Characterization of Linezolid-Analogue L3-Resistance Mutation in Staphylococcus aureus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Minimum Inhibitory Concentration (MIC) Determination
2.2. Cytotoxicity Assay
2.3. Isolation of Resistant and Revertant Mutant 10f
2.4. Bacterial DNA Extraction
2.5. Polymerase Chain Reaction (PCR) Amplification of Individual 23S rRNA, rplD and rplC Genes
2.6. Modelling
3. Results
3.1. 10f Antimicrobial Activity
3.2. Isolation of S. aureus Mutant Resistant to 10f
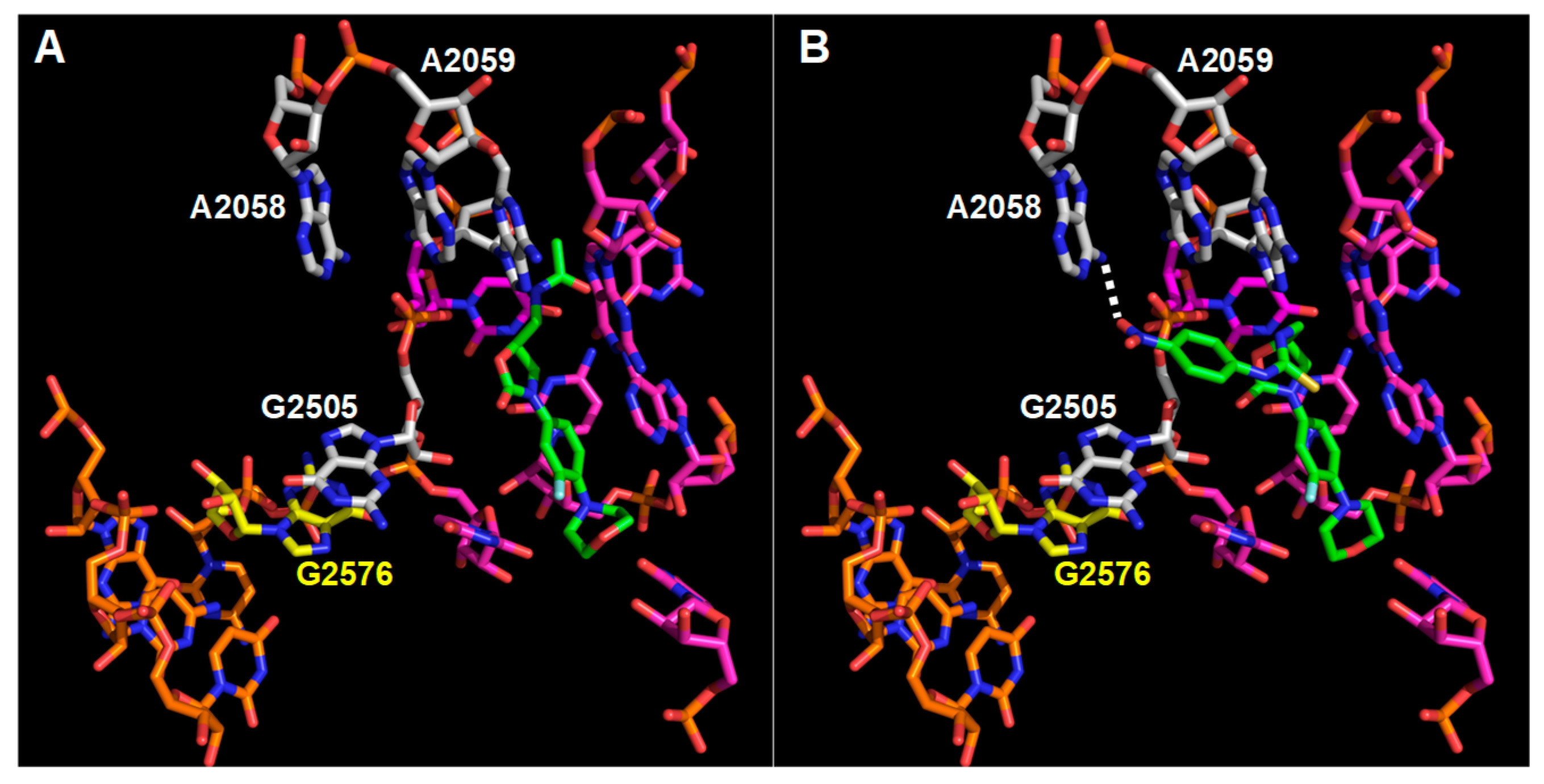
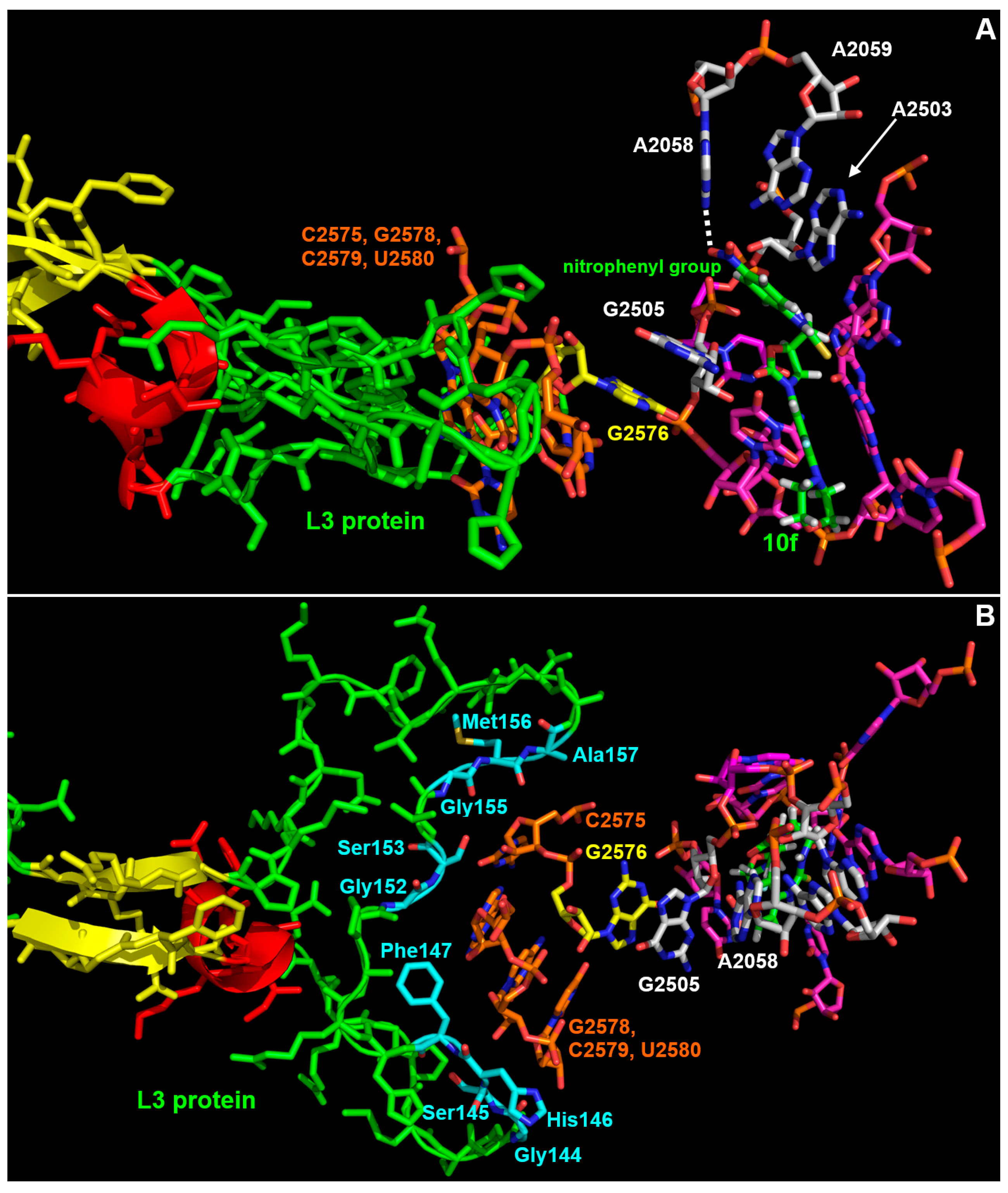
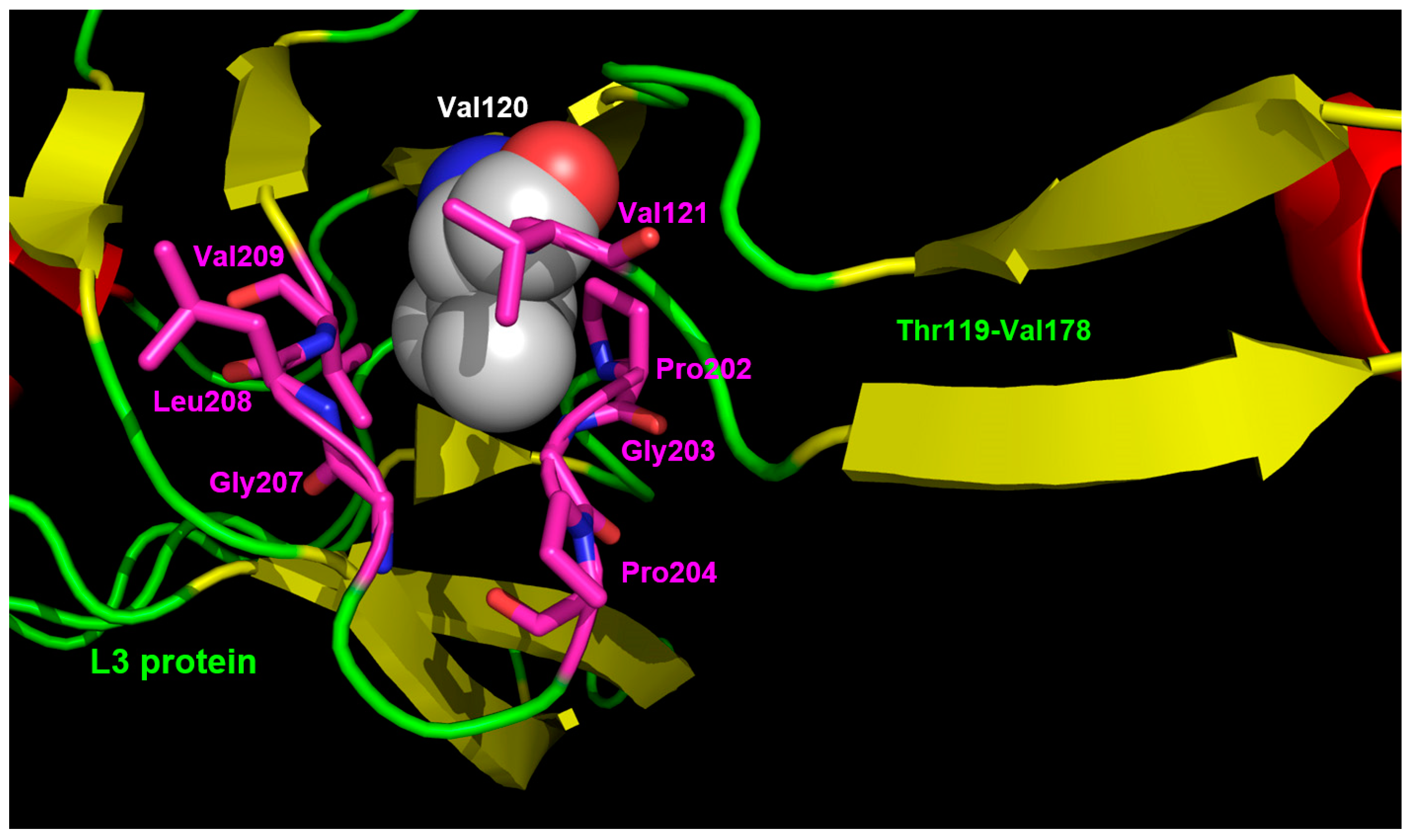
3.3. Interaction Model between 10f and the Bacterial Ribosome
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Manfredi, R. Update on the Appropriate Use of Linezolid in Clinical Practice. Ther. Clin. Risk Manag. 2006, 2, 455–464. [Google Scholar] [CrossRef] [Green Version]
- Boak, L.M.; Rayner, C.R.; Grayson, M.L.; Paterson, D.L.; Spelman, D.; Khumra, S.; Capitano, B.; Forrest, A.; Li, J.; Nation, R.L.; et al. Clinical Population Pharmacokinetics and Toxicodynamics of Linezolid. Antimicrob. Agents Chemother. 2014, 58, 2334–2343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besier, S.; Ludwig, A.; Zander, J.; Brade, V.; Wichelhaus, T.A. Linezolid Resistance in Staphylococcus aureus: Gene Dosage Effect, Stability, Fitness Costs, and Cross-Resistances. Antimicrob. Agents Chemother. 2008, 52, 1570–1572. [Google Scholar] [CrossRef] [Green Version]
- Sadowy, E. Linezolid Resistance Genes and Genetic Elements Enhancing Their Dissemination in Enterococci and Streptococci. Plasmid 2018, 99, 89–98. [Google Scholar] [CrossRef]
- Toh, S.-M.; Xiong, L.; Arias, C.A.; Villegas, M.V.; Lolans, K.; Quinn, J.; Mankin, A.S. Acquisition of a Natural Resistance Gene Renders a Clinical Strain of Methicillin-Resistant Staphylococcus aureus Resistant to the Synthetic Antibiotic Linezolid: Linezolid Resistance through Ribosome Modification. Mol. Microbiol. 2007, 64, 1506–1514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Rosa, M.; Zanfardino, A.; Notomista, E.; Wichelhaus, T.A.; Saturnino, C.; Varcamonti, M.; Soriente, A. Novel Promising Linezolid Analogues: Rational Design, Synthesis and Biological Evaluation. Eur. J. Med. Chem. 2013, 69, 779–785. [Google Scholar] [CrossRef] [PubMed]
- Zanfardino, A.; Bosso, A.; Gallo, G.; Pistorio, V.; Di Napoli, M.; Gaglione, R.; Dell’Olmo, E.; Varcamonti, M.; Notomista, E.; Arciello, A.; et al. Human Apolipoprotein E as a Reservoir of Cryptic Bioactive Peptides: The Case of ApoE 133-167. J. Pept. Sci. 2018, 24, e3095. [Google Scholar] [CrossRef] [PubMed]
- Vitiello, G.; Zanfardino, A.; Tammaro, O.; Di Napoli, M.; Caso, M.F.; Pezzella, A.; Varcamonti, M.; Silvestri, B.; D’Errico, G.; Costantini, A.; et al. Bioinspired Hybrid Eumelanin–TiO2 Antimicrobial Nanostructures: The Key Role of Organo–Inorganic Frameworks in Tuning Eumelanin’s Biocide Action Mechanism through Membrane Interaction. RSC Adv. 2018, 8, 28275–28283. [Google Scholar] [CrossRef] [Green Version]
- Pitcher, D.G.; Saunders, N.A.; Owen, R.J. Rapid Extraction of Bacterial Genomic DNA with Guanidium Thiocyanate. Lett. Appl. Microbiol. 1989, 8, 151–156. [Google Scholar] [CrossRef]
- Meka, V.G.; Pillai, S.K.; Sakoulas, G.; Wennersten, C.; Venkataraman, L.; DeGirolami, P.C.; Eliopoulos, G.M.; Moellering, R.C., Jr.; Gold, H.S. Linezolid Resistance in Sequential Staphylococcus aureus Isolates Associated with a T2500A Mutation in the 23S RRNA Gene and Loss of a Single Copy of RRNA. J. Infect. Dis. 2004, 190, 311–317. [Google Scholar] [CrossRef] [Green Version]
- Pillai, S.K.; Sakoulas, G.; Wennersten, C.; Eliopoulos, G.M.; Moellering, R.C., Jr.; Ferraro, M.J.; Gold, H.S. Linezolid Resistance in Staphylococcus Aureus: Characterization and Stability of Resistant Phenotype. J. Infect. Dis. 2002, 186, 1603–1607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prunier, A.-L.; Trong, H.N.; Tande, D.; Segond, C.; Leclercq, R. Mutation of L4 Ribosomal Protein Conferring Unusual Macrolide Resistance in Two Independent Clinical Isolates of Staphylococcus aureus. Microb. Drug Resist. 2005, 11, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Schulze, H.; Nierhaus, K.H. Minimal Set of Ribosomal Components for Reconstitution of the Peptidyltransferase Activity. EMBO J. 1982, 1, 609–613. [Google Scholar] [CrossRef] [PubMed]
- Gamalinda, M.; Ohmayer, U.; Jakovljevic, J.; Kumcuoglu, B.; Woolford, J.; Mbom, B.; Lin, L.; Woolford, J.L. A Hierarchical Model for Assembly of Eukaryotic 60S Ribosomal Subunit Domains. Genes Dev. 2014, 28, 198–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Shem, A.; Garreau De Loubresse, N.; Melnikov, S.; Jenner, L.; Yusupova, G.; Yusupov, M. The Structure of the Eukaryotic Ribosome at 3.0 Å Resolution. Science 2011, 334, 1524–1529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolter, N.; Smith, A.M.; Farrell, D.J.; Schaffner, W.; Moore, M.; Whitney, C.G.; Jorgensen, J.H.; Klugman, K.P. Novel Mechanism of Resistance to Oxazolidinones, Macrolides, and Chloramphenicol in Ribosomal Protein L4 of the Pneumococcus. Antimicrob. Agents Chemother. 2005, 49, 3554–3557. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, S.; Shen, J.; Kadlec, K.; Wang, Y.; Brenner Michael, G.; Feßler, A.T.; Vester, B. Lincosamides, Streptogramins, Phenicols, and Pleuromutilins: Mode of Action and Mechanisms of Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a027037. [Google Scholar] [CrossRef] [Green Version]
- Miller, K.; Dunsmore, C.J.; Fishwick, C.W.G.; Chopra, I. Linezolid and Tiamulin Cross-Resistance in Staphylococcus aureus Mediated by Point Mutations in the Peptidyl Transferase Center. Antimicrob. Agents Chemother. 2008, 52, 1737–1742. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Trallero, E.; Tamayo, E.; Montes, M.; García-Arenzana, J.M.; Iriarte, V. In Vitro Activities of Retapamulin and 16 Other Antimicrobial Agents against Recently Obtained Streptococcus Pyogenes Isolates. Antimicrob. Agents Chemother. 2011, 55, 2406–2408. [Google Scholar] [CrossRef] [Green Version]
- Locke, J.B.; Hilgers, M.; Shaw, K.J. Mutations in Ribosomal Protein L3 Are Associated with Oxazolidinone Resistance in Staphylococci of Clinical Origin. Antimicrob. Agents Chemother. 2009, 53, 5275–5278. [Google Scholar] [CrossRef] [Green Version]
- Eyal, Z.; Matzov, D.; Krupkin, M.; Wekselman, I.; Paukner, S.; Zimmerman, E.; Rozenberg, H.; Bashan, A.; Yonath, A. Structural Insights into Species-Specific Features of the Ribosome from the Pathogen Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2015, 112, E5805–E5814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zurenko, G.E.; Yagi, B.H.; Schaadt, R.D.; Allison, J.W.; Kilburn, J.O.; Glickman, S.E.; Hutchinson, D.K.; Barbachyn, M.R.; Brickner, S.J. In Vitro Activities of U-100592 and U-100766, Novel Oxazolidinone Antibacterial Agents. Antimicrob. Agents Chemother. 1996, 40, 839–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pi, R.; Liu, Q.; Jiang, Q.; Gao, Q. Characterization of Linezolid-Resistance-Associated Mutations in Mycobacterium Tuberculosis through WGS. J. Antimicrob. Chemother. 2019, 74, 1795–1798. [Google Scholar] [CrossRef] [PubMed]
- Klitgaard, R.N.; Ntokou, E.; Nørgaard, K.; Biltoft, D.; Hansen, L.H.; Trædholm, N.M.; Kongsted, J.; Vester, B. Mutations in the Bacterial Ribosomal Protein L3 and Their Association with Antibiotic Resistance. Antimicrob. Agents Chemother. 2015, 59, 3518–3528. [Google Scholar] [CrossRef] [Green Version]


| Primer | Sequence |
|---|---|
| For-rrn-all | 5′-CCCGCACGAAAGGCGTAACG-3′ |
| Rev-rrn1 | 5′-CGCTTCTTCTTGATTTAAACTTTC-3′ |
| Rev-rrn2 | 5′-GGCTTTGGCGGTATAGACG-3′ |
| Rev-rrn3 | 5′-GGCAGATGCTCTCCCAGC-3′ |
| Rev-rrn4 | 5′-CAACGTGTAAATCATTTCGATC-3′ |
| Rev-rrn5 | 5′-GGCTCCACAGGTAGGACTCG-3′ |
| Rev-rrn6 | 5′-CCTGAGCCAGGATCAAAC-3′ |
| Rev-rrn-all | 5′-GTTGGGAAATCTCATCTTG-3′ |
| rplC-For | 5′-ATGACCAAAGGAATCTTAGG-3′ |
| rplC-Rev | 5′-TTATTTATTACCTTTTTTAATTGAAG-3′ |
| rplD-For | 5′-ATGGCTAATTATGATGTT-3′ |
| rplD-Rev | 5′-TTATCCGAGCACCTCCTC-3′ |
| Strain | MIC 10f (µg/mL) | MIC Linezolid (µg/mL) |
|---|---|---|
| S. aureus ATCC 29213 | 2 | 1 |
| S. aureus ATCC 6538P | 1 | 1 |
| MRSA WKZ-2 | 2 | 1 |
| E. faecalis ATCC 29212 | 2 | 1 |
| E. faecium ATCC 14434 | 1 | 2 |
| S. epidermidis ATCC 12228 | 0.5 | 0.5 |
| Strain | MIC 10f (µg/mL) | MIC Linezolid (µg/mL) |
|---|---|---|
| S. aureus ATCC 6538P | 1 | 1 |
| S. aureus 10fR | 16 | 1 |
| S. aureus 10frev | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zanfardino, A.; Di Napoli, M.; Migliore, F.; Hay Mele, B.; Soriente, A.; De Rosa, M.; Notomista, E.; Varcamonti, M. Characterization of Linezolid-Analogue L3-Resistance Mutation in Staphylococcus aureus. Microorganisms 2023, 11, 700. https://doi.org/10.3390/microorganisms11030700
Zanfardino A, Di Napoli M, Migliore F, Hay Mele B, Soriente A, De Rosa M, Notomista E, Varcamonti M. Characterization of Linezolid-Analogue L3-Resistance Mutation in Staphylococcus aureus. Microorganisms. 2023; 11(3):700. https://doi.org/10.3390/microorganisms11030700
Chicago/Turabian StyleZanfardino, Anna, Michela Di Napoli, Federica Migliore, Bruno Hay Mele, Annunziata Soriente, Margherita De Rosa, Eugenio Notomista, and Mario Varcamonti. 2023. "Characterization of Linezolid-Analogue L3-Resistance Mutation in Staphylococcus aureus" Microorganisms 11, no. 3: 700. https://doi.org/10.3390/microorganisms11030700
APA StyleZanfardino, A., Di Napoli, M., Migliore, F., Hay Mele, B., Soriente, A., De Rosa, M., Notomista, E., & Varcamonti, M. (2023). Characterization of Linezolid-Analogue L3-Resistance Mutation in Staphylococcus aureus. Microorganisms, 11(3), 700. https://doi.org/10.3390/microorganisms11030700








