Prevalence and Genetic Diversity of Legionella spp. in Hotel Water-Supply Systems in Latvia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Culturing of Legionella
2.3. DNA Extraction
2.4. Sequence-Based Typing
2.5. Whole Genome Sequencing
2.6. Data Analysis
3. Results
3.1. Prevalence of Legionella
3.2. Temperature of Hot Water
3.3. Levels of Colonization
3.4. Serotyping
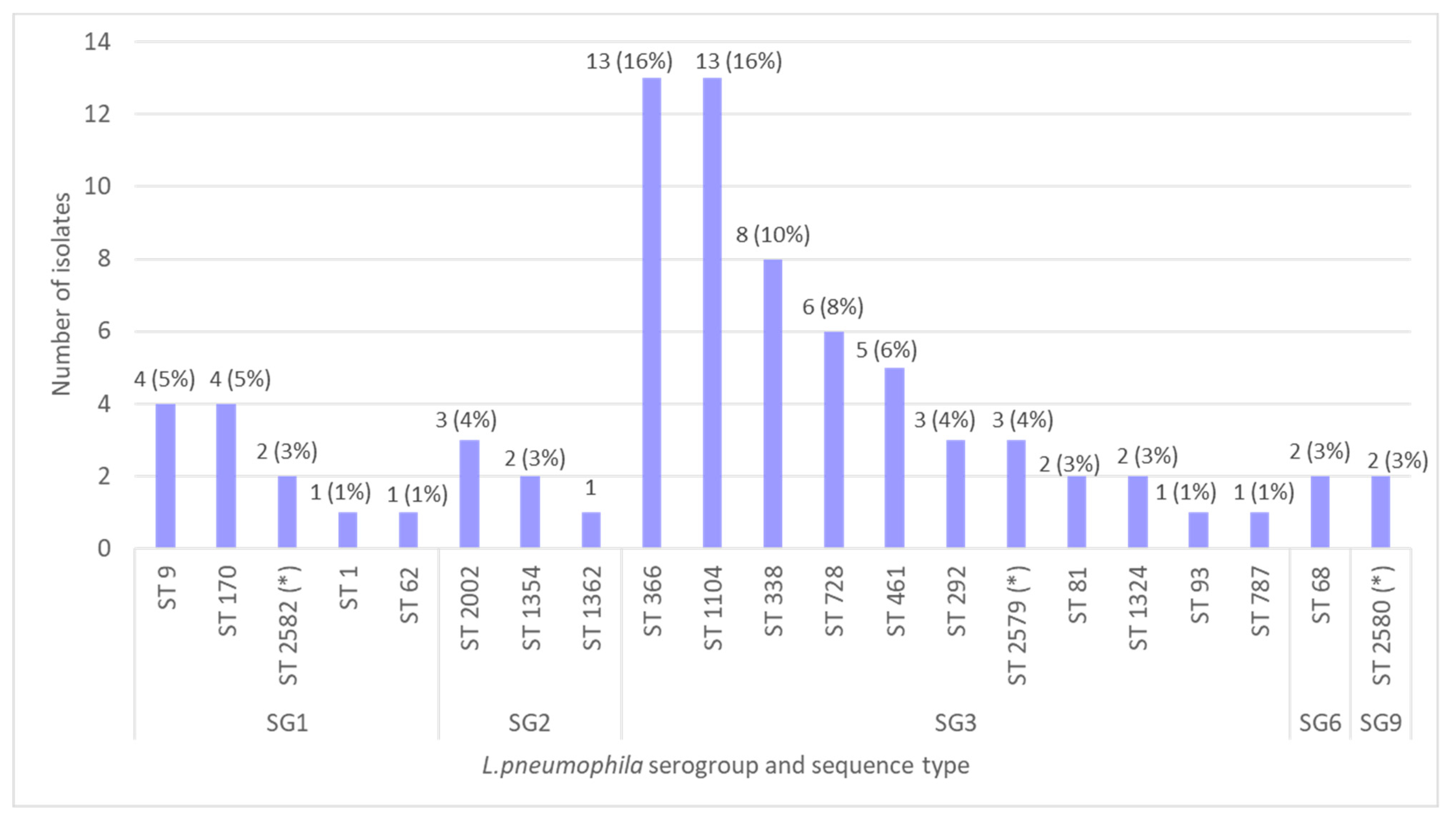
3.5. Sequence-Based Typing
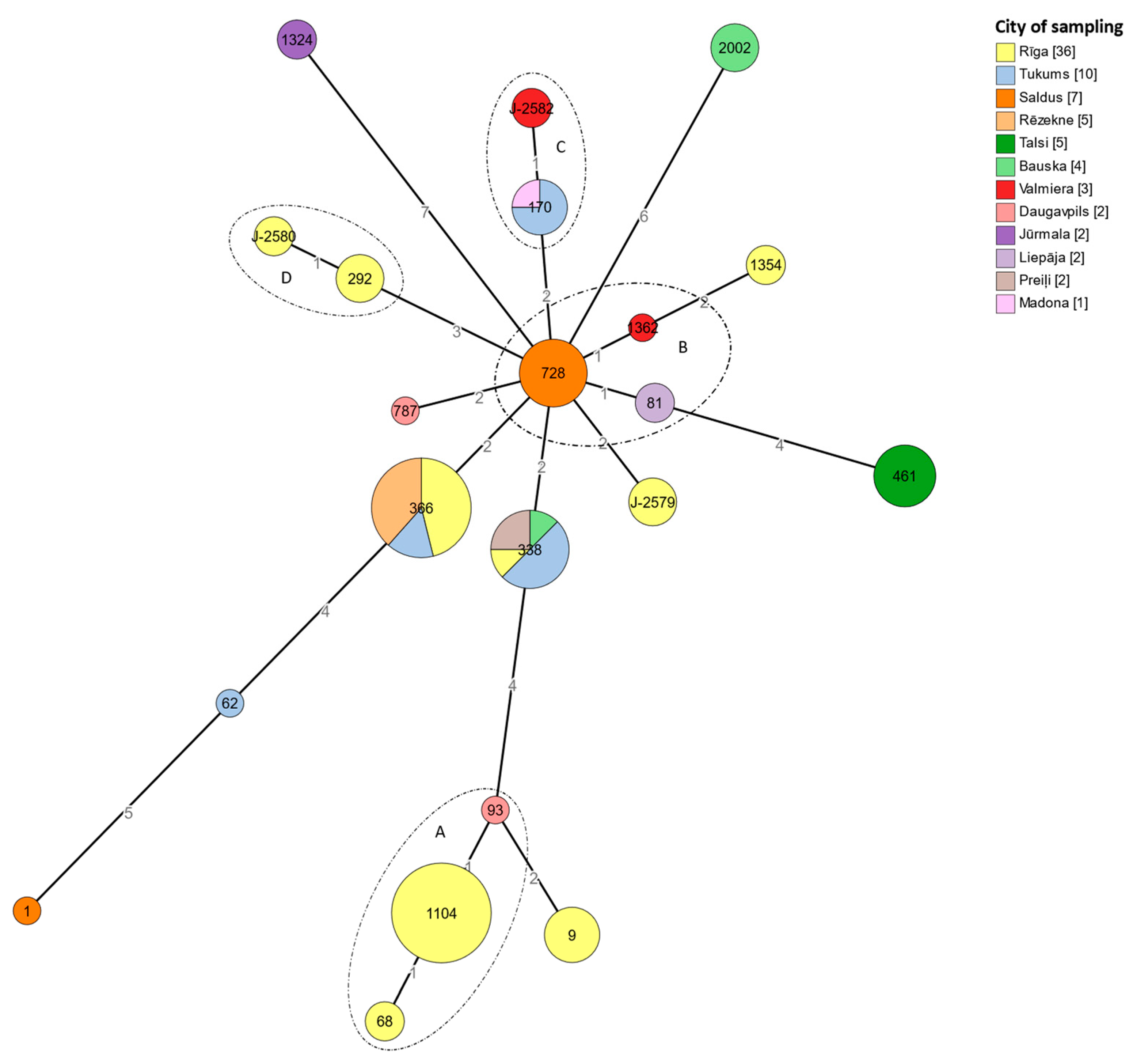
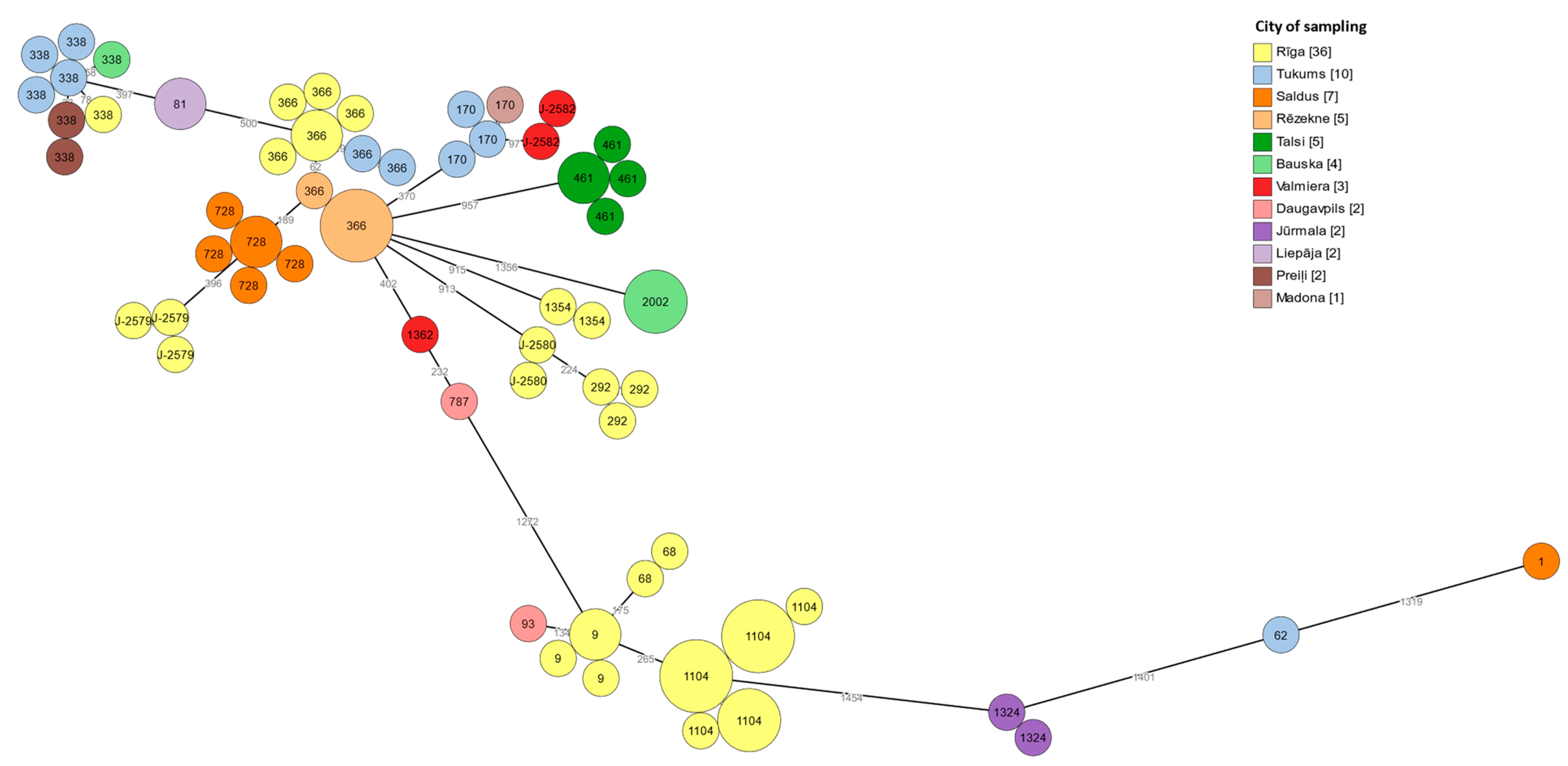
3.6. cgMLST Typing
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kessler, M.A.; Osman, F.; Marx, J.; Pop-Vicas, A.; Safdar, N. Hospital-acquired Legionella pneumonia outbreak at an academic medical center: Lessons learned. Am. J. Infect. Control 2021, 49, 1014–1020. [Google Scholar] [CrossRef] [PubMed]
- Paranjape, K.; Bédard, É.; Whyte, L.G.; Ronholm, J.; Prévost, M.; Faucher, S.P. Presence of Legionella spp. in cooling towers: The role of microbial diversity, Pseudomonas, and continuous chlorine application. Water Res. 2019, 169, 115252. [Google Scholar] [CrossRef]
- Mathys, W.; Stanke, J.; Harmuth, M.; Junge-Mathys, E. Occurrence of Legionella in hot water systems of single-family residences in suburbs of two German cities with special reference to solar and district heating. Int. J. Hyg. Environ. Health 2008, 211, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Fields, B.S.; Benson, R.F.; Besser, R.E. Legionella and Legionnaires’ Disease: 25 Years of Investigation. Clin. Microbiol. Rev. 2002, 15, 506–526. [Google Scholar] [CrossRef] [Green Version]
- Phin, N.; Parry-Ford, F.; Harrison, T.; Stagg, H.R.; Zhang, N.; Kumar, K.; Lortholary, O.; Zumla, A.; Abubakar, I. Epidemiology and clinical management of Legionnaires’ disease. Lancet Infect. Dis. 2014, 14, 1011–1021. [Google Scholar] [CrossRef] [PubMed]
- Ashbolt, N.J. Environmental (Saprozoic) Pathogens of Engineered Water Systems: Understanding Their Ecology for Risk Assessment and Management. Pathogens 2015, 4, 390–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuris, A.M.; Lafferty, K.D.; Sokolow, S.H. Sapronosis: A distinctive type of infectious agent. Trends Parasitol. 2014, 30, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Gattuso, G.; Rizzo, R.; Lavoro, A.; Spoto, V.; Porciello, G.; Montagnese, C.; Cinà, D.; Cosentino, A.; Lombardo, C.; Mezzatesta, M.L.; et al. Overview of the Clinical and Molecular Features of Legionella pneumophila: Focus on Novel Surveillance and Diagnostic Strategies. Antibiotics 2022, 11, 370. [Google Scholar] [CrossRef]
- Cervero-Aragó, S.; Sommer, R.; Araujo, R.M. Effect of UV irradiation (253.7 nm) on free Legionella and Legionella associated with its amoebae hosts. Water Res. 2014, 67, 299–309. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Legionnaires’ Disease—Annual Epidemiological Report for 2020. 10 May 2022. Available online: https://www.ecdc.europa.eu/en/publications-data/legionnaires-disease-annual-epidemiological-report-2020 (accessed on 15 November 2022).
- Graham, F.F.; Finn, N.; White, P.; Hales, S.; Baker, M.G. Global Perspective of Legionella Infection in Community-Acquired Pneumonia: A Systematic Review and Meta-Analysis of Observational Studies. Int. J. Environ. Res. Public Health 2022, 19, 1907. [Google Scholar] [CrossRef]
- Guyard, C.; Low, D.E. Legionella infections and travel associated legionellosis. Travel Med. Infect. Dis. 2011, 9, 176–186. [Google Scholar] [CrossRef]
- EN ISO 11731:2017; Water Quality—Enumeration of Legionella. European Committee for Standartization: Brussels, Belgium, 2017.
- Gaia, V.; Fry, N.K.; Afshar, B.; Lück, P.C.; Meugnier, H.; Etienne, J.; Peduzzi, R.; Harrison, T.G. Consensus Sequence-Based Scheme for Epidemiological Typing of Clinical and Environmental Isolates of Legionella pneumophila. J. Clin. Microbiol. 2005, 43, 2047–2052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Department of Health. Health Protection Agency 1988–2005. Sequence-Based Typing (SBT) Database for Legionella pneumophila Website. Available online: https://discovery.nationalarchives.gov.uk/details/r/C14933394 (accessed on 27 February 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Prjibelski, A.D.; Puglia, G.D.; Antipov, D.; Bushmanova, E.; Giordano, D.; Mikheenko, A.; Vitale, D.; Lapidus, A. Extending rnaSPAdes functionality for hybrid transcriptome assembly. BMC Bioinform. 2020, 21, 302. [Google Scholar] [CrossRef] [PubMed]
- Mentasti, M.; Underwood, A.; Lück, C.; Kozak-Muiznieks, N.A.; Harrison, T.G.; Fry, N.K. Extension of the Legionella pneumophila sequence-based typing scheme to include strains carrying a variant of the N-acylneuraminate cytidylyltransferase gene. Clin. Microbiol. Infect. 2014, 20, O435–O441. [Google Scholar] [CrossRef] [Green Version]
- Gordon, M.; Yakunin, E.; Valinsky, L.; Chalifa-Caspi, V.; Moran-Gilad, J. A bioinformatics tool for ensuring the backwards compatibility of Legionella pneumophila typing in the genomic era. Clin. Microbiol. Infect. 2017, 23, 306–310. [Google Scholar] [CrossRef] [Green Version]
- Moran-Gilad, J.; Prior, K.; Yakunin, E.; Harrison, T.G.; Underwood, A.; Lazarovitch, T.; Valinsky, L.; Lück, C.; Krux, F.; Agmon, V.; et al. Design and application of a core genome multilocus sequence typing scheme for investigation of Legionnaires’ disease incidents. Eurosurveillance 2015, 20, 21186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, M.; Machado, M.P.; Silva, D.N.; Rossi, M.; Moran-Gilad, J.; Santos, S.; Ramirez, M.; Carriço, J.A. chewBBACA: A complete suite for gene-by-gene schema creation and strain identification. Microb. Genom. 2018, 4, e000166. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadakis, A.; Keramarou, M.; Chochlakis, D.; Sandalakis, V.; Mouchtouri, V.A.; Psaroulaki, A. Legionella spp. Colonization in Water Systems of Hotels Linked with Travel-Associated Legionnaires’ Disease. Water 2021, 13, 2243. [Google Scholar] [CrossRef]
- De Filippis, P.; Mozzetti, C.; Amicosante, M.; D’Alò, G.L.; Messina, A.; Varrenti, D.; Giammattei, R.; Di Giorgio, F.; Corradi, S.; D’Auria, A.; et al. Occurrence of Legionella in showers at recreational facilities. J. Water Health 2017, 15, 402–409. [Google Scholar] [CrossRef]
- Doménech-Sánchez, A.; Laso, E.; Berrocal, C.I.; Albertí, S. Environmental surveillance of Legionella in tourist facilities of the Balearic Islands, Spain, 2006 to 2010 and 2015 to 2018. Eurosurveillance 2022, 27, 2100769. [Google Scholar] [CrossRef] [PubMed]
- Yakunin, E.; Kostyal, E.; Agmon, V.; Grotto, I.; Valinsky, L.; Moran-Gilad, J. A Snapshot of the Prevalence and Molecular Diversity of Legionella pneumophila in the Water Systems of Israeli Hotels. Pathogens 2020, 9, 414. [Google Scholar] [CrossRef] [PubMed]
- Bešić, A.; Karakaš, S.; Obradović, Z.; Mušović, A.; Hrapović, E. Travel-related epidemiological studies of legionellosis in Federation of Bosnia and Herzegovina. Health Technol. 2021, 11, 971–979. [Google Scholar] [CrossRef]
- Doménech-Sánchez, A.; Laso, E.; Albertí, S. Determination of Legionella spp. prevalence in Spanish hotels in five years. Are tourists really at risk? Travel Med. Infect. Dis. 2022, 46, 102269. [Google Scholar] [CrossRef] [PubMed]
- Rasheduzzaman; Singh, R.; Haas, C.N.; Gurian, P.L. Required water temperature in hotel plumbing to control Legionella growth. Water Res. 2020, 182, 115943. [Google Scholar] [CrossRef] [PubMed]
- De Giglio, O.; Fasano, F.; Diella, G.; Lopuzzo, M.; Napoli, C.; Apollonio, F.; Brigida, S.; Calia, C.; Campanale, C.; Marzella, A.; et al. Legionella and legionellosis in touristic-recreational facilities: Influence of climate factors and geostatistical analysis in Southern Italy (2001–2017). Environ. Res. 2019, 178, 108721. [Google Scholar] [CrossRef]
- Keše, D.; Obreza, A.; Rojko, T.; Kišek, T. Legionella pneumophila—Epidemiology and Characterization of Clinical Isolates, Slovenia, 2006–2020. Diagnostics 2021, 11, 1201. [Google Scholar] [CrossRef]
- Lévesque, S.; Lalancette, C.; Bernard, K.; Pacheco, A.L.; Dion, R.; Longtin, J.; Tremblay, C. Molecular Typing of Legionella pneumophila Isolates in the Province of Quebec from 2005 to 2015. PLoS ONE 2016, 11, e0163818. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Zhao, S.; Cai, X.; Mu, D.; Zhang, X.; Kang, J.; Zhao, L.; Chen, Y. Sequence-based typing of clinical and environmental Legionella pneumophila isolates in Shenyang, China. Enfermedades Infecc. Microbiol. Clin. 2021, 39, 383–389. [Google Scholar] [CrossRef]
- Herwaldt, L.A.; Marra, A.R. Legionella . Curr. Opin. Infect. Dis. 2018, 31, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Cazalet, C.; Rusniok, C.; Brüggemann, H.; Zidane, N.; Magnier, A.; Ma, L.; Tichit, M.; Jarraud, S.; Bouchier, C.; Vandenesch, F.; et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 2004, 36, 1165–1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vekens, E.; Soetens, O.; De Mendonça, R.; Echahidi, F.; Roisin, S.; Deplano, A.; Eeckhout, L.; Achtergael, W.; Piérard, D.; Denis, O.; et al. Sequence-based typing of Legionella pneumophila serogroup 1 clinical isolates from Belgium between 2000 and 2010. Eurosurveillance 2012, 17, 20302. [Google Scholar] [CrossRef] [PubMed]
- Pancer, K. Sequence-based typing of Legionella pneumophila strains isolated from hospital water distribution systems as a com-plementary element of risk assessment of legionellosis in Poland. Ann. Agric. Environ. Med. AAEM 2013, 20, 436–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozak-Muiznieks, N.A.; Lucas, C.E.; Brown, E.; Pondo, T.; Taylor, T.H., Jr.; Frace, M.; Miskowski, D.; Winchell, J.M. Prevalence of Sequence Types among Clinical and Environmental Isolates of Legionella pneumophila Serogroup 1 in the United States from 1982 to 2012. J. Clin. Microbiol. 2014, 52, 201–211. [Google Scholar] [CrossRef] [Green Version]
- Sreenath, K.; Chaudhry, R.; Vinayaraj, E.V.; Dey, A.B.; Kabra, S.K.; Thakur, B.; Guleria, R. Distribution of Virulence Genes and Sequence-Based Types Among Legionella pneumophila Isolated from the Water Systems of a Tertiary Care Hospital in India. Front. Public Health 2020, 8, 596463. [Google Scholar] [CrossRef]
- Harrison, T.G.; Afshar, B.; Doshi, N.; Fry, N.; Lee, J.V. Distribution of Legionella pneumophila serogroups, monoclonal antibody subgroups and DNA sequence types in recent clinical and environmental isolates from England and Wales (2000–2008). Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 781–791. [Google Scholar] [CrossRef]
- Ginevra, C.; Forey, F.; Campèse, C.; Reyrolle, M.; Che, D.; Etienne, J.; Jarraud, S. Lorraine Strain of Legionella pneumophila Serogroup 1, France. Emerg. Infect. Dis. 2008, 14, 673–675. [Google Scholar] [CrossRef]
- Boer, J.W.D.; Bruin, J.P.; Verhoef, L.P.B.; Van der Zwaluw, K.; Jansen, R.; Yzerman, E.P.F. Genotypic comparison of clinical Legionella isolates and patient-related environmental isolates in The Netherlands, 2002–2006. Clin. Microbiol. Infect. 2008, 14, 459–466. [Google Scholar] [CrossRef] [Green Version]
- Wüthrich, D.; Gautsch, S.; Spieler-Denz, R.; Dubuis, O.; Gaia, V.; Moran-Gilad, J.; Hinic, V.; Seth-Smith, H.M.; Nickel, C.H.; Tschudin-Sutter, S.; et al. Air-conditioner cooling towers as complex reservoirs and continuous source of Legionella pneumophila infection evidenced by a genomic analysis study in 2017, Switzerland. Eurosurveillance 2019, 24, 1800192. [Google Scholar] [CrossRef] [Green Version]
- van Belkum, A.; Tassios, P.; Dijkshoorn, L.; Haeggman, S.; Cookson, B.; Fry, N.; Fussing, V.; Green, J.; Feil, E.; Gerner-Smidt, P.; et al. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 2007, 13, 1–46. [Google Scholar] [CrossRef] [PubMed]

| Location of Hotel | Total Tested | Legionella spp. Positive | ||
|---|---|---|---|---|
| No. of Hotels (%) | No. of Samples (%) | No. of Hotels (%) | No. of Samples (%) | |
| Riga | 54 (67.5) | 683 (81.9) | 28 (51.9) | 162 (23.7) |
| Other cities and towns | 26 (32.5) | 151 (18.1) | 19 (73.1) | 73 (48.3) |
| Total | 80 (100) | 834 (100) | 47 (58.8) | 235 (28.2) |
| Water Source in Hotels | Total | ||
|---|---|---|---|
| Level of Colonization, CFU/L | Surface Water | Underground Aquifers | |
| Min | 50 | 50 | 50 |
| Max | 1.1 × 104 | 1.1 × 104 | 1.1 × 104 |
| Average | 1.0 × 103 ± 1.3 × 102 | 1.6 × 103 ± 2.6 × 102 | 1.2 × 103 ± 1.2 × 102 |
| Temperature, °C | Before Flushing | After Flushing |
|---|---|---|
| Min | 16.2 | 27.7 |
| Max | 62.9 | 68.8 |
| Average | 35.7 ± 0.7 | 49.8 ± 0.4 |
| Mode | 27.0 | 47.0 |
| Level of Colonization, CFU/L | Before Flushing | After Flushing |
|---|---|---|
| Min | 50 | 50 |
| Max | 1.1 × 104 | 9.0 × 103 |
| Average | 1.7 × 103 ± 2.8 × 102 | 1.2 × 103 ± 1.8 × 102 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valciņa, O.; Pūle, D.; Ķibilds, J.; Lazdāne, A.; Trofimova, J.; Makarova, S.; Konvisers, G.; Ķimse, L.; Krūmiņa, A.; Bērziņš, A. Prevalence and Genetic Diversity of Legionella spp. in Hotel Water-Supply Systems in Latvia. Microorganisms 2023, 11, 596. https://doi.org/10.3390/microorganisms11030596
Valciņa O, Pūle D, Ķibilds J, Lazdāne A, Trofimova J, Makarova S, Konvisers G, Ķimse L, Krūmiņa A, Bērziņš A. Prevalence and Genetic Diversity of Legionella spp. in Hotel Water-Supply Systems in Latvia. Microorganisms. 2023; 11(3):596. https://doi.org/10.3390/microorganisms11030596
Chicago/Turabian StyleValciņa, Olga, Daina Pūle, Juris Ķibilds, Andžela Lazdāne, Jūlija Trofimova, Svetlana Makarova, Genadijs Konvisers, Laima Ķimse, Angelika Krūmiņa, and Aivars Bērziņš. 2023. "Prevalence and Genetic Diversity of Legionella spp. in Hotel Water-Supply Systems in Latvia" Microorganisms 11, no. 3: 596. https://doi.org/10.3390/microorganisms11030596
APA StyleValciņa, O., Pūle, D., Ķibilds, J., Lazdāne, A., Trofimova, J., Makarova, S., Konvisers, G., Ķimse, L., Krūmiņa, A., & Bērziņš, A. (2023). Prevalence and Genetic Diversity of Legionella spp. in Hotel Water-Supply Systems in Latvia. Microorganisms, 11(3), 596. https://doi.org/10.3390/microorganisms11030596






