Identification of Z-Tyr-Ala-CHN2, a Cathepsin L Inhibitor with Broad-Spectrum Cell-Specific Activity against Coronaviruses, including SARS-CoV-2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Viruses
2.2. Antiviral Compounds
2.3. Antibodies, Cellular Counterstains, and Staining Buffers for Immunofluorescence and LysoTracker Assays
2.4. Phenotypic Screening and In Vitro Assays
2.4.1. SARS-CoV-2 Antiviral Assay in VeroE6-eGFP Cells
2.4.2. SARS-CoV-2 Viral RNA Copy Determination in A549-hACE2 Cells
2.4.3. Immunofluorescence-Based Antiviral Assays
2.4.4. Immunofluorescence-Based Antiviral Assay Using LysoTracker Dye to Detect Accumulation of Acidic Organelles
2.4.5. Time-of-Addition (TOA) Assay
2.4.6. Pseudotyped Virus Neutralization Assay in VeroE6 Cells
2.4.7. Antiviral Assessment in Caco-2 Cells by Visual CPE Scoring
2.4.8. Chemiluminescence-Based and Colorimetric Cytotoxicity Assays
2.4.9. Cathepsin L Inhibition Assay
2.4.10. Air–Liquid Interface (ALI) Cultures
2.4.11. Dose-Response Curve Fitting and EC50 and CC50 Calculations
3. Results
3.1. Z-Tyr-Ala-CHN2 Shows Antiviral Activity against SARS-CoV-2 in VeroE6-eGFP Cells
3.2. Confirmation of Z-Tyr-Ala-CHN2 Activity in a Viral RNA Yield Assay
3.3. Assessment of Broad-Spectrum Antiviral Activity of Z-Tyr-Ala-CHN2 against SARS-CoV-1 and HCoV-229E
3.4. Antiviral Activity of Z-Tyr-Ala-CHN2 across Different Cellular Backgrounds
3.5. Assessment of the Mechanism of Action of Z-Tyr-Ala-CHN2
3.5.1. TOA Assay
3.5.2. Pseudotyped Virus Neutralization Assay in VeroE6 Cells
3.5.3. Z-Tyr-Ala-CHN2 Is a Cathepsin L Inhibitor That Does Not Cause Disruption of Lysosome Trafficking
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Control and Prevention. Interim Clinical Considerations for Use of COVID-19 Vaccines Currently Approved or Authorized in the United States. Available online: https://www.cdc.gov/vaccines/covid-19/clinical-considerations/covid-19-vaccines-us.html (accessed on 28 February 2022).
- Tagoe, E.T.; Sheikh, N.; Morton, A.; Nonvignon, J.; Sarker, A.R.; Williams, L.; Megiddo, I. COVID-19 vaccination in lower-middle income countries: National stakeholder views on challenges, barriers, and potential solutions. Front. Public Health. 2021, 9, 709127. [Google Scholar] [CrossRef]
- Menni, C.; May, A.; Polidori, L.; Louca, P.; Wolf, J.; Capdevila, J.; Hu, C.; Ourselin, S.; Steves, C.J.; Valdes, A.M.; et al. COVID-19 vaccine waning and effectiveness and side-effects of boosters: A prospective community study from the ZOE COVID Study. Lancet Infect. Dis. 2022, 22, 1002–1010. [Google Scholar] [CrossRef]
- Wang, R.; Chen, J.; Wei, G.W. Mechanisms of SARS-CoV-2 evolution revealing vaccine-resistant mutations in Europe and America. J. Phys. Chem. Lett. 2021, 12, 11850–11857. [Google Scholar] [CrossRef] [PubMed]
- Singanayagam, A.; Hakki, S.; Dunning, J.; Madon, K.J.; Crone, M.A.; Koycheva, A.; Derqui-Fernandez, N.; Barnett, J.L.; Whitfield, M.G.; Varro, R.; et al. Community transmission and viral load kinetics of the SARS-CoV-2 delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: A prospective, longitudinal, cohort study. Lancet Infect. Dis. 2022, 22, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Dejnirattisai, W.; Shaw, R.H.; Supasa, P.; Liu, C.; Stuart, A.S.; Pollard, A.J.; Liu, X.; Lambe, T.; Crook, D.; Stuart, D.I.; et al. Reduced neutralisation of SARS-CoV-2 omicron B.1.1.529 variant by post-immunisation serum. Lancet 2022, 399, 234–236. [Google Scholar] [CrossRef] [PubMed]
- Cele, S.; Jackson, L.; Khoury, D.S.; Khan, K.; Moyo-Gwete, T.; Tegally, H.; San, J.E.; Cromer, D.; Scheepers, C.; Amoako, D.; et al. SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. medRxiv 2021. preprint. [Google Scholar] [CrossRef]
- Science Brief: COVID-19 Vaccines and Vaccination. Available online: https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/fully-vaccinated-people.html (accessed on 21 October 2022).
- Pegu, A.; O’Connell, S.E.; Schmidt, S.D.; O’Dell, S.; Talana, C.A.; Lai, L.; Albert, J.; Anderson, E.; Bennett, H.; Corbett, K.S.; et al. Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science 2021, 373, 1372–1377. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Choi, A.; Koch, M.; Ma, L.; Hill, A.; Nunna, N.; Huang, W.; Oestreicher, J.; Colpitts, T.; Bennett, H.; et al. Preliminary Analysis of Safety and Immunogenicity of a SARS-CoV-2 Variant Vaccine Booster. medRxiv 2021. preprint. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Pourkarim, F.; Pourtaghi-Anvarian, S.; Rezaee, H. Molnupiravir: A new candidate for COVID-19 treatment. Pharmacol. Res. Perspect. 2022, 10, e00909. [Google Scholar] [CrossRef]
- Jayk Bernal, A.; Gomes da Silva, M.M.; Musungaie, D.B.; Kovalchuk, E.; Gonzalez, A.; Delos Reyes, V.; Martin-Quiros, A.; Caraco, Y.; Williams-Diaz, A.; Brown, M.L.; et al. Molnupiravir for oral treatment of COVID-19 in nonhospitalized patients. N. Engl. J. Med. 2022, 386, 509–520. [Google Scholar] [CrossRef]
- Tam, D.; Qarawi, A.; Luu, M.; Turnage, M.; Tran, L.; Tawfik, G.; Minh, L.; Huy, N.; Iiyama, T.; Kita, K.; et al. Favipiravir and its potentials in COVID-19 pandemic: An update. Asian Pac. J. Trop. Med. 2021, 14, 433–439. [Google Scholar] [CrossRef]
- Lescure, F.X.; Honda, H.; Fowler, R.A.; Lazar, J.S.; Shi, G.; Wung, P.; Patel, N.; Hagino, O.; Sarilumab, C.-G.S.G. Sarilumab in patients admitted to hospital with severe or critical COVID-19: A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Respir. Med. 2021, 9, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Salama, C.; Han, J.; Yau, L.; Reiss, W.G.; Kramer, B.; Neidhart, J.D.; Criner, G.J.; Kaplan-Lewis, E.; Baden, R.; Pandit, L.; et al. Tocilizumab in patients hospitalized with COVID-19 pneumonia. N. Engl. J. Med. 2021, 384, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration. Coronavirus (COVID-19) Update: FDA Authorizes First Oral Antiviral for Treatment of COVID-19. Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-oral-antiviral-treatment-covid-19 (accessed on 31 January 2021).
- Abdelnabi, R.; Foo, C.S.; Kaptein, S.J.F.; Zhang, X.; Do, T.N.D.; Langendries, L.; Vangeel, L.; Breuer, J.; Pang, J.; Williams, R.; et al. The combined treatment of Molnupiravir and Favipiravir results in a potentiation of antiviral efficacy in a SARS-CoV-2 hamster infection model. EBioMedicine 2021, 72, 103595. [Google Scholar] [CrossRef]
- Eloy, P.; Le Grand, R.; Malvy, D.; Guedj, J. Combined treatment of molnupiravir and favipiravir against SARS-CoV-2 infection: One + zero equals two? EBioMedicine 2021, 74, 103663. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, S. How SARS-CoV-2 (COVID-19) spreads within infected hosts-what we know so far. Emerg. Top. Life Sci. 2020, 4, 371–378. [Google Scholar] [CrossRef]
- Xiu, S.; Dick, A.; Ju, H.; Mirzaie, S.; Abdi, F.; Cocklin, S.; Zhan, P.; Liu, X. Inhibitors of SARS-CoV-2 entry: Current and future opportunities. J. Med. Chem. 2020, 63, 12256–12274. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Padmanabhan, P.; Desikan, R.; Dixit, N.M. Targeting TMPRSS2 and Cathepsin B/L together may be synergistic against SARS-CoV-2 infection. PLoS Comput. Biol. 2020, 16, e1008461. [Google Scholar] [CrossRef]
- Kreutzberger, A.J.B.; Sanyal, A.; Ojha, R.; Pyle, J.D.; Vapalahti, O.; Balistreri, G.; Kirchhausen, T. Synergistic block of SARS-CoV-2 infection by combined drug inhibition of the host entry factors PIKfyve kinase and TMPRSS2 protease. J. Virol. 2021, 95, e0097521. [Google Scholar] [CrossRef] [PubMed]
- Clinicaltrials.Gov. Efficacy of Nafamostat in COVID-19 Patients (RACONA Study) (RACONA). NCT04352400. Available online: https://clinicaltrials.gov/ct2/show/NCT04352400 (accessed on 9 February 2022).
- Frueh, F.W.; Maneval, D.C.; Bohm, R.P.; Dufour, J.P.; Blair, R.V.; Powell, K.; Aye, P.P.; Golden, N.A.; Roy, C.J.; Spencer, S.; et al. An orally available cathepsin L inhibitor protects lungs against SARS-CoV-2-induced diffuse alveolar damage in African green monkeys. Biorxiv 2021. [Google Scholar] [CrossRef]
- Hoffmann, M.; Schroeder, S.; Kleine-Weber, H.; Muller, M.A.; Drosten, C.; Pohlmann, S. Nafamostat mesylate blocks activation of SARS-CoV-2: New treatment option for COVID-19. Antimicrob. Agents Chemother. 2020, 64, e00754-20. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.R.; Alugubelli, Y.R.; Ma, Y.; Vatansever, E.C.; Scott, D.A.; Qiao, Y.; Yu, G.; Xu, S.; Liu, W.R. MPI8 is potent against SARS-CoV-2 by inhibiting dually and selectively the SARS-CoV-2 main protease and the host Cathepsin, L. ChemMedChem 2022, 17, e202100456. [Google Scholar] [CrossRef]
- Mellott, D.M.; Tseng, C.T.; Drelich, A.; Fajtova, P.; Chenna, B.C.; Kostomiris, D.H.; Hsu, J.; Zhu, J.; Taylor, Z.W.; Tat, V.; et al. A clinical-stage cysteine protease inhibitor blocks SARS-CoV-2 infection of human and monkey cells. ACS Chem. Biol. 2021, 16, 642–650. [Google Scholar] [CrossRef]
- Shah, P.P.; Wang, T.; Kaletsky, R.L.; Myers, M.C.; Purvis, J.E.; Jing, H.; Huryn, D.M.; Greenbaum, D.C.; Smith, A.B., 3rd; Bates, P.; et al. A small-molecule oxocarbazate inhibitor of human cathepsin L blocks severe acute respiratory syndrome and ebola pseudotype virus infection into human embryonic kidney 293T cells. Mol. Pharmacol. 2010, 78, 319–324. [Google Scholar] [CrossRef]
- Zhou, Y.; Vedantham, P.; Lu, K.; Agudelo, J.; Carrion, R., Jr.; Nunneley, J.W.; Barnard, D.; Pohlmann, S.; McKerrow, J.H.; Renslo, A.R.; et al. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral. Res. 2015, 116, 76–84. [Google Scholar] [CrossRef]
- Hoffmann, M.; Hofmann-Winkler, H.; Smith, J.C.; Krüger, N.; Arora, P.; Sørensen, L.K.; Søgaard, O.S.; Hasselstrøm, J.B.; Winkler, M.; Hempel, T.; et al. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine 2021, 65, 103255. [Google Scholar] [CrossRef]
- Aulner, N.; Danckaert, A.; Ihm, J.; Shum, D.; Shorte, S.L. Next-generation phenotypic screening in early drug discovery for infectious diseases. Trends Parasitol. 2019, 35, 559–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rietdijk, J.; Tampere, M.; Pettke, A.; Georgiev, P.; Lapins, M.; Warpman-Berglund, U.; Spjuth, O.; Puumalainen, M.R.; Carreras-Puigvert, J. A phenomics approach for antiviral drug discovery. BMC Biol. 2021, 19, 156. [Google Scholar] [CrossRef] [PubMed]
- Ivens, T.; Van den Eynde, C.; Van Acker, K.; Nijs, E.; Dams, G.; Bettens, E.; Ohagen, A.; Pauwels, R.; Hertogs, K. Development of a homogeneous screening assay for automated detection of antiviral agents active against severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods 2005, 129, 56–63. [Google Scholar] [CrossRef]
- Bojkova, D.; Widera, M.; Ciesek, S.; Wass, M.N.; Michaelis, M.; Cinatl, J., Jr. Reduced interferon antagonism but similar drug sensitivity in Omicron variant compared to Delta variant of SARS-CoV-2 isolates. Cell Res. 2022, 32, 319–321. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Hurdiss, D.L.; Drabek, D.; Mykytyn, A.Z.; Kaiser, F.K.; González-Hernández, M.; Muñoz-Santos, D.; Lamers, M.M.; van Haperen, R.; Li, W.; et al. An ACE2-blocking antibody confers broad neutralization and protection against Omicron and other SARS-CoV-2 variants of concern. Sci. Immunol. 2022, 7, eabp9312. [Google Scholar] [CrossRef] [PubMed]
- Mykytyn, A.Z.; Breugem, T.I.; Riesebosch, S.; Schipper, D.; van den Doel, P.B.; Rottier, R.J.; Lamers, M.M.; Haagmans, B.L. SARS-CoV-2 entry into human airway organoids is serine protease-mediated and facilitated by the multibasic cleavage site. Elife 2021, 10, e64508. [Google Scholar] [CrossRef]
- Chiu, W.; Verschueren, L.; Van den Eynde, C.; Buyck, C.; De Meyer, S.; Jochmans, D.; Bojkova, D.; Ciesek, S.; Cinatl, J.; De Jonghe, S.; et al. Development and optimization of a high-throughput screening assay for in vitro anti-SARS-CoV-2 activity: Evaluation of 5676 Phase 1 Passed Structures. J. Med. Virol. 2022, 94, 3101–3111. [Google Scholar] [CrossRef] [PubMed]
- Cornelissen, F.; Cik, M.; Gustin, E. Phaedra, a protocol-driven system for analysis and validation of high-content imaging and flow cytometry. J. Biomol. Screen. 2012, 17, 496–506. [Google Scholar] [CrossRef] [Green Version]
- Kaname, Y.; Tani, H.; Kataoka, C.; Shiokawa, M.; Taguwa, S.; Abe, T.; Moriishi, K.; Kinoshita, T.; Matsuura, Y. Acquisition of complement resistance through incorporation of CD55/decay-accelerating factor into viral particles bearing baculovirus GP64. J. Virol. 2010, 84, 3210–3219. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Li, W.; Drabek, D.; Okba, N.M.A.; van Haperen, R.; Osterhaus, A.; van Kuppeveld, F.J.M.; Haagmans, B.L.; Grosveld, F.; Bosch, B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020, 11, 2251. [Google Scholar] [CrossRef]
- Kokic, G.; Hillen, H.S.; Tegunov, D.; Dienemann, C.; Seitz, F.; Schmitzova, J.; Farnung, L.; Siewert, A.; Hobartner, C.; Cramer, P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021, 12, 279. [Google Scholar] [CrossRef]
- de Vries, M.; Mohamed, A.S.; Prescott, R.A.; Valero-Jimenez, A.M.; Desvignes, L.; O’Connor, R.; Steppan, C.; Devlin, J.C.; Ivanova, E.; Herrera, A.; et al. A comparative analysis of SARS-CoV-2 antivirals characterizes 3CL(pro) inhibitor PF-00835231 as a potential new treatment for COVID-19. J. Virol. 2021, 95, e01819-20. [Google Scholar] [CrossRef]
- Heinen, N.; Klohn, M.; Steinmann, E.; Pfaender, S. In vitro lung models and their application to study SARS-CoV-2 pathogenesis and disease. Viruses 2021, 13, 792. [Google Scholar] [CrossRef] [PubMed]
- National Institutes of Health COVID-19 Treatment Guidelines. Therapeutic Management of of Nonhospitalized Adults with COVID-19. Available online: https://www.covid19treatmentguidelines.nih.gov/management/clinical-management-of-adults/nonhospitalized-adults--therapeutic-management/ (accessed on 12 December 2022).
- Eymieux, S.; Rouille, Y.; Terrier, O.; Seron, K.; Blanchard, E.; Rosa-Calatrava, M.; Dubuisson, J.; Belouzard, S.; Roingeard, P. Ultrastructural modifications induced by SARS-CoV-2 in Vero cells: A kinetic analysis of viral factory formation, viral particle morphogenesis and virion release. Cell Mol. Life Sci. 2021, 78, 3565–3576. [Google Scholar] [CrossRef] [PubMed]
- Ogando, N.S.; Dalebout, T.J.; Zevenhoven-Dobbe, J.C.; Limpens, R.; van der Meer, Y.; Caly, L.; Druce, J.; de Vries, J.J.C.; Kikkert, M.; Barcena, M.; et al. SARS-coronavirus-2 replication in Vero E6 cells: Replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020, 101, 925–940. [Google Scholar] [CrossRef] [PubMed]
- Whitt, M.A. Generation of VSV pseudotypes using recombinant DeltaG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J. Virol. Methods 2010, 169, 365–374. [Google Scholar] [CrossRef] [Green Version]
- Crawford, C.; Mason, R.W.; Wikstrom, P.; Shaw, E. The design of peptidyldiazomethane inhibitors to distinguish between the cysteine proteinases calpain II, cathepsin L and cathepsin B. Biochem. J. 1988, 253, 751–758. [Google Scholar] [CrossRef] [Green Version]
- Mason, R.W.; Bartholomew, L.T.; Hardwick, B.S. The use of benzyloxycarbonyl[125I]iodotyrosylalanyldiazomethane as a probe for active cysteine proteinases in human tissues. Biochem. J. 1989, 263, 945–949. [Google Scholar] [CrossRef] [Green Version]
- Tummino, T.A.; Rezelj, V.V.; Fischer, B.; Fischer, A.; O’Meara, M.J.; Monel, B.; Vallet, T.; White, K.M.; Zhang, Z.; Alon, A.; et al. Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2. Science 2021, 373, 541–547. [Google Scholar] [CrossRef]
- Boike, L.; Henning, N.J.; Nomura, D.K. Advances in covalent drug discovery. Nat. Rev. Drug. Discov. 2022, 21, 881–898. [Google Scholar] [CrossRef]
- Remdesivir US Prescribing Information. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/214787Orig1s000lbl.pdf (accessed on 4 April 2022).
- Organisation, W.H. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 16 January 2023).
- de Lartigue, J.; Polson, H.; Feldman, M.; Shokat, K.; Tooze, S.A.; Urbe, S.; Clague, M.J. PIKfyve regulation of endosome-linked pathways. Traffic 2009, 10, 883–893. [Google Scholar] [CrossRef] [Green Version]
- Rutherford, A.C.; Traer, C.; Wassmer, T.; Pattni, K.; Bujny, M.V.; Carlton, J.G.; Stenmark, H.; Cullen, P.J. The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J. Cell Sci. 2006, 119, 3944–3957. [Google Scholar] [CrossRef] [Green Version]
- Steuten, K.; Kim, H.; Widen, J.C.; Babin, B.M.; Onguka, O.; Lovell, S.; Bolgi, O.; Cerikan, B.; Neufeldt, C.J.; Cortese, M.; et al. Challenges for targeting SARS-CoV-2 proteases as a therapeutic strategy for COVID-19. ACS Infect. Dis. 2021, 7, 1457–1468. [Google Scholar] [CrossRef] [PubMed]
- Koch, J.; Uckeley, Z.M.; Doldan, P.; Stanifer, M.; Boulant, S.; Lozach, P.Y. TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells. EMBO J. 2021, 40, e107821. [Google Scholar] [CrossRef] [PubMed]
- Kawase, M.; Shirato, K.; van der Hoek, L.; Taguchi, F.; Matsuyama, S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012, 86, 6537–6545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abu-Farha, M.; Thanaraj, T.A.; Qaddoumi, M.G.; Hashem, A.; Abubaker, J.; Al-Mulla, F. The role of lipid metabolism in COVID-19 virus infection and as a drug target. Int. J. Mol. Sci. 2020, 21, 3544. [Google Scholar] [CrossRef]
- Bonam, S.R.; Wang, F.; Muller, S. Lysosomes as a therapeutic target. Nat. Rev. Drug. Discov. 2019, 18, 923–948. [Google Scholar] [CrossRef] [Green Version]
- Burkard, C.; Verheije, M.H.; Wicht, O.; van Kasteren, S.I.; van Kuppeveld, F.J.; Haagmans, B.L.; Pelkmans, L.; Rottier, P.J.; Bosch, B.J.; de Haan, C.A. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014, 10, e1004502. [Google Scholar] [CrossRef] [Green Version]
- Chu, V.C.; McElroy, L.J.; Chu, V.; Bauman, B.E.; Whittaker, G.R. The avian coronavirus infectious bronchitis virus undergoes direct low-pH-dependent fusion activation during entry into host cells. J. Virol. 2006, 80, 3180–3188. [Google Scholar] [CrossRef] [Green Version]
- Muller, C.; Hardt, M.; Schwudke, D.; Neuman, B.W.; Pleschka, S.; Ziebuhr, J. Inhibition of cytosolic phospholipase A2 alpha impairs an early step of coronavirus replication in cell culture. J. Virol. 2018, 92, e01463-17. [Google Scholar] [CrossRef] [Green Version]
- Yang, N.; Shen, H.M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020, 16, 1724–1731. [Google Scholar] [CrossRef]
- Bertram, S.; Dijkman, R.; Habjan, M.; Heurich, A.; Gierer, S.; Glowacka, I.; Welsch, K.; Winkler, M.; Schneider, H.; Hofmann-Winkler, H.; et al. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 2013, 87, 6150–6160. [Google Scholar] [CrossRef] [Green Version]
- Shirato, K.; Kanou, K.; Kawase, M.; Matsuyama, S. Clinical isolates of human coronavirus 229E bypass the endosome for cell entry. J. Virol. 2017, 91, e01387-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakayama, T.; Lee, I.T.; Jiang, S.; Matter, M.S.; Yan, C.H.; Overdevest, J.B.; Wu, C.T.; Goltsev, Y.; Shih, L.C.; Liao, C.K.; et al. Determinants of SARS-CoV-2 entry and replication in airway mucosal tissue and susceptibility in smokers. Cell Rep. Med. 2021, 2, 100421. [Google Scholar] [CrossRef] [PubMed]
- Steinman, J.B.; Lum, F.M.; Ho, P.P.; Kaminski, N.; Steinman, L. Reduced development of COVID-19 in children reveals molecular checkpoints gating pathogenesis illuminating potential therapeutics. Proc. Natl. Acad. Sci. USA 2020, 117, 24620–24626. [Google Scholar] [CrossRef] [PubMed]
- Bridges, J.P.; Vladar, E.K.; Huang, H.; Mason, R.J. Respiratory epithelial cell responses to SARS-CoV-2 in COVID-19. Thorax 2022, 77, 203–209. [Google Scholar] [CrossRef]
- Schreiner, T.; Allnoch, L.; Beythien, G.; Marek, K.; Becker, K.; Schaudien, D.; Stanelle-Bertram, S.; Schaumburg, B.; Mounogou Kouassi, N.; Beck, S.; et al. SARS-CoV-2 infection dysregulates cilia and basal cell homeostasis in the respiratory epithelium of hamsters. Int. J. Mol. Sci. 2022, 23, 5124. [Google Scholar] [CrossRef]
- Zhao, M.M.; Yang, W.L.; Yang, F.Y.; Zhang, L.; Huang, W.J.; Hou, W.; Fan, C.F.; Jin, R.H.; Feng, Y.M.; Wang, Y.C.; et al. Cathepsin L plays a key role.e.e in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct. Target. Ther. 2021, 6, 134. [Google Scholar] [CrossRef]
- Nie, X.; Qian, L.; Sun, R.; Huang, B.; Dong, X.; Xiao, Q.; Zhang, Q.; Lu, T.; Yue, L.; Chen, S.; et al. Multi-organ proteomic landscape of COVID-19 autopsies. Cell 2021, 184, 775–791.e714. [Google Scholar] [CrossRef]
- Hashimoto, R.; Sakamoto, A.; Deguchi, S.; Yi, R.; Sano, E.; Hotta, A.; Takahashi, K.; Yamanaka, S.; Takayama, K. Dual inhibition of TMPRSS2 and cathepsin B prevents SARS-CoV-2 infection in iPS cells. Mol. Ther.-Nucleic Acids 2021, 26, 1107–1114. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. COVID Data Tracker. Available online: https://covid.cdc.gov/covid-data-tracker/#variant-proportions (accessed on 20 May 2022).
- Padmanabhan, P.; Dixit, N.M. Evidence of increased Cathepsin B/L and decreased TMPRSS2 usage for cell entry by the SARS-CoV-2 Omicron variant. Biorxiv 2022. [Google Scholar] [CrossRef]
- Peacock, T.P.; Brown, J.C.; Zhou, J.; Thakur, N.; Sukhova, K.; Newman, J.; Kugathasan, R.; Yan, A.W.C.; Furnon, W.; De Lorenzo, G.; et al. The altered entry pathway and antigenic distance of the SARS-CoV-2 Omicron variant map to separate domains of spike protein. Biorxiv 2022. [Google Scholar] [CrossRef]
- Yamamoto, M.; Tomita, K.; Hirayama, Y.; Inoue, J.-i.; Kawaguchi, Y.; Gohda, J. SARS-CoV-2 Omicron spike H655Y mutation is responsible for enhancement of the endosomal entry pathway and reduction of cell surface entry pathways. Biorxiv 2022. [Google Scholar] [CrossRef]
- Willett, B.J.; Grove, J.; MacLean, O.A.; Wilkie, C.; De Lorenzo, G.; Furnon, W.; Cantoni, D.; Scott, S.; Logan, N.; Ashraf, S.; et al. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat. Microbiol. 2022, 7, 1161–1179. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.A.T.; Giannakoulias, S.G.; Barrett, T.M.; Liu, C.; Petersson, E.J. Rational design of thioamide peptides as selective inhibitors of cysteine protease cathepsin L. Chem. Sci. 2021, 12, 10825–10835. [Google Scholar] [CrossRef] [PubMed]
- Kwon, C.W.; Yeo, S.; Chang, P.S. Characterization and molecular docking study of cathepsin L inhibitory peptides (SnuCalCpIs) from Calotropis procera R. Br. Sci. Rep. 2022, 12, 5825. [Google Scholar] [CrossRef] [PubMed]
- Pang, N.Y.; Pang, A.S.; Chow, V.T.; Wang, D.Y. Understanding neutralising antibodies against SARS-CoV-2 and their implications in clinical practice. Mil. Med. Res. 2021, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Essa, R.Z.; Wu, Y.S.; Batumalaie, K.; Sekar, M.; Poh, C.L. Antiviral peptides against SARS-CoV-2: Therapeutic targets, mechanistic antiviral activity, and efficient delivery. Pharmacol. Rep. 2022, 74, 1166–1181. [Google Scholar] [CrossRef]
- Khater, I.; Nassar, A. Potential antiviral peptides targeting the SARS-CoV-2 spike protein. BMC Pharmacol. Toxicol. 2022, 23, 91. [Google Scholar] [CrossRef]
- Zannella, C.; Chianese, A.; Greco, G.; Santella, B.; Squillaci, G.; Monti, A.; Doti, N.; Sanna, G.; Manzin, A.; Morana, A.; et al. Design of Three Residues Peptides against SARS-CoV-2 Infection. Viruses 2022, 14, 2103. [Google Scholar] [CrossRef]
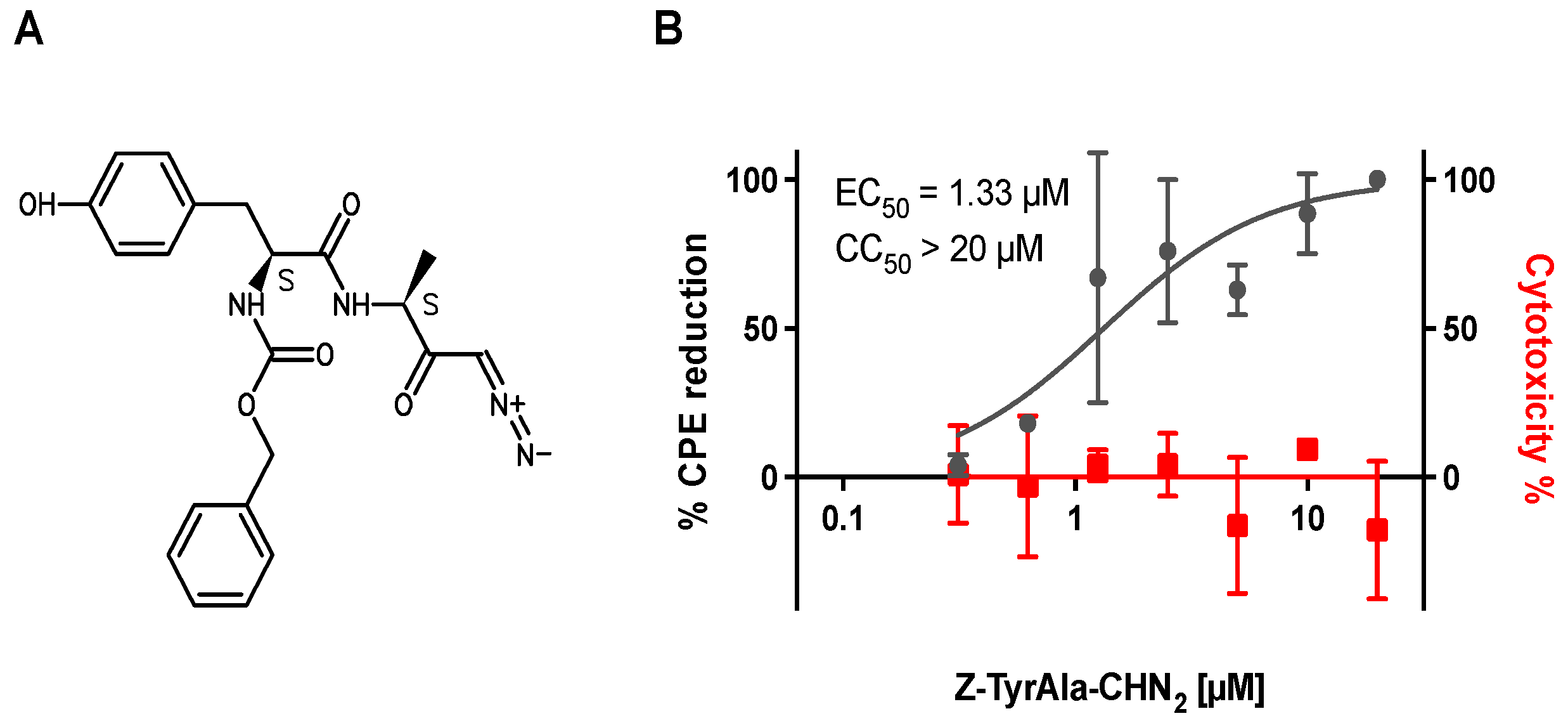
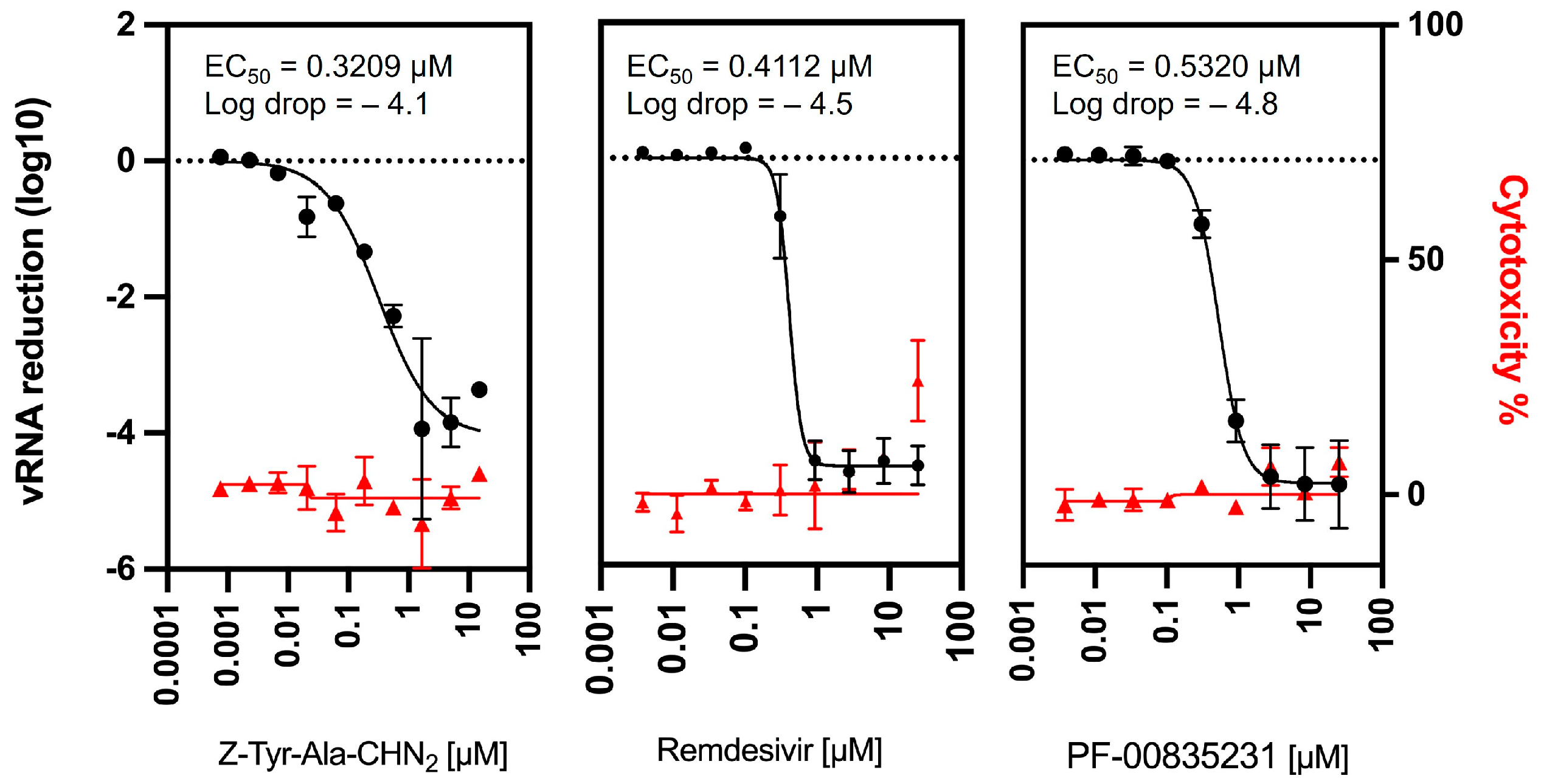
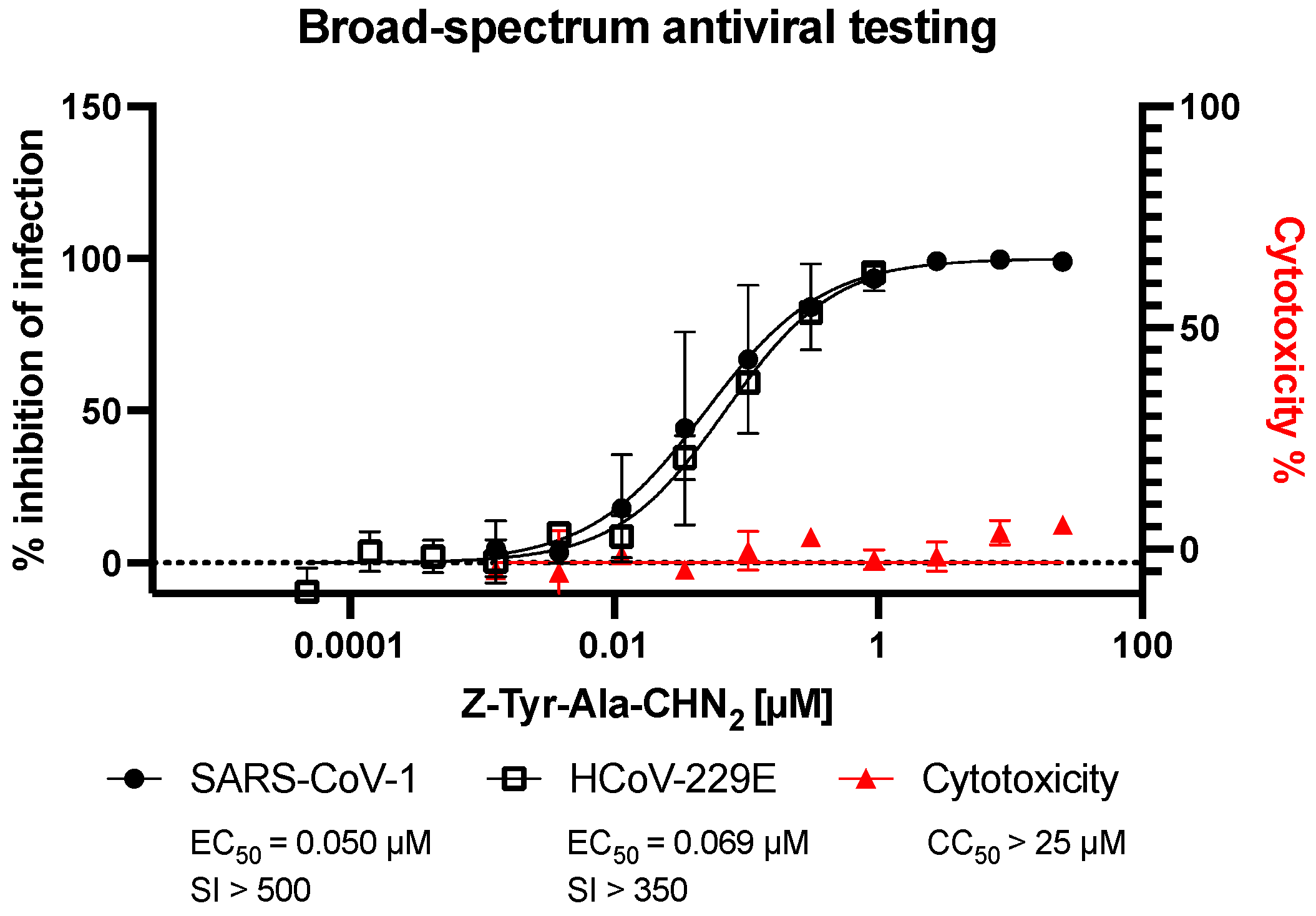
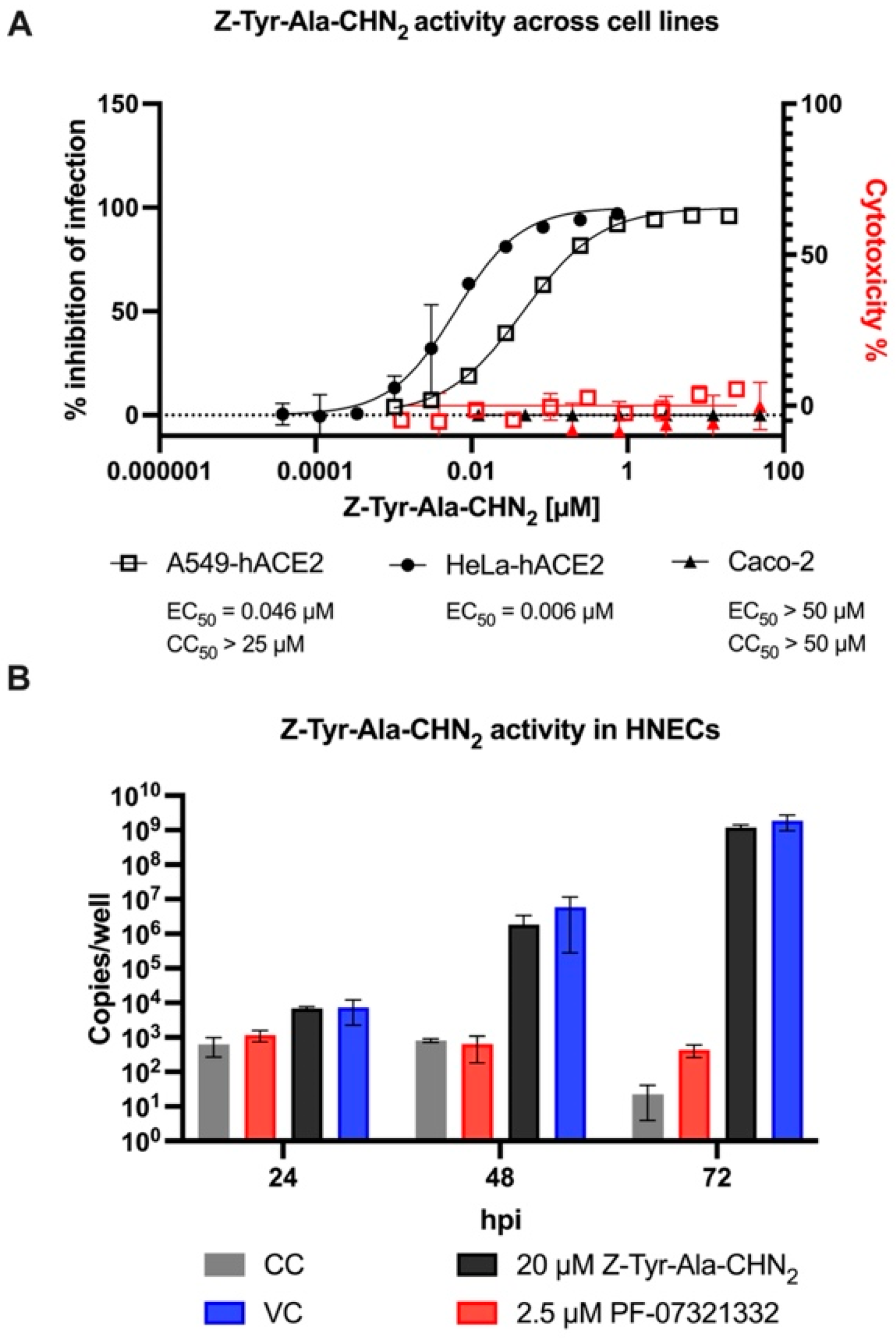



| Z-Tyr-Ala-CHN2 | Remdesivir | PF-00835231 | K777 | Molnu-Piravir | ||
|---|---|---|---|---|---|---|
| EC50 ± SD (µM) | Assay Type | |||||
| VeroE6-eGFP SARS-CoV-2 (n = 2) | CPE reduction (GFP) | 1.33 ± 0.49 | 1.42 ± 0.15 (n = 4) | >50 | 0.54 ± 0.14 | NT |
| A549-hACE2 SARS-CoV-2 (n = 3) | HCI (spike stain) | 0.05 ± 0.003 | 0.18 ± 0.01 | 0.18 ± 0.008 | 0.004 ± 0.0002 | NT |
| HeLa-hACE2 SARS-CoV-2 (n = 3) | HCI (dsRNA stain) | 0.01 ± 0.001 | 0.11 ± 0.01 | 0.16 ± 0.02 | 0.0006 ± 0.00005 | 1.01 ± 0.11 |
| A549-hACE2 SARS-CoV-1 (n = 3) | HCI (spike stain) | 0.05 ± 0.016 | 0.33 ± 0.02 | 0.55 ± 0.04 | 0.003 ± 0.0005 | NT |
| HeLa-hACE2 HCoV-229E (n = 2) | HCI (dsRNA stain) | 0.07 ± 0.01 | 0.11 ± 0.009 | 0.47 ± 0.04 | 0.002 ± 0.0004 | 1.32 ± 0.18 |
| Caco-2 SARS-CoV-2 (n = 6) | CPE reduction (manual scoring) | >50 | 0.77 ± 0.26 | NT | NT | NT |
| HNECs SARS-CoV-2 (n = 2) | vRNA (qPCR) | >20 | NT | 0.18 ± 0.14 * | NT | NT |
| CC50 (µM) | ||||||
| A549-hACE2 (n = 2–3) | Toxicity (ATPlite) | >25 | >25 | >12.5 | >0.05 | >50 |
| VeroE6-eGFP (n = 3) | Toxicity (ATPlite) | >20 (n = 3) | >100 (n = 3) | >20 (n = 2) | ~50 (n = 3) | >100 (n = 3) |
| SI | ||||||
| A549-hACE2 SARS-CoV-2 | >500 | >138 | >69 | >12.5 | NT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doijen, J.; Temmerman, K.; Van den Eynde, C.; Diels, A.; Van den Broeck, N.; Van Gool, M.; Heo, I.; Jaensch, S.; Zwaagstra, M.; Diosa Toro, M.; et al. Identification of Z-Tyr-Ala-CHN2, a Cathepsin L Inhibitor with Broad-Spectrum Cell-Specific Activity against Coronaviruses, including SARS-CoV-2. Microorganisms 2023, 11, 717. https://doi.org/10.3390/microorganisms11030717
Doijen J, Temmerman K, Van den Eynde C, Diels A, Van den Broeck N, Van Gool M, Heo I, Jaensch S, Zwaagstra M, Diosa Toro M, et al. Identification of Z-Tyr-Ala-CHN2, a Cathepsin L Inhibitor with Broad-Spectrum Cell-Specific Activity against Coronaviruses, including SARS-CoV-2. Microorganisms. 2023; 11(3):717. https://doi.org/10.3390/microorganisms11030717
Chicago/Turabian StyleDoijen, Jordi, Koen Temmerman, Christel Van den Eynde, Annick Diels, Nick Van den Broeck, Michiel Van Gool, Inha Heo, Steffen Jaensch, Marleen Zwaagstra, Mayra Diosa Toro, and et al. 2023. "Identification of Z-Tyr-Ala-CHN2, a Cathepsin L Inhibitor with Broad-Spectrum Cell-Specific Activity against Coronaviruses, including SARS-CoV-2" Microorganisms 11, no. 3: 717. https://doi.org/10.3390/microorganisms11030717





