R.; Ramnani, P. Microbial keratinases and their prospective applications: An overview. Appl. Microbiol. Biotechnol. 2006, 70, 21–33. [Google Scholar] [CrossRef]
- Chatuvedi, V.; Agrawal, K.; Verma, P. Chicken feathers: A treasure cove of useful metabolites and value-added products. J. Sustain. 2021, 4, 231–243. [Google Scholar]
- Hmidet, N.; Ali Nel, H.; Zouari-Fakhfakh, N.; Haddar, A.; Nasri, M.; Sellemi-Kamoun, A. Chicken feathers: A complex substrate for the co-production of alpha-amylase and proteases by B. licheniformis NH1. J. Ind. Microbiol. Biotechnol. 2010, 37, 983–990. [Google Scholar] [CrossRef]
- Yahaya, R.S.R.; Normi, Y.M.; Phang, L.Y.; Ahmad, S.A.; Abdullah, J.O.; Sabri, S. Molecular strategies to increase keratinase production in heterologous expression systems for industrial applications. Appl. Microbiol. Biotechnol. 2021, 105, 3955–3969. [Google Scholar] [CrossRef]
- Jagadeesan, Y.; Meenakshisundaram, S.; Saravanan, V.; Balaiah, A. Sustainable production, biochemical and molecular characterization of thermo-and-solvent stable alkaline serine keratinase from novel Bacillus pumilus AR57 for promising poultry solid waste management. Int. J. Biol. Macromol. 2020, 163, 135–146. [Google Scholar] [CrossRef]
- Anna, S.; Veronika, L.; Svetlana, T.; Alexander, O. Patented Keratinolytic Enzymes for Industrial Application: An Overview. Recent Pat. Biotechnol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Nair, A.; Sarma, S.J. The impact of carbon and nitrogen catabolite repression in microorganisms. Microbiol. Res. 2021, 251, 126831. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Beg, Q.K.; Lorenz, P. Bacterial alkaline proteases: Molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 2002, 59, 15–32. [Google Scholar] [PubMed]
- Efem, S.E. Clinical observations on the wound healing properties of honey. Br. J. Surg. 1988, 75, 679–681. [Google Scholar] [CrossRef]
- Subrahmanyam, M. Topical application of honey in treatment of burns. Br. J. Surg. 1991, 78, 497–498. [Google Scholar] [CrossRef]
- Wahdan, H.A. Causes of the antimicrobial activity of honey. Infection 1998, 26, 26–31. [Google Scholar] [CrossRef]
- Lashani, E.; Davoodabadi, A.; Soltan Dallal, M.M. Some probiotic properties of Lactobacillus species isolated from honey and their antimicrobial activity against foodborne pathogens. Vet. Res. Forum. 2020, 11, 121–126. [Google Scholar]
- Sabaté, D.C.; Carrillo, L.; Audisio, M.C. Inhibition of Paenibacillus larvae and Ascosphaera apis by Bacillus subtilis isolated from honeybee gut and honey samples. Res. Microbiol. 2009, 160, 193–199. [Google Scholar] [CrossRef]
- Clements-Decker, T.; Kode, M.; Khan, S.; Khan, W. Underexplored bacteria as reservoirs of novel antimicrobial lipopeptides. Front. Chem. 2022, 10, 1025979. [Google Scholar] [CrossRef]
- Antonioli Júnior, R.; Poloni, J.F.; Pinto, É.S.M.; Dorn, M. Interdisciplinary Overview of Lipopeptide and Protein-Containing Biosurfactants. Genes 2022, 14, 76. [Google Scholar] [CrossRef]
- Díaz, P.R.; Torres, M.; Petroselli, G.; Erra-Balsells, R.; Audisio, M.C. Antibacterial activity of Bacillus licheniformis B6 against viability and biofilm formation of foodborne pathogens of health importance. World J. Microbiol. Biotechnol. 2022, 38, 181. [Google Scholar] [CrossRef]
- Wang, H.; Fewer, D.P.; Holm, L.; Rouhiainen, L.; Sivonen, K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc. Natl. Acad. Sci. USA 2014, 111, 9259–9264. [Google Scholar] [CrossRef]
- Aleti, G.; Sessitsch, A.; Brader, G. Genome mining: Prediction of lipopeptides and polyketides from Bacillus and related Firmicutes. Comput. Struct. Biotechnol. J. 2015, 13, 192–203. [Google Scholar] [CrossRef]
- Ramirez-Olea, H.; Reyes-Ballesteros, B.; Chavez-Santoscoy, R.A. Potential application of the probiotic Bacillus licheniformis as an adjuvant in the treatment of diseases in humans and animals: A systematic review. Front. Microbiol. 2022, 13, 993451. [Google Scholar] [CrossRef]
- Madslien, E.H.; Rønning, H.T.; Lindbäck, T.; Hassel, B.; Andersson, M.A.; Granum, P.E. Lichenysin is produced by most Bacillus licheniformis strains. J. Appl. Microbiol. 2013, 115, 1068–1080. [Google Scholar] [CrossRef]
- Begley, M.; Cotter, P.D.; Hill, C.; Ross, R.P. Identification of a novel two-peptide lantibiotic, lichenicidin, following rational genome mining for LanM proteins. Appl. Environ. Microbiol. 2009, 75, 5451–5460. [Google Scholar] [CrossRef]
- Bagameri, L.; Baci, G.M.; Dezmirean, D.S. Royal Jelly as a Nutraceutical Natural Product with a Focus on Its Antibacterial Activity. Pharmaceutics 2022, 14, 1142. [Google Scholar] [CrossRef]
- Anderson, K.E.; Sheehan, T.H.; Mott, B.M.; Maes, P.; Snyder, L.; Schwan, M.R.; Walton, A.; Jones, B.M.; Corby-Harris, V. Microbial ecology of the hive and pollination landscape: Bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PLoS ONE 2013, 8, e83125. [Google Scholar] [CrossRef]
- James, N.; Umesh, M.; Sarojini, S.; Shanmugam, S.; Nasif, O.; Alharbi, S.A.; Lan Chi, N.T.; Brindhadevi, K. Unravelling the potential plant growth activity of halotolerant Bacillus licheniformis NJ04 isolated from soil and its possible use as a green bioinoculant on Solanum lycopersicum L. Environ. Res. 2023, 216, 114620. [Google Scholar] [CrossRef]
- Yakimov, M.M.; Timmis, K.N.; Wray, V.; Fredrickson, H.L. Characterization of a new lipopeptide surfactant produced by thermotolerant and halotolerant subsurface Bacillus licheniformis BAS50. Appl. Environ. Microbiol. 1995, 61, 1706–1713. [Google Scholar] [CrossRef]
- Eppelmann, K.; Doekel, S.; Marahiel, M.A. Engineered biosynthesis of the peptide antibiotic bacitracin in the surrogate host Bacillus subtilis. J. Biol. Chem. 2001, 276, 34824–34831. [Google Scholar] [CrossRef] [PubMed]
- May, J.J.; Wendrich, T.M.; Marahiel, M.A. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 2001, 276, 7209–7217. [Google Scholar] [CrossRef] [PubMed]
- Coburn, P.S.; Gilmore, M.S. The Enterococcus faecalis cytolysin: A novel toxin active against eukaryotic and prokaryotic cells. Cell Microbiol. 2003, 5, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Koumoutsi, A.; Chen, X.H.; Henne, A.; Liesegang, H.; Hitzeroth, G.; Franke, P.; Vater, J.; Borriss, R. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 2004, 186, 1084–1096. [Google Scholar] [CrossRef]
- Wescombe, P.A.; Tagg, J.R. Purification and characterization of streptin, a type A1 lantibiotic produced by Streptococcus pyogenes. Appl. Environ. Microbiol. 2003, 69, 2737–2747. [Google Scholar] [CrossRef]
- Umezawa, H.; Aoyagi, T.; Nishikiori, T.; Okuyama, A.; Yamagishi, Y.; Hamada, M.; Takeuchi, T. Plipastatins: New inhibitors of phospholipase A2, produced by Bacillus cereus BMG302-fF67. I. Taxonomy, production, isolation and preliminary characterization. J. Antibiot. 1986, 39, 737–744. [Google Scholar] [CrossRef]
- Hegemann, J.D.; Zimmermann, M.; Xie, X.; Marahiel, M.A. Lasso peptides: An intriguing class of bacterial natural products. Acc. Chem. Res. 2015, 48, 1909–1919. [Google Scholar] [CrossRef]
- Allenby, N.E.; Watts, C.A.; Homuth, G.; Pragai, Z.; Wipat, A.; Ward, A.C.; Harwood, C.R. Phosphate starvation induces the sporulation killing factor of Bacillus subtilis. J. Bacteriol. 2006, 188, 5299–5303. [Google Scholar] [CrossRef]
- Babasaki, K.; Takao, T.; Shimonishi, Y.; Kurahashi, K. 1985. Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: Isolation, structural analysis, and biogenesis. J. Biochem. 1985, 98, 585–603. [Google Scholar] [CrossRef]
- Patel, P.S.; Huang, S.; Fisher, S.; Pirnik, D.; Aklonis, C.; Dean, L.; Meyers, E.; Fernandes, P.; Mayerl, F. Bacillaene, a novel inhibitor of procaryotic protein synthesis produced by Bacillus subtilis: Production, taxonomy, isolation, physico-chemical characterization and biological activity. J. Antibiot. 1995, 48, 997–1003. [Google Scholar] [CrossRef]
- Rogers, H.J.; Newton, G.G.; Abraham, E.P. Production and purification of bacilysin. Biochem. J. 1965, 97, 573–578. [Google Scholar] [CrossRef]
- Arima, K.; Kakinuma, A.; Tamura, G. Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: Isolation, characterization and its inhibition of fibrin clot formation. Biochem. Biophys. Res. Commun. 1968, 31, 488–494. [Google Scholar] [CrossRef]
- Paik, S.H.; Chakicherla, A.; Hansen, J.N. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J. Biol. Chem. 1998, 273, 23134–23142. [Google Scholar] [CrossRef] [Green Version]
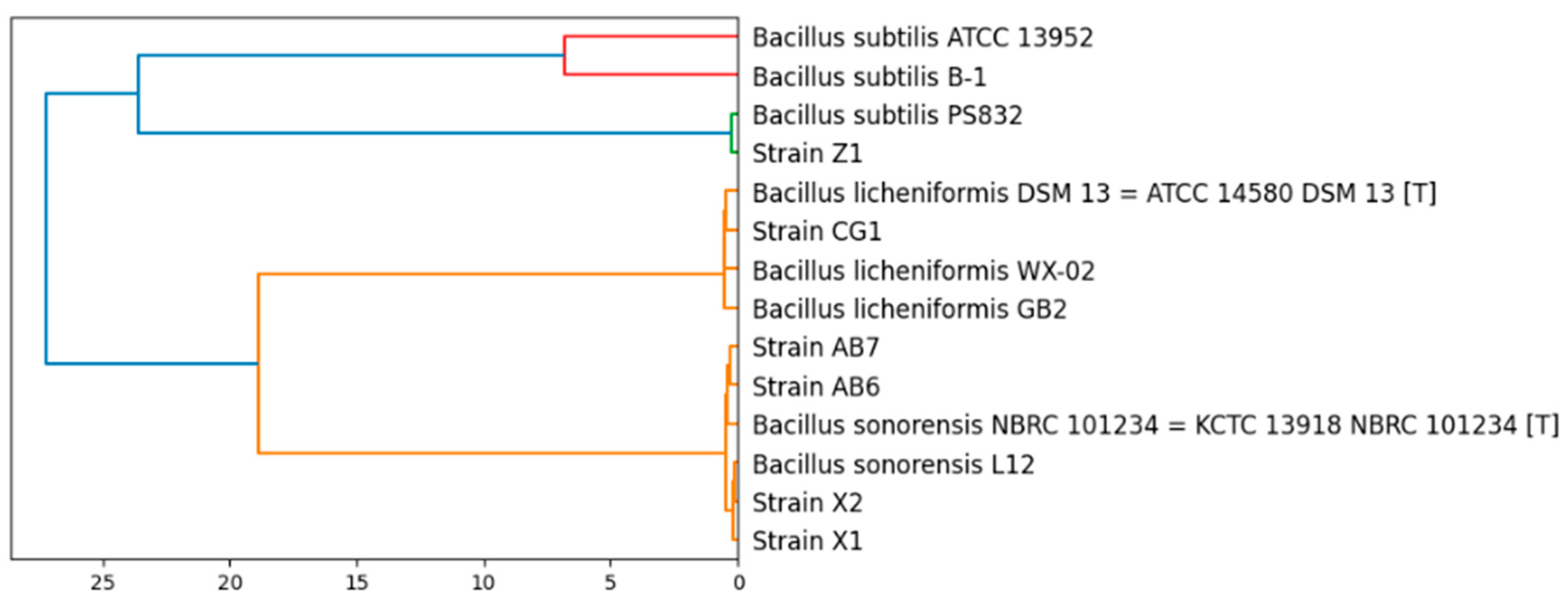


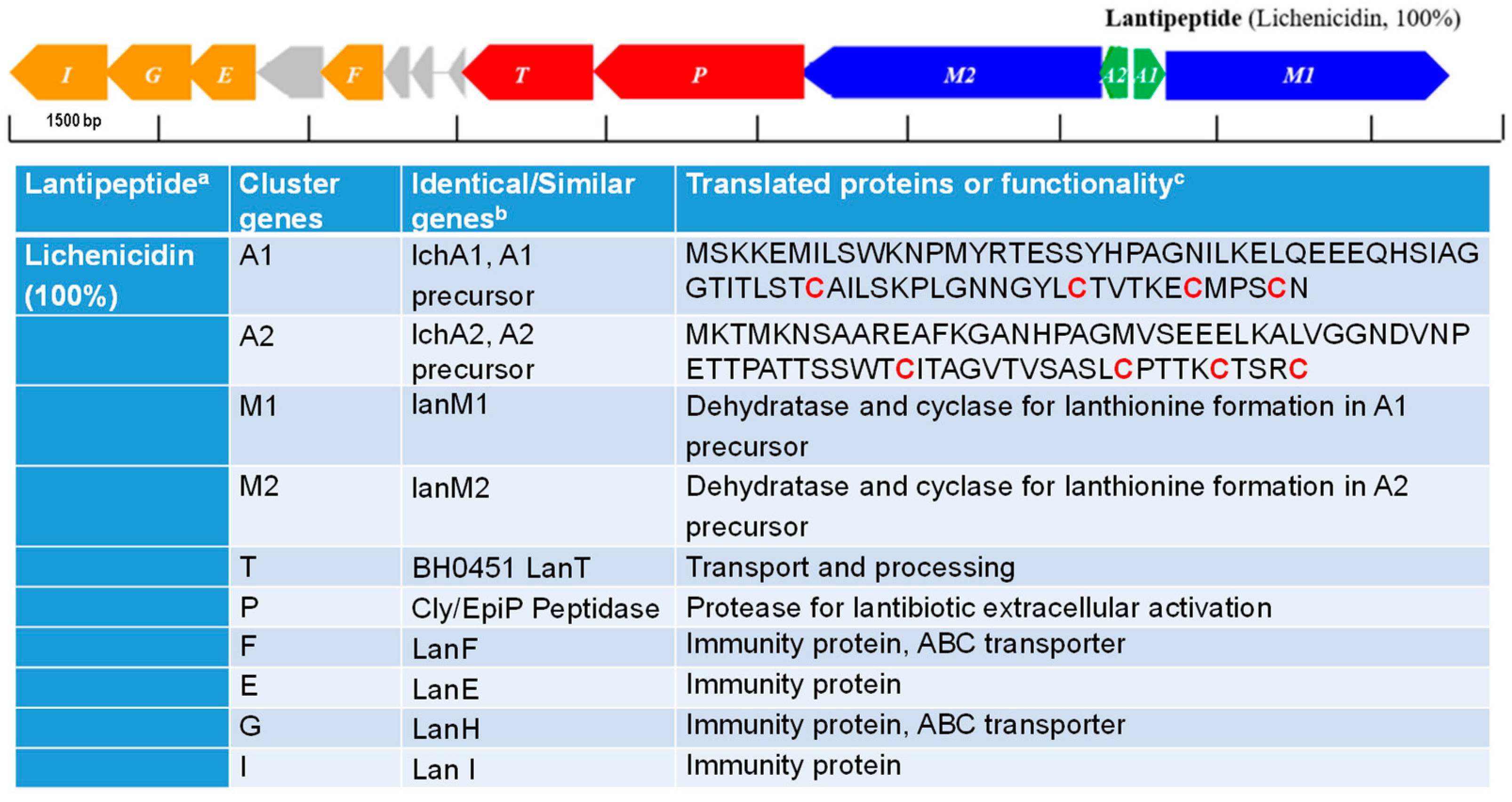
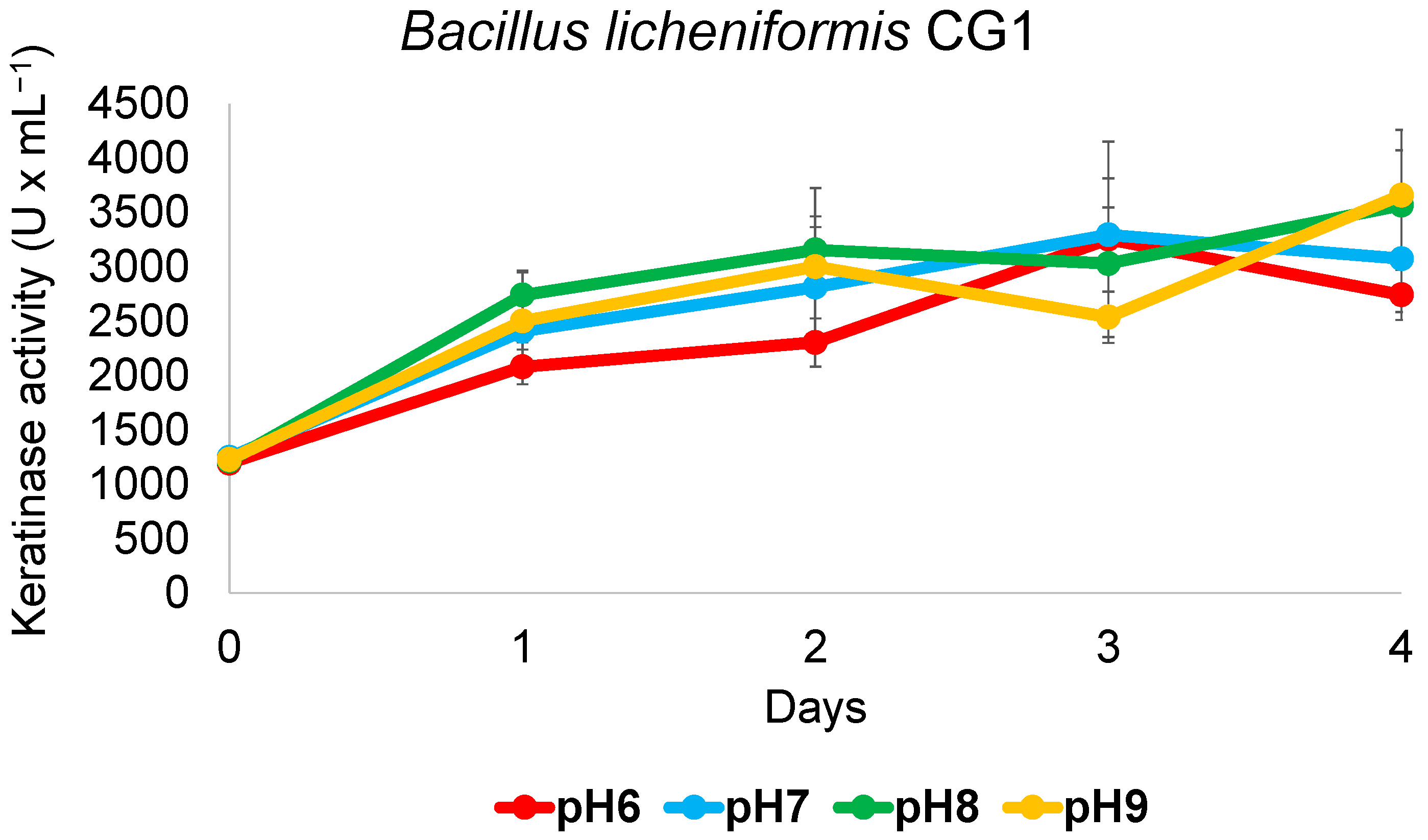
| Strain | Organism | Source | Location | BioSample | References |
|---|---|---|---|---|---|
| O29 | Lactobacillus kunkeei | Bee pollen | Verney Junction, BUCKINGHAMSHIRE | SAMN11831833 | [15] |
| E1 | Pediococcus acidilactici | Raw honey | Saffron Walden, ESSEX | SAMN11831832 | [15] |
| AD1 | Streptomyces drozdowiczii | Raw honey | Woking, SURREY | SAMN20207146 | [14] |
| AD2 | Streptomyces griseoaurantiacus | Raw honey | Woking, SURREY | SAMN20207147 | [14] |
| AN1 | Streptomyces albus | Bee pollen | West Byfleet, SURREY | SAMN20207148 | [14] |
| Z1 | Bacillus subtilis | Raw honey | Woking, SURREY | SAMN31988539 | This study |
| CG1 | Bacillus licheniformis | Raw honey | Shere, SURREY | SAMN31988540 | This study |
| AB6 | Bacillus sonorensis | Raw honey | West Byfleet, SURREY | SAMN31988543 | This study |
| AB7 | Bacillus sonorensis | Raw honey | West Byfleet, SURREY | SAMN31988544 | This study |
| X1 | Bacillus sonorensis | Royal jelly | Woking, SURREY | SAMN31988541 | This study |
| X2 | Bacillus sonorensis | Royal jelly | Woking, SURREY | SAMN31988542 | This study |
| Strain | Genome Accession | Bases | Contigs | Encoded Proteins | Secreted Proteins |
|---|---|---|---|---|---|
| Bacillus subtilis Z1 | JAPQWB000000000 | 4316935 | 305 | 4280 | 399 |
| Bacillus licheniformis CG1 | JAPQWC000000000 | 4228198 | 208 | 4252 | 407 |
| Bacillus sonorensis AB6 | JAPQWF000000000 | 4767461 | 595 | 4532 | 441 |
| Bacillus sonorensis AB7 | JAPQWG000000000 | 4580150 | 233 | 4440 | 437 |
| Bacillus sonorensis X1 | JAPQWD000000000 | 4903862 | 502 | 4770 | 447 |
| Bacillus sonorensis X2 | JAPQWE000000000 | 4809962 | 323 | 4807 | 445 |
| Strain | Anti- Micrococcus Activity | Anti- Mycobacterium Activity | Max. Keratinase Activity (U/mL) |
|---|---|---|---|
| Bacillus subtilis Z1 | Positive | Negative | 240 |
| Bacillus licheniformis CG1 | Positive | Positive | 3800 |
| Bacillus sonorensis AB6 | Positive | Negative | 240 |
| Bacillus sonorensis AB7 | Positive | Negative | 1450 |
| Bacillus sonorensis X1 | Positive | Negative | 290 |
| Bacillus sonorensis X2 | Positive | Negative | 260 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martín-González, D.; Bordel, S.; Solis, S.; Gutierrez-Merino, J.; Santos-Beneit, F. Characterization of Bacillus Strains from Natural Honeybee Products with High Keratinolytic Activity and Antimicrobial Potential. Microorganisms 2023, 11, 456. https://doi.org/10.3390/microorganisms11020456
Martín-González D, Bordel S, Solis S, Gutierrez-Merino J, Santos-Beneit F. Characterization of Bacillus Strains from Natural Honeybee Products with High Keratinolytic Activity and Antimicrobial Potential. Microorganisms. 2023; 11(2):456. https://doi.org/10.3390/microorganisms11020456
Chicago/Turabian StyleMartín-González, Diego, Sergio Bordel, Selvin Solis, Jorge Gutierrez-Merino, and Fernando Santos-Beneit. 2023. "Characterization of Bacillus Strains from Natural Honeybee Products with High Keratinolytic Activity and Antimicrobial Potential" Microorganisms 11, no. 2: 456. https://doi.org/10.3390/microorganisms11020456
APA StyleMartín-González, D., Bordel, S., Solis, S., Gutierrez-Merino, J., & Santos-Beneit, F. (2023). Characterization of Bacillus Strains from Natural Honeybee Products with High Keratinolytic Activity and Antimicrobial Potential. Microorganisms, 11(2), 456. https://doi.org/10.3390/microorganisms11020456






