Regulation of Surfactant Protein Gene Expression by Aspergillus fumigatus in NCl-H441 Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Fungal Strains
2.3. Cell Treatments
2.4. Microscopy
2.5. Real-Time qPCR
2.6. Statistics
3. Results
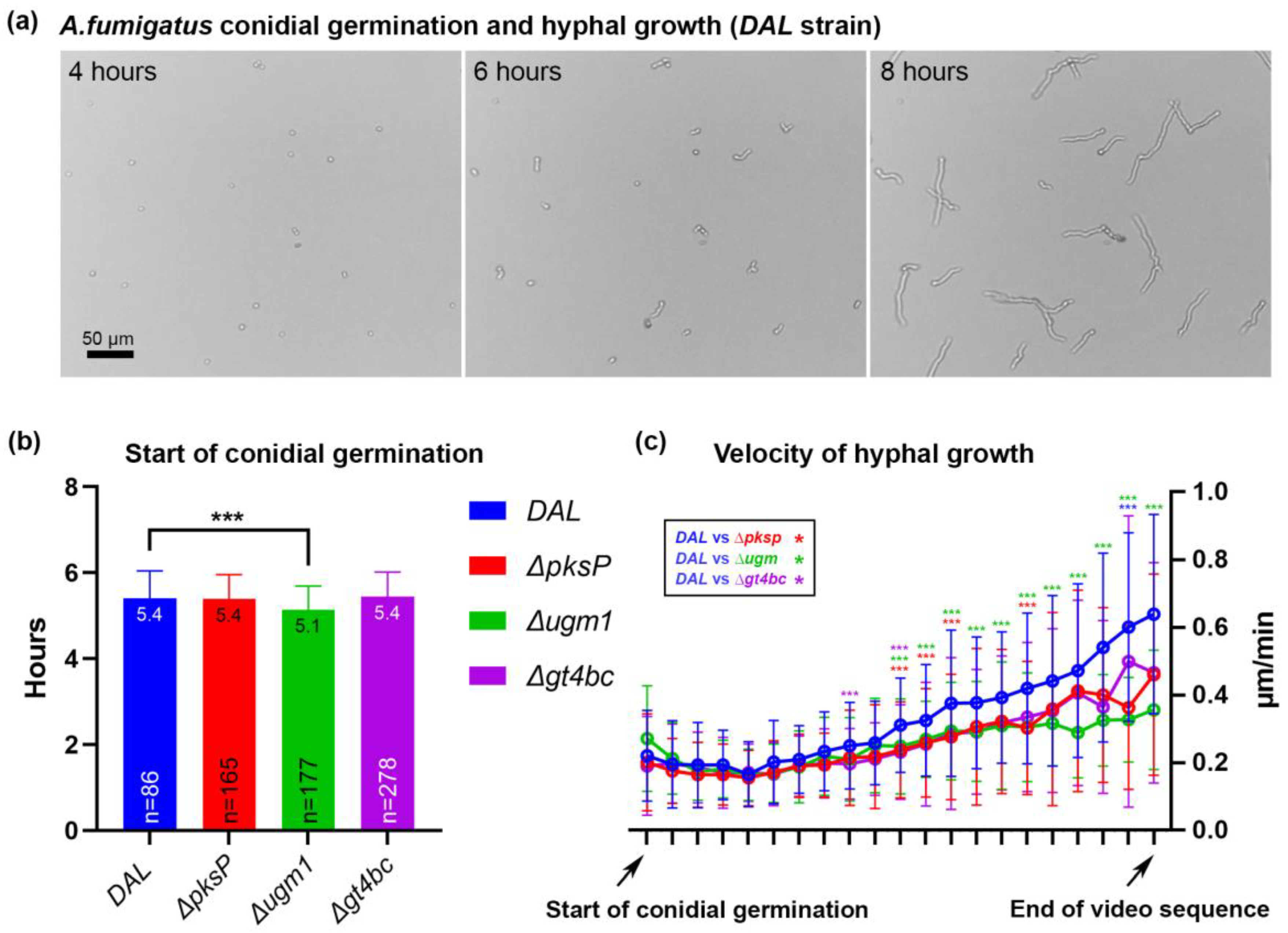
3.1. Kinetics of Fungal Growth
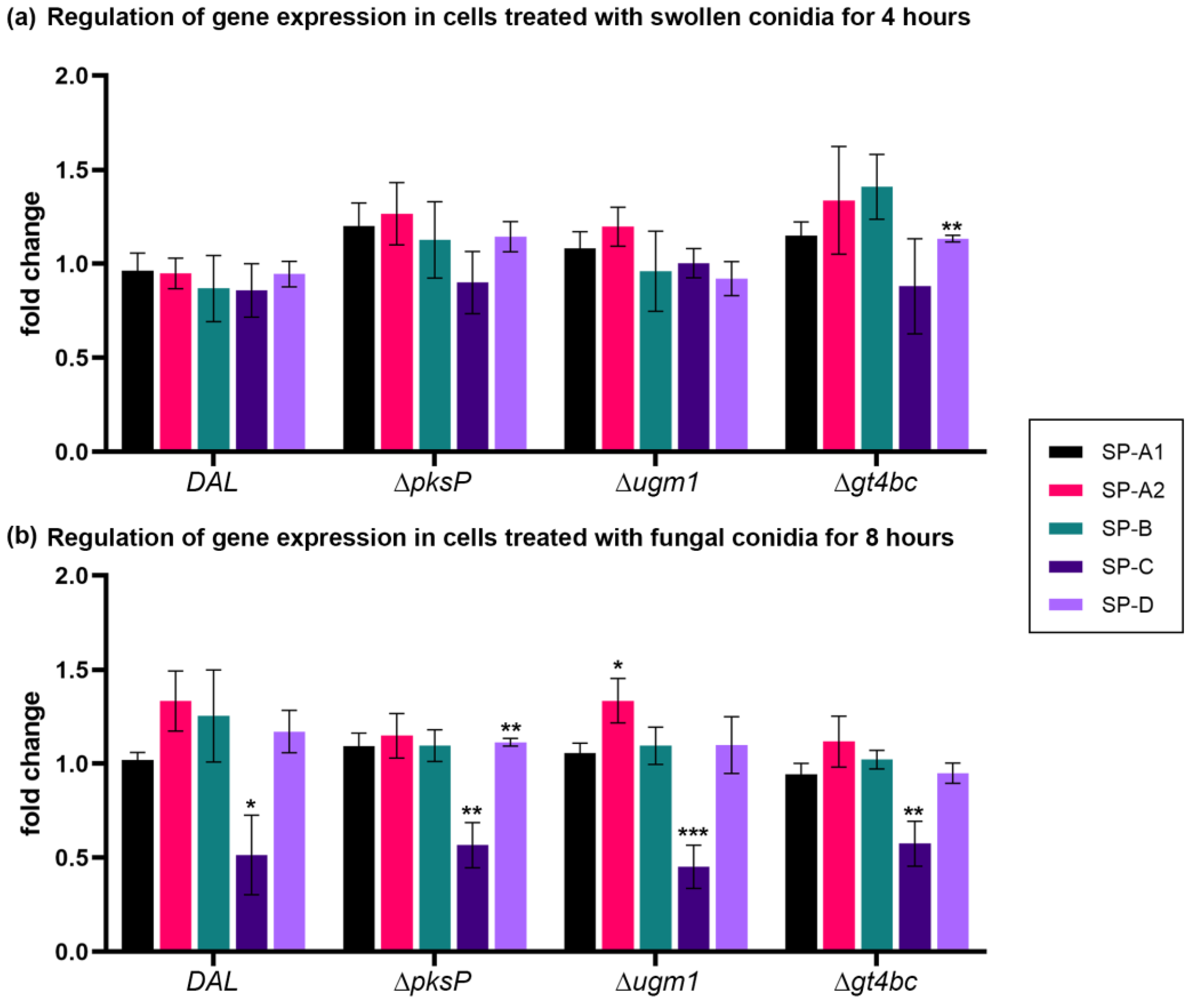
3.2. Infection of Human NCI-H441 Cells with A. fumigatus Conidia
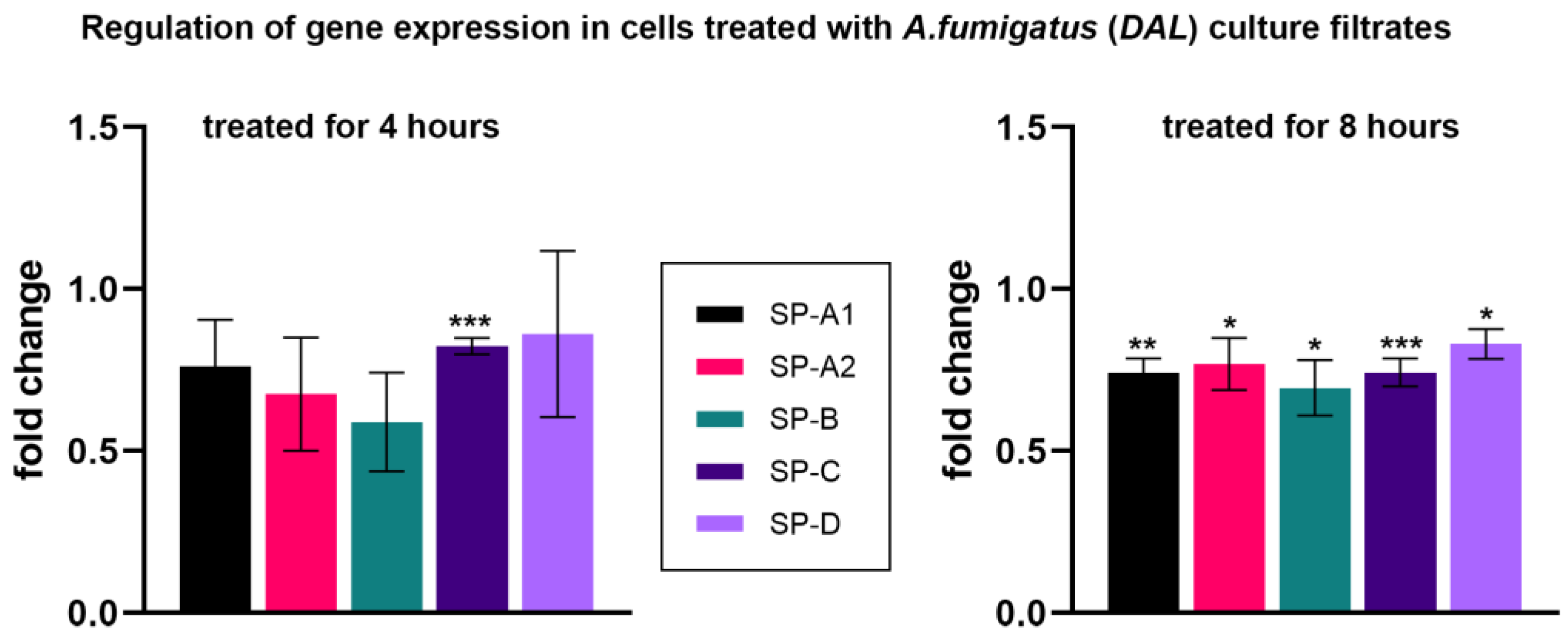
3.3. Treatment of Human NCI-H441 Cells with A. fumigatus Culture Filtrates
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rhodes, J.C. Aspergillus fumigatus: Growth and virulence. Med. Mycol. 2006, 44 (Suppl. S1), S77–S81. [Google Scholar] [CrossRef] [PubMed]
- Kwon-Chung, K.J.; Sugui, J.A. Aspergillus fumigatus—What Makes the Species a Ubiquitous Human Fungal Pathogen? PLoS Pathog. 2013, 9, e1003743. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.-P.; Chamilos, G. Aspergillus fumigatus and Aspergillosis in 2019. Clin. Microbiol. Rev. 2019, 33, e00140-18. [Google Scholar] [CrossRef] [PubMed]
- Osherov, N. Interaction of the pathogenic mold Aspergillus fumigatus with lung epithelial cells. Front. Microbiol. 2012, 3, 346. [Google Scholar] [CrossRef] [PubMed]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef]
- Abad, A.; Fernández-Molina, J.V.; Bikandi, J.; Ramírez, A.; Margareto, J.; Sendino, J.; Hernando, F.L.; Pontón, J.; Garaizar, J.; Rementeria, A. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev. Iberoam. Micol. 2010, 27, 155–182. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.; Fontaine, T.; Heddergott, C.; Robinet, P.; Aimanianda, V.; Beau, R.; Beauvais, A.; Mouyna, I.; Prevost, M.-C.; Fekkar, A.; et al. Biosynthesis of cell wall mannan in the conidium and the mycelium of Aspergillus fumigatus. Cell. Microbiol. 2016, 18, 1881–1891. [Google Scholar] [CrossRef]
- Madan, T.; Eggleton, P.; Kishore, U.; Strong, P.; Aggrawal, S.S.; Sarma, P.U.; Reid, K.B. Binding of pulmonary surfactant proteins A and D to Aspergillus fumigatus conidia enhances phagocytosis and killing by human neutrophils and alveolar macrophages. Infect. Immun. 1997, 65, 3171–3179. [Google Scholar] [CrossRef]
- Latgé, J.-P.; Beauvais, A.; Chamilos, G. The Cell Wall of the Human Fungal Pathogen Aspergillus fumigatus: Biosynthesis, Organization, Immune Response, and Virulence. Annu. Rev. Microbiol. 2017, 71, 99–116. [Google Scholar] [CrossRef]
- Baltussen, T.J.H.; Zoll, J.; Verweij, P.E.; Melchers, W.J.G. Molecular Mechanisms of Conidial Germination in Aspergillus spp. Microbiol. Mol. Biol. Rev. 2020, 84. [Google Scholar] [CrossRef]
- Beauvais, A.; Fontaine, T.; Aimanianda, V.; Latgé, J.-P. Aspergillus cell wall and biofilm. Mycopathologia 2014, 178, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Heinekamp, T.; Thywißen, A.; Macheleidt, J.; Keller, S.; Valiante, V.; Brakhage, A.A. Aspergillus fumigatus melanins: Interference with the host endocytosis pathway and impact on virulence. Front. Microbiol. 2012, 3, 440. [Google Scholar] [CrossRef] [PubMed]
- Bayry, J.; Beaussart, A.; Dufrêne, Y.F.; Sharma, M.; Bansal, K.; Kniemeyer, O.; Aimanianda, V.; Brakhage, A.A.; Kaveri, S.V.; Kwon-Chung, K.J.; et al. Surface structure characterization of Aspergillus fumigatus conidia mutated in the melanin synthesis pathway and their human cellular immune response. Infect. Immun. 2014, 82, 3141–3153. [Google Scholar] [CrossRef] [PubMed]
- Jahn, B.; Koch, A.; Schmidt, A.; Wanner, G.; Gehringer, H.; Bhakdi, S.; Brakhage, A.A. Isolation and characterization of a pigmentless-conidium mutant of Aspergillus fumigatus with altered conidial surface and reduced virulence. Infect. Immun. 1997, 65, 5110–5117. [Google Scholar] [CrossRef]
- Briard, B.; Muszkieta, L.; Latgé, J.-P.; Fontaine, T. Galactosaminogalactan of Aspergillus fumigatus, a bioactive fungal polymer. Mycologia 2016, 108, 572–580. [Google Scholar] [CrossRef]
- Gravelat, F.N.; Beauvais, A.; Liu, H.; Lee, M.J.; Snarr, B.D.; Chen, D.; Xu, W.; Kravtsov, I.; Hoareau, C.M.Q.; Vanier, G.; et al. Aspergillus galactosaminogalactan mediates adherence to host constituents and conceals hyphal β-glucan from the immune system. PLoS Pathog. 2013, 9, e1003575. [Google Scholar] [CrossRef]
- Sheppard, D.C. Molecular mechanism of Aspergillus fumigatus adherence to host constituents. Curr. Opin. Microbiol. 2011, 14, 375–379. [Google Scholar] [CrossRef]
- Speth, C.; Rambach, G.; Lass-Flörl, C.; Howell, P.L.; Sheppard, D.C. Galactosaminogalactan (GAG) and its multiple roles in Aspergillus pathogenesis. Virulence 2019, 10, 976–983. [Google Scholar] [CrossRef]
- Vivek-Ananth, R.P.; Mohanraj, K.; Vandanashree, M.; Jhingran, A.; Craig, J.P.; Samal, A. Comparative systems analysis of the secretome of the opportunistic pathogen Aspergillus fumigatus and other Aspergillus species. Sci. Rep. 2018, 8, 6617. [Google Scholar] [CrossRef]
- Krappmann, S. How to invade a susceptible host: Cellular aspects of aspergillosis. Curr. Opin. Microbiol. 2016, 34, 136–146. [Google Scholar] [CrossRef]
- Murayama, T.; Amitani, R.; Ikegami, Y.; Nawada, R.; Lee, W.J.; Kuze, F. Suppressive effects of Aspergillus fumigatus culture filtrates on human alveolar macrophages and polymorphonuclear leucocytes. Eur. Respir. J. 1996, 9, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Yokota, K.; Shimada, H.; Kamaguchi, A.; Sakaguchi, O. Studies on the toxin of Aspergillus fumigatus. VII. Purification and some properities of hemolytic toxin (asp-hemolysin) from culture filtrates and mycelia. Microbiol. Immunol. 1977, 21, 11–22. [Google Scholar] [CrossRef]
- Kauffman, H.F.; Tomee, J.F.; van de Riet, M.A.; Timmerman, A.J.; Borger, P. Protease-dependent activation of epithelial cells by fungal allergens leads to morphologic changes and cytokine production. J. Allergy Clin. Immunol. 2000, 105, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Sharon, H.; Amar, D.; Levdansky, E.; Mircus, G.; Shadkchan, Y.; Shamir, R.; Osherov, N. PrtT-regulated proteins secreted by Aspergillus fumigatus activate MAPK signaling in exposed A549 lung cells leading to necrotic cell death. PLoS ONE 2011, 6, e17509. [Google Scholar] [CrossRef] [PubMed]
- Kamei, K.; Watanabe, A. Aspergillus mycotoxins and their effect on the host. Med. Mycol. 2005, 43 (Suppl. S1), S95–S99. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, A.; Kamei, K.; Sekine, T.; Waku, M.; Nishimura, K.; Miyaji, M.; Kuriyama, T. Immunosuppressive substances in Aspergillus fumigatus culture filtrate. J. Infect. Chemother. 2003, 9, 114–121. [Google Scholar] [CrossRef]
- Amitani, R.; Taylor, G.; Elezis, E.N.; Llewellyn-Jones, C.; Mitchell, J.; Kuze, F.; Cole, P.J.; Wilson, R. Purification and characterization of factors produced by Aspergillus fumigatus which affect human ciliated respiratory epithelium. Infect. Immun. 1995, 63, 3266–3271. [Google Scholar] [CrossRef] [PubMed]
- Robinson, B.W.; Venaille, T.J.; Mendis, A.H.; McAleer, R. Allergens as proteases: An aspergillus fumigatus proteinase directly induces human epithelial cell detachment. J. Allergy Clin. Immunol. 1990, 86, 726–731. [Google Scholar] [CrossRef]
- Cañadas, O.; Olmeda, B.; Alonso, A.; Pérez-Gil, J. Lipid-Protein and Protein-Protein Interactions in the Pulmonary Surfactant System and Their Role in Lung Homeostasis. Int. J. Mol. Sci. 2020, 21, 3708. [Google Scholar] [CrossRef]
- Agudelo, C.W.; Samaha, G.; Garcia-Arcos, I. Alveolar lipids in pulmonary disease. A review. Lipids Health Dis. 2020, 19, 122. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Mallampalli, R.K. The Role of Surfactant in Lung Disease and Host Defense against Pulmonary Infections. Ann. Am. Thorac. Soc. 2015, 12, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Crouch, E.; Wright, J.R. Surfactant proteins a and d and pulmonary host defense. Annu. Rev. Physiol. 2001, 63, 521–554. [Google Scholar] [CrossRef] [PubMed]
- Selman, M.; Lin, H.-M.; Montaño, M.; Jenkins, A.L.; Estrada, A.; Lin, Z.; Wang, G.; DiAngelo, S.L.; Guo, X.; Umstead, T.M.; et al. Surfactant protein A and B genetic variants predispose to idiopathic pulmonary fibrosis. Hum. Genet. 2003, 113, 542–550. [Google Scholar] [CrossRef] [PubMed]
- Nogee, L.M.; Dunbar, A.E.; Wert, S.E.; Askin, F.; Hamvas, A.; Whitsett, J.A. A mutation in the surfactant protein C gene associated with familial interstitial lung disease. N. Engl. J. Med. 2001, 344, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Augusto, L.A.; Li, J.; Synguelakis, M.; Johansson, J.; Chaby, R. Structural basis for interactions between lung surfactant protein C and bacterial lipopolysaccharide. J. Biol. Chem. 2002, 277, 23484–23492. [Google Scholar] [CrossRef] [PubMed]
- Haczku, A.; Atochina, E.N.; Tomer, Y.; Chen, H.; Scanlon, S.T.; Russo, S.; Xu, J.; Panettieri, R.A., Jr.; Beers, M.F. Aspergillus fumigatus-induced allergic airway inflammation alters surfactant homeostasis and lung function in BALB/c mice. Am. J. Respir. Cell Mol. Biol. 2001, 25, 45–50. [Google Scholar] [CrossRef]
- Glasser, S.W.; Witt, T.L.; Senft, A.P.; Baatz, J.E.; Folger, D.; Maxfield, M.D.; Akinbi, H.T.; Newton, D.A.; Prows, D.R.; Korfhagen, T.R. Surfactant protein C-deficient mice are susceptible to respiratory syncytial virus infection. Am. J. Physiol. Lung Cell. Mol. Physiol. 2009, 297, L64–L72. [Google Scholar] [CrossRef]
- Madan, T.; Reid, K.B.M.; Clark, H.; Singh, M.; Nayak, A.; Sarma, P.U.; Hawgood, S.; Kishore, U. Susceptibility of mice genetically deficient in SP-A or SP-D gene to invasive pulmonary aspergillosis. Mol. Immunol. 2010, 47, 1923–1930. [Google Scholar] [CrossRef]
- Augusto, L.; Le Blay, K.; Auger, G.; Blanot, D.; Chaby, R. Interaction of bacterial lipopolysaccharide with mouse surfactant protein C inserted into lipid vesicles. Am. J. Physiol. Lung Cell. Mol. Physiol. 2001, 281, L776–L785. [Google Scholar] [CrossRef]
- Guagliardo, R.; Pérez-Gil, J.; de Smedt, S.; Raemdonck, K. Pulmonary surfactant and drug delivery: Focusing on the role of surfactant proteins. J. Control. Release 2018, 291, 116–126. [Google Scholar] [CrossRef]
- Wong, S.S.W.; Rani, M.; Dodagatta-Marri, E.; Ibrahim-Granet, O.; Kishore, U.; Bayry, J.; Latgé, J.-P.; Sahu, A.; Madan, T.; Aimanianda, V. Fungal melanin stimulates surfactant protein D-mediated opsonization of and host immune response to Aspergillus fumigatus spores. J. Biol. Chem. 2018, 293, 4901–4912. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.S.W.; Dellière, S.; Schiefermeier-Mach, N.; Lechner, L.; Perkhofer, S.; Bomme, P.; Fontaine, T.; Schlosser, A.G.; Sorensen, G.L.; Madan, T.; et al. Surfactant protein D inhibits growth, alters cell surface polysaccharide exposure and immune activation potential of Aspergillus fumigatus. Cell Surf. 2022, 8, 100072. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-S.; Shin, K.-S. Transcription Factor HSF1 Suppresses the Expression of Surfactant Protein D in Cells Infected with Aspergillus fumigatus. Pathogens 2021, 10, 709. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-S. Transcription Factor PU.1 Inhibits Aspergillus fumigatus Infection via Surfactant Protein-D. Biomed. Sci. Lett. 2018, 24, 175–182. [Google Scholar] [CrossRef]
- Atochina, E.N.; Beers, M.F.; Tomer, Y.; Scanlon, S.T.; Russo, S.J.; Panettieri, R.A., Jr.; Haczku, A. Attenuated allergic airway hyperresponsiveness in C57BL/6 mice is associated with enhanced surfactant protein (SP)-D production following allergic sensitization. Respir. Res. 2003, 4, 15. [Google Scholar] [CrossRef]
- Thau, N.; Monod, M.; Crestani, B.; Rolland, C.; Tronchin, G.; Latgé, J.P.; Paris, S. rodletless mutants of Aspergillus fumigatus. Infect. Immun. 1994, 62, 4380–4388. [Google Scholar] [CrossRef]
- Schiefermeier-Mach, N.; Perkhofer, S.; Heinrich, L.; Haller, T. Stimulation of surfactant exocytosis in primary alveolar type II cells by A. fumigatus. Med. Mycol. 2021, 59, 168–179. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Floros, J.; Wang, G.; Mikerov, A.N. Genetic complexity of the human innate host defense molecules, surfactant protein A1 (SP-A1) and SP-A2--impact on function. Crit. Rev. Eukaryot. Gene Expr. 2009, 19, 125–137. [Google Scholar] [CrossRef]
- Lamarre, C.; Beau, R.; Balloy, V.; Fontaine, T.; Wong Sak Hoi, J.; Guadagnini, S.; Berkova, N.; Chignard, M.; Beauvais, A.; Latgé, J.-P. Galactofuranose attenuates cellular adhesion of Aspergillus fumigatus. Cell. Microbiol. 2009, 11, 1612–1623. [Google Scholar] [CrossRef]
- Rucka, Z.; Vanhara, P.; Koutna, I.; Tesarova, L.; Potesilova, M.; Stejskal, S.; Simara, P.; Dolezel, J.; Zvonicek, V.; Coufal, O.; et al. Differential effects of insulin and dexamethasone on pulmonary surfactant-associated genes and proteins in A549 and H441 cells and lung tissue. Int. J. Mol. Med. 2013, 32, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Miakotina, O.L.; Goss, K.L.; Snyder, J.M. Insulin utilizes the PI 3-kinase pathway to inhibit SP-A gene expression in lung epithelial cells. Respir. Res. 2002, 3, 26. [Google Scholar] [CrossRef] [PubMed]
- Schiefermeier-Mach, N.; Haller, T.; Geley, S.; Perkhofer, S. Migrating Lung Monocytes Internalize and Inhibit Growth of Aspergillus fumigatus Conidia. Pathogens 2020, 9, 983. [Google Scholar] [CrossRef]
- Cooper, J.R.; Abdullatif, M.B.; Burnett, E.C.; Kempsell, K.E.; Conforti, F.; Tolley, H.; Collins, J.E.; Davies, D.E. Long Term Culture of the A549 Cancer Cell Line Promotes Multilamellar Body Formation and Differentiation towards an Alveolar Type II Pneumocyte Phenotype. PLoS ONE 2016, 11, e0164438. [Google Scholar] [CrossRef]
- Daly, P.; Verhaegen, S.; Clynes, M.; Kavanagh, K. Culture filtrates of Aspergillus fumigatus induce different modes of cell death in human cancer cell lines. Mycopathologia 1999, 146, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Chiang, L.Y.; Sheppard, D.C.; Gravelat, F.N.; Patterson, T.F.; Filler, S.G. Aspergillus fumigatus stimulates leukocyte adhesion molecules and cytokine production by endothelial cells in vitro and during invasive pulmonary disease. Infect. Immun. 2008, 76, 3429–3438. [Google Scholar] [CrossRef]
- Tomee, J.F.; Wierenga, A.T.; Hiemstra, P.S.; Kauffman, H.K. Proteases from Aspergillus fumigatus induce release of proinflammatory cytokines and cell detachment in airway epithelial cell lines. J. Infect. Dis. 1997, 176, 300–303. [Google Scholar] [CrossRef]
- Mulugeta, S.; Beers, M.F. Surfactant protein C: Its unique properties and emerging immunomodulatory role in the lung. Microbes Infect. 2006, 8, 2317–2323. [Google Scholar] [CrossRef]
- Glasser, S.W.; Maxfield, M.D.; Ruetschilling, T.L.; Akinbi, H.T.; Baatz, J.E.; Kitzmiller, J.A.; Page, K.; Xu, Y.; Bao, E.L.; Korfhagen, T.R. Persistence of LPS-induced lung inflammation in surfactant protein-C-deficient mice. Am. J. Respir. Cell Mol. Biol. 2013, 49, 845–854. [Google Scholar] [CrossRef]
- Glasser, S.W.; Senft, A.P.; Whitsett, J.A.; Maxfield, M.D.; Ross, G.F.; Richardson, T.R.; Prows, D.R.; Xu, Y.; Korfhagen, T.R. Macrophage dysfunction and susceptibility to pulmonary Pseudomonas aeruginosa infection in surfactant protein C-deficient mice. J. Immunol. 2008, 181, 621–628. [Google Scholar] [CrossRef]
- Augusto, L.A.; Synguelakis, M.; Espinassous, Q.; Lepoivre, M.; Johansson, J.; Chaby, R. Cellular antiendotoxin activities of lung surfactant protein C in lipid vesicles. Am. J. Respir. Crit. Care Med. 2003, 168, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Milad, N.; Morissette, M.C. Revisiting the role of pulmonary surfactant in chronic inflammatory lung diseases and environmental exposure. Eur. Respir. Rev. 2021, 30, 210077. [Google Scholar] [CrossRef] [PubMed]
- Raychaudhuri, B.; Abraham, S.; Bonfield, T.L.; Malur, A.; Deb, A.; DiDonato, J.A.; Kavuru, M.S.; Thomassen, M.J. Surfactant blocks lipopolysaccharide signaling by inhibiting both mitogen-activated protein and IkappaB kinases in human alveolar macrophages. Am. J. Respir. Cell Mol. Biol. 2004, 30, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Rauwolf, K.K.; Hoertnagl, C.; Lass-Floerl, C.; Groll, A.H. Interaction in vitro of pulmonary surfactant with antifungal agents used for treatment and prevention of invasive aspergillosis. J. Antimicrob. Chemother. 2022, 77, 695–698. [Google Scholar] [CrossRef]
| NCI-H441 cells infected with A. fumigatus conidia for 4 h | ||||||||||
| SP-A1 | SP-A2 | SP-B | SP-C | SP-D | ||||||
| Strains | Mean ± SE | p-value | Mean ± SE | p-value | Mean ± SE | p-value | Mean ±SE | p-value | Mean ± SE | p-value |
| DAL vs. control | 0.96 ± 0.10 | 0.706 | 0.95 ± 0.08 | 0.557 | 0.87 ± 0.18 | 0.496 | 0.86 ± 0.10 | 0.218 | 0.95 ± 0.07 | 0.464 |
| ΔpksP vs. control | 1.20 ± 0.12 | 0.170 | 1.27 ± 0.17 | 0.185 | 1.13 ± 0.20 | 0.567 | 0.90 ± 0.12 | 0.439 | 1.14 ± 0.08 | 0.146 |
| Δugm1 vs. control | 1.08 ± 0.09 | 0.403 | 1.20 ± 0.10 | 0.130 | 0.96 ± 0.21 | 0.862 | 1.00 ± 0.06 | 0.966 | 0.92 ± 0.09 | 0.428 |
| Δgt4bc vs. control | 1.15 ± 0.07 | 0.115 | 1.34 ± 0.29 | 0.304 | 1.41 ± 0.17 | 0.076 | 0.88 ± 0.18 | 0.538 | 1.13 ± 0.02 | ** 0.002 |
| NCI-H441 cells infected with A. fumigatus conidia for 8 h | ||||||||||
| SP-A1 | SP-A2 | SP-B | SP-C | SP-D | ||||||
| Strains | Mean ± SE | p-value | Mean ± SE | p-value | Mean ± SE | p-value | Mean ± SE | p- value | Mean ± SE | p-value |
| DAL vs. control | 1.02 ± 0.04 | 0.691 | 1.33 ± 0.16 | 0.105 | 1.25 ± 0.25 | 0.359 | 0.51 ± 0.21 | * 0.051 | 1.17 ± 0.11 | 0.206 |
| ΔpksP vs. control | 1.09 ± 0.07 | 0.263 | 1.15 ± 0.12 | 0.279 | 1.10 ± 0.09 | 0.323 | 0.57 ± 0.12 | ** 0.007 | 1.11 ± 0.02 | ** 0.005 |
| Δugm1 vs. control | 1.06 ± 0.05 | 0.356 | 1.34 ± 0.12 | * 0.047 | 1.10 ± 0.10 | 0.395 | 0.45 ± 0.12 | *** < 0.001 | 1.10 ± 0.15 | 0.550 |
| Δgt4bc vs. control | 0.94 ± 0.06 | 0.380 | 1.12 ± 0.14 | 0.435 | 1.02 ± 0.05 | 0.689 | 0.58 ± 0.12 | ** 0.007 | 0.95 ± 0.05 | 0.403 |
| NCI-H441 cells infected with A. fumigatus culture filtrates for 4 and 8 h | ||||||||||
| SP-A1 | SP-A2 | SP-B | SP-C | SP-D | ||||||
| Time | Mean ± SE | p-value | Mean ± SE | p-value | Mean ± SE | p-value | Mean ± SE | p-value | Mean ± SE | p-value |
| 4 h | 0.76 ± 0.15 | 0.176 | 0.68 ± 0.18 | 0.137 | 0.59 ± 0.15 | 0.054 | 0.82 ± 0.03 | *** <0.001 | 0.86 ± 0.26 | 0.619 |
| 8 h | 0.74 ± 0.04 | ** 0.004 | 0.77 ± 0.08 | * 0.046 | 0.69 ± 0.09 | * 0.024 | 0.74 ± 0.05 | *** <0.001 | 0.83 ± 0.05 | * 0.022 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schiefermeier-Mach, N.; Heinrich, L.; Lechner, L.; Perkhofer, S. Regulation of Surfactant Protein Gene Expression by Aspergillus fumigatus in NCl-H441 Cells. Microorganisms 2023, 11, 1011. https://doi.org/10.3390/microorganisms11041011
Schiefermeier-Mach N, Heinrich L, Lechner L, Perkhofer S. Regulation of Surfactant Protein Gene Expression by Aspergillus fumigatus in NCl-H441 Cells. Microorganisms. 2023; 11(4):1011. https://doi.org/10.3390/microorganisms11041011
Chicago/Turabian StyleSchiefermeier-Mach, Natalia, Lea Heinrich, Lukas Lechner, and Susanne Perkhofer. 2023. "Regulation of Surfactant Protein Gene Expression by Aspergillus fumigatus in NCl-H441 Cells" Microorganisms 11, no. 4: 1011. https://doi.org/10.3390/microorganisms11041011
APA StyleSchiefermeier-Mach, N., Heinrich, L., Lechner, L., & Perkhofer, S. (2023). Regulation of Surfactant Protein Gene Expression by Aspergillus fumigatus in NCl-H441 Cells. Microorganisms, 11(4), 1011. https://doi.org/10.3390/microorganisms11041011






