Adaption of Pseudomonas ogarae F113 to the Rhizosphere Environment—The AmrZ-FleQ Hub
Abstract
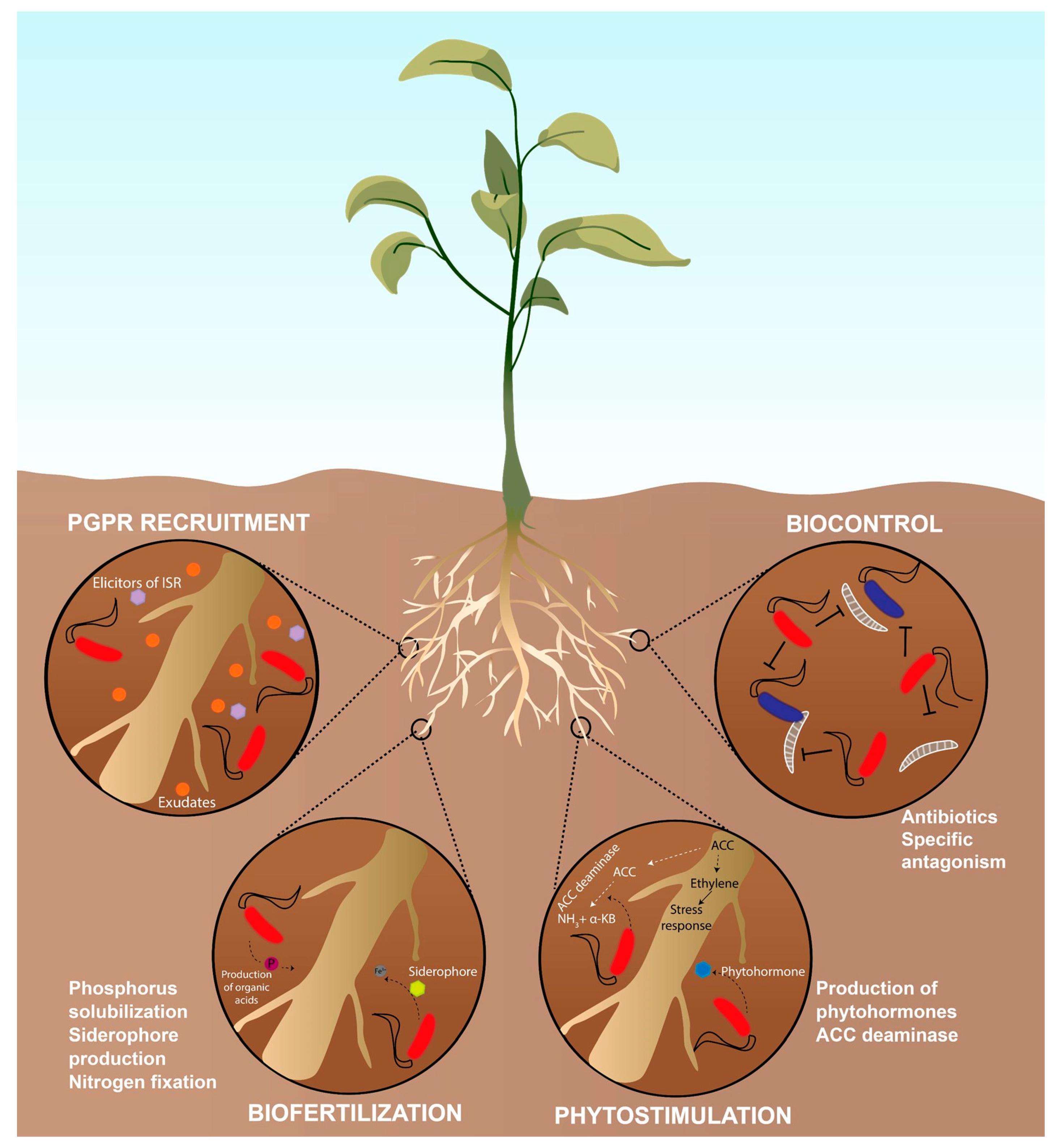
:1. The Rhizosphere and the Plant Growth Promoting Rhizobacteria (PGPR)
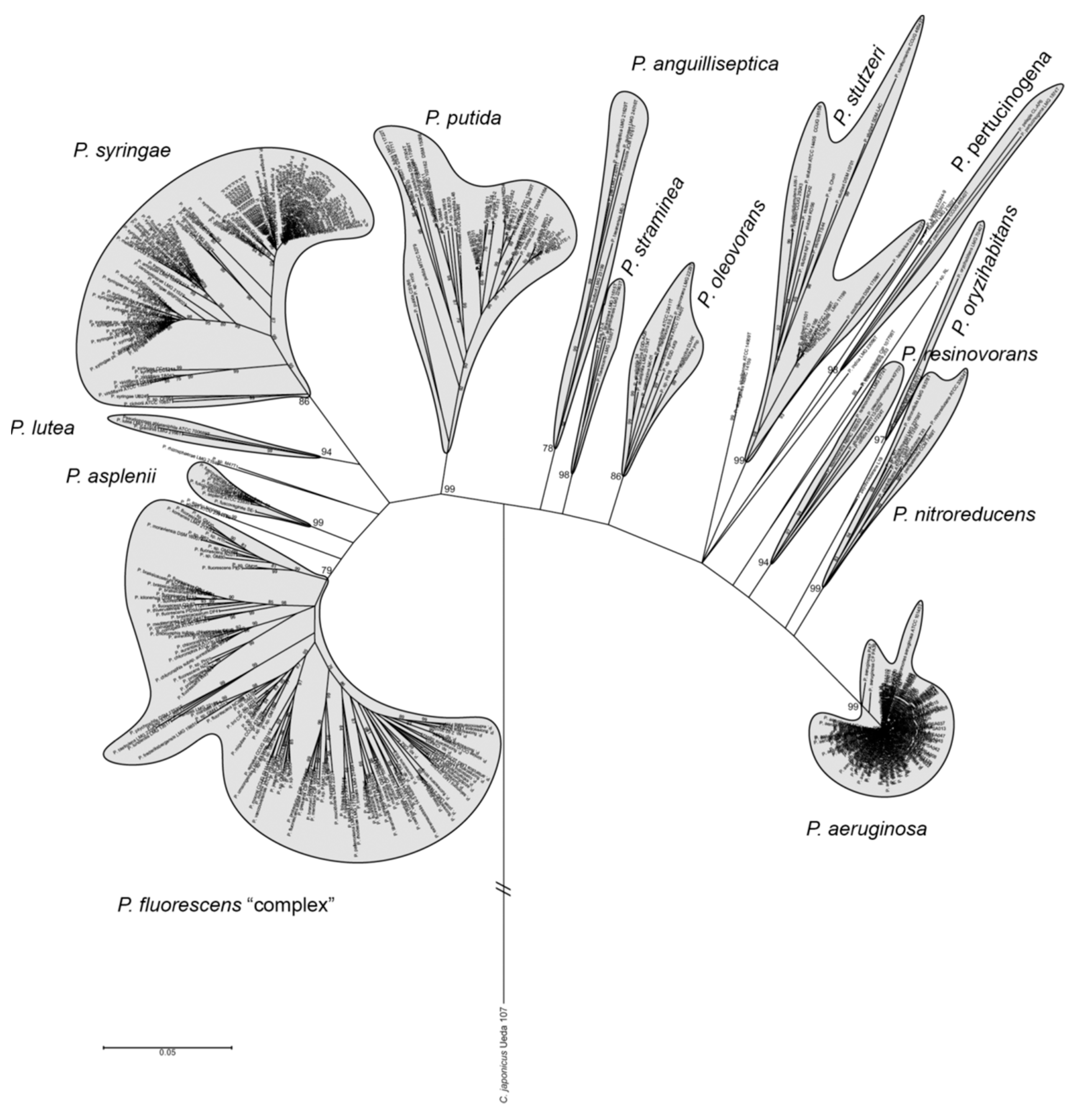
2. Pseudomonas as PGPR
3. Bacterial Lifestyles and Rhizosphere Colonization: Traits Involved in Rhizosphere Colonization
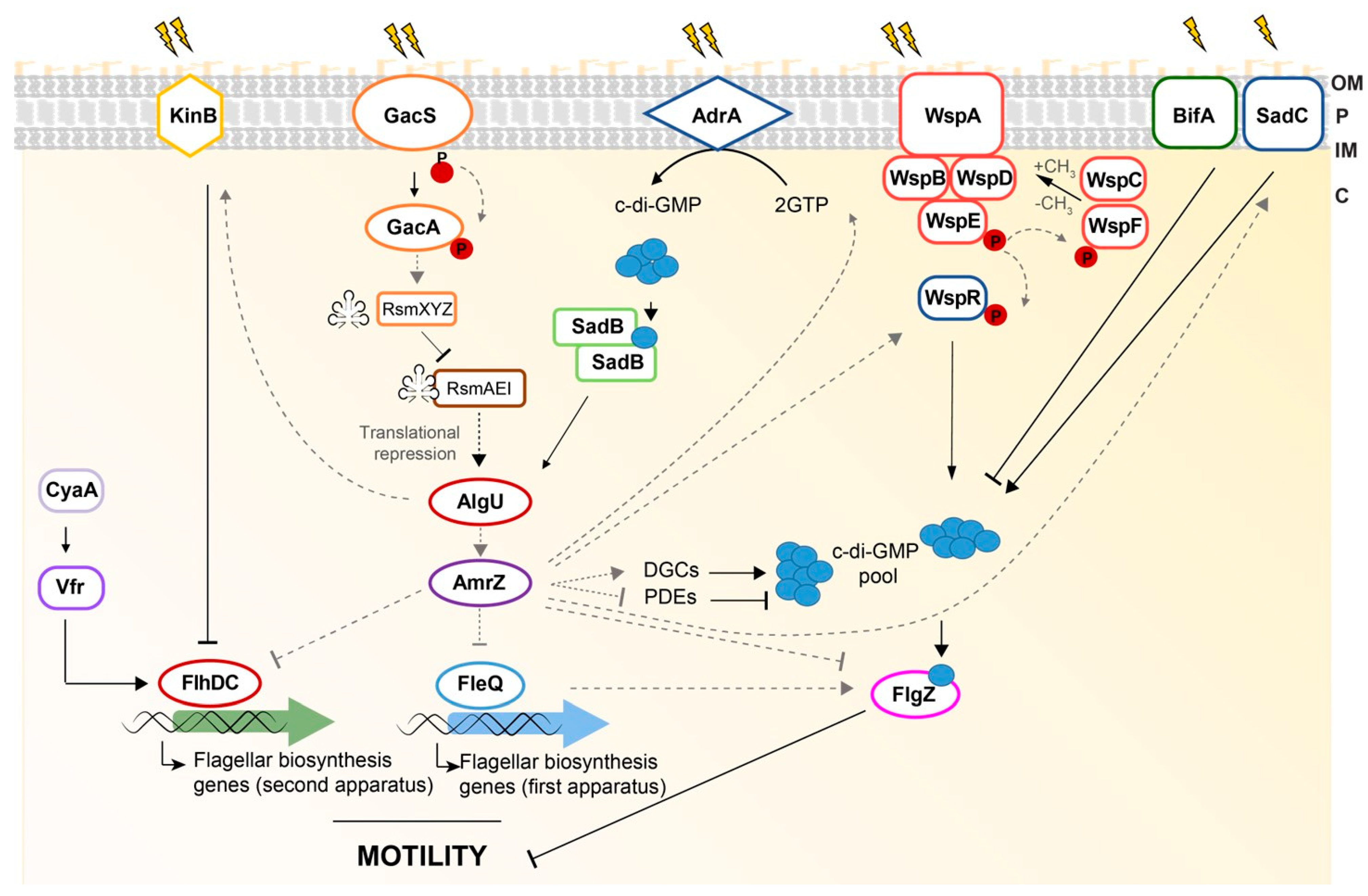
4. Motility
5. Chemotaxis
6. Microcolony and Biofilm Formation: The Extracellular Matrix (ECM)
7. Regulation of Rhizosphere Adaption: The AmrZ-FleQ Hub
8. AmrZ
9. FleQ
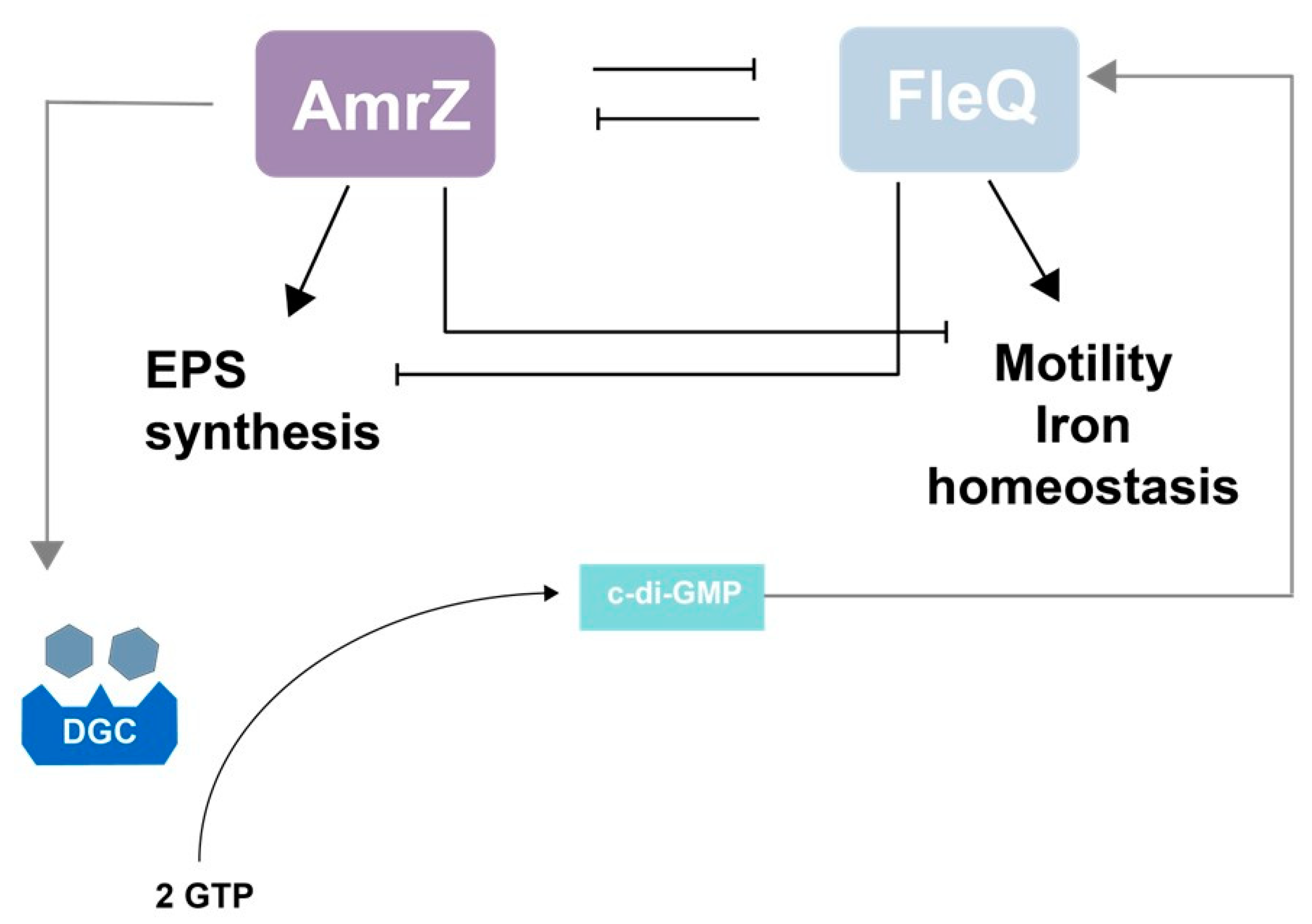
10. The AmrZ-FleQ Hub
11. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hiltner, L. Uber nevere Erfahrungen und Probleme auf dem Gebiet der Boden Bakteriologie und unter besonderer Beurchsichtigung der Grundungung und Broche. Arbeit. Deut. Landw. Ges. Berl. 1904, 98, 59–78. [Google Scholar]
- Lynch, J.; de Leij, F. Rhizosphere; eLS, John Wiley & Sons, Ltd.: Chichester, UK, 2012. [Google Scholar]
- Bazin, M.; Markham, P.; Scott, E.; Lynch, J. Population dynamics and rhizosphere interactions. In The Rhizosphere; John Wiley & Sons, Ltd.: Chichester, UK, 1990; pp. 99–127. [Google Scholar]
- Wang, N.R.; Haney, C.H. Harnessing the genetic potential of the plant microbiome. Biochemist 2020, 42, 20–25. [Google Scholar] [CrossRef]
- Fitzpatrick, C.R.; Salas-González, I.; Conway, J.M.; Finkel, O.M.; Gilbert, S.; Russ, D.; Teixeira, P.J.P.L.; Dangl, J.L. The Plant Microbiome: From Ecology to Reductionism and Beyond. Annu. Rev. Microbiol. 2020, 74, 81–100. [Google Scholar] [CrossRef]
- Jones, D.L.; Nguyen, C.; Finlay, R.D. Carbon flow in the rhizosphere: Carbon trading at the soil–root interface. Plant Soil 2009, 321, 5–33. [Google Scholar] [CrossRef]
- Walker, T.S.; Bais, H.P.; Grotewold, E.; Vivanco, J.M. Root exudation and rhizosphere biology. Plant Physiol. 2003, 132, 44–51. [Google Scholar] [CrossRef] [Green Version]
- Bais, H.P.; Park, S.-W.; Weir, T.L.; Callaway, R.M.; Vivanco, J.M. How plants communicate using the underground information superhighway. Trends Plant Sci. 2004, 9, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Venturi, V.; Keel, C. Signaling in the rhizosphere. Trends Plant Sci. 2016, 21, 187–198. [Google Scholar] [CrossRef]
- Bloemberg, G.V.; Lugtenberg, B.J. Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr. Opin. Plant Biol. 2001, 4, 343–350. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Vismans, G.; Yu, K.; Song, Y.; de Jonge, R.; Burgman, W.P.; Burmølle, M.; Herschend, J.; Bakker, P.A.; Pieterse, C.M. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018, 12, 1496–1507. [Google Scholar] [CrossRef] [Green Version]
- Schroth, M.N.; Hancock, J.G. Disease-suppressive soil and root-colonizing bacteria. Science 1982, 216, 1376–1381. [Google Scholar] [CrossRef]
- Gray, E.; Smith, D. Intracellular and extracellular PGPR: Commonalities and distinctions in the plant–bacterium signaling processes. Soil Biol. Biochem. 2005, 37, 395–412. [Google Scholar] [CrossRef]
- Kloepper, J.W. Plant growth-promoting rhizobacteria on radishes. In Proceedings of the 4th International Conference on Plant Pathogenic Bacter, Station de Pathologie Vegetale et Phytobacteriologie, INRA, Angers, France, 27 August–2 September 1978; pp. 879–882. [Google Scholar]
- Jacobsen, B.; Zidack, N.; Larson, B. The role of Bacillus-based biological control agents in integrated pest management systems: Plant diseases. Phytopathology 2004, 94, 1272–1275. [Google Scholar] [CrossRef] [Green Version]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef]
- Beattie, G.A. Plant-associated bacteria: Survey, molecular phylogeny, genomics and recent advances. In Plant-Associated Bacteria; Springer: Dordrecht, The Netherlands, 2007; pp. 1–56. [Google Scholar]
- Pastor-Bueis, R.; Mulas, R.; Gómez, X.; González-Andrés, F. Innovative liquid formulation of digestates for producing a biofertilizer based on Bacillus siamensis: Field testing on sweet pepper. J. Plant Nutr. Soil Sci. 2017, 180, 748–758. [Google Scholar] [CrossRef]
- Shirley, M.; Avoscan, L.; Bernaud, E.; Vansuyt, G.; Lemanceau, P. Comparison of iron acquisition from Fe–pyoverdine by strategy I and strategy II plants. Botany 2011, 89, 731–735. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Rahman, M.A.; Guo, J.; Kang, Z.; Babar, M.A. Comparative physiological and metabolic analysis reveals a complex mechanism involved in drought tolerance in chickpea (Cicer arietinum L.) induced by PGPR and PGRs. Sci. Rep. 2019, 9, 2097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz, N.; Armada, E.; Duque, E.; Roldán, A.; Azcón, R. Contribution of arbuscular mycorrhizal fungi and/or bacteria to enhancing plant drought tolerance under natural soil conditions: Effectiveness of autochthonous or allochthonous strains. J. Plant Physiol. 2015, 174, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef]
- Glick, B.R. The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 1995, 41, 109–117. [Google Scholar] [CrossRef]
- Riley, M.A.; Wertz, J.E. Bacteriocins: Evolution, ecology, and application. Annu. Rev. Microbiol. 2002, 56, 117–137. [Google Scholar] [CrossRef] [Green Version]
- Neeraja, C.; Anil, K.; Purushotham, P.; Suma, K.; Sarma, P.; Moerschbacher, B.M.; Podile, A.R. Biotechnological approaches to develop bacterial chitinases as a bioshield against fungal diseases of plants. Crit. Rev. Biotechnol. 2010, 30, 231–241. [Google Scholar] [CrossRef]
- Złoch, M.; Thiem, D.; Gadzała-Kopciuch, R.; Hrynkiewicz, K. Synthesis of siderophores by plant-associated metallotolerant bacteria under exposure to Cd2+. Chemosphere 2016, 156, 312–325. [Google Scholar] [CrossRef]
- Kamilova, F.; Validov, S.; Azarova, T.; Mulders, I.; Lugtenberg, B. Enrichment for enhanced competitive plant root tip colonizers selects for a new class of biocontrol bacteria. Environ. Microbiol. 2005, 7, 1809–1817. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.; Bakker, P.; Pieterse, C. Induction and expression of PGPR-mediated induced resistance against pathogens. IOBC/Wprs Bull. 1998, 21, 103–110. [Google Scholar]
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 35, 1044–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanier, R.Y.; Palleroni, N.J.; Doudoroff, M. The aerobic pseudomonads a taxonomic study. Microbiology 1966, 43, 159–271. [Google Scholar] [CrossRef] [Green Version]
- Palleroni, N.J. Prokaryote taxonomy of the 20th century and the impact of studies on the genus Pseudomonas: A personal view. Microbiology 2003, 149, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Palleroni, N.J. The Pseudomonas story. Environ. Microbiol. 2010, 12, 1377–1383. [Google Scholar] [CrossRef]
- Parte, A.C. LPSN—List of prokaryotic names with standing in nomenclature (bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol. 2018, 68, 1825–1829. [Google Scholar] [CrossRef]
- Palleroni, N.J.; Genus, I. Pseudomonas. In Bergey’s Manual of Systematic Bacteriology, 1st ed.; Bergey, D.H., Krieg, N.R., Holt, J.G., Eds.; The Williams and Wilkins Co.: Baltimore, MD, USA, 1984; Volume 1, pp. 141–199. [Google Scholar]
- Raaijmakers, J.M.; Mazzola, M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 2012, 50, 403–424. [Google Scholar] [CrossRef]
- Ramos, J.L. Pseudomonas Volume 1: Genomics, Life Style and Molecular Architecture; Springer: New York, NY, USA, 2004. [Google Scholar]
- Morris, C.E.; Sands, D.C.; Vinatzer, B.A.; Glaux, C.; Guilbaud, C.; Buffiere, A.; Yan, S.; Dominguez, H.; Thompson, B.M. The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J. 2008, 2, 321–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waters, C.M.; Goldberg, J.B. Pseudomonas aeruginosa in cystic fibrosis: A chronic cheater. Proc. Natl. Acad. Sci. USA 2019, 116, 6525–6527. [Google Scholar] [CrossRef] [Green Version]
- Xin, X.-F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Meier-Kolthoff, J.P.; Göker, M.; Martín, M.; Rivilla, R.; Redondo-Nieto, M. Genomic and Genetic Diversity within the Pseudomonas fluorescens Complex. PLoS ONE 2016, 11, e0150183. [Google Scholar] [CrossRef] [Green Version]
- Wasi, S.; Tabrez, S.; Ahmad, M. Use of Pseudomonas spp. for the bioremediation of environmental pollutants: A review. Environ. Monit. Assess. 2013, 185, 8147–8155. [Google Scholar] [CrossRef] [PubMed]
- Olivera, E.R.; Carnicero, D.; Jodra, R.; Miñambres, B.; García, B.; Abraham, G.A.; Gallardo, A.; Román, J.S.; García, J.L.; Naharro, G. Genetically engineered Pseudomonas: A factory of new bioplastics with broad applications. Environ. Microbiol. 2001, 3, 612–618. [Google Scholar] [CrossRef]
- Nikel, P.I.; De Lorenzo, V. Pseudomonas putida as a functional chassis for industrial biocatalysis: From native biochemistry to trans-metabolism. Metab. Eng. 2018, 50, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Mulet, M.; Lalucat, J.; García-Valdés, E. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 2010, 12, 1513–1530. [Google Scholar]
- Gomila, M.; Peña, A.; Mulet, M.; Lalucat, J.; García-Valdés, E. Phylogenomics and systematics in Pseudomonas. Front. Microbiol. 2015, 6, 214. [Google Scholar] [CrossRef] [Green Version]
- Lalucat, J.; Mulet, M.; Gomila, M.; García-Valdés, E. Genomics in Bacterial Taxonomy: Impact on the Genus Pseudomonas. Genes 2020, 11, 139. [Google Scholar] [CrossRef] [Green Version]
- Seaton, S.C.; Silby, M.W. Genetics and functional genomics of the Pseudomonas fluorescens Group. In Genomics of Plant-Associated Bacteria; Springer: Berlin/Heidelberg, Germany, 2014; pp. 99–125. [Google Scholar]
- Weller, D.M.; Landa, B.; Mavrodi, O.; Schroeder, K.; De La Fuente, L.; Blouin Bankhead, S.; Allende Molar, R.; Bonsall, R.; Mavrodi, D.; Thomashow, L. Role of 2, 4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in the defense of plant roots. Plant Biol. 2007, 9, 4–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couillerot, O.; Prigent-Combaret, C.; Caballero-Mellado, J.; Moënne-Loccoz, Y. Pseudomonas fluorescens and closely-related fluorescent pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett. Appl. Microbiol. 2009, 48, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Loper, J.E.; Hassan, K.A.; Mavrodi, D.V.; Davis II, E.W.; Lim, C.K.; Shaffer, B.T.; Elbourne, L.D.; Stockwell, V.O.; Hartney, S.L.; Breakwell, K. Comparative genomics of plant-associated Pseudomonas spp.: Insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012, 8, e1002784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrido-Sanz, D.; Arrebola, E.; Martínez-Granero, F.; García-Méndez, S.; Muriel, C.; Blanco-Romero, E.; Martín, M.; Rivilla, R.; Redondo-Nieto, M. Classification of isolates from the Pseudomonas fluorescens complex into phylogenomic groups based in group-specific markers. Front. Microbiol. 2017, 8, 413. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.H.; Browne, P.; Prigent-Combaret, C.; Combes-Meynet, E.; Morrissey, J.P.; O’Gara, F. Biochemical and genomic comparison of inorganic phosphate solubilization in Pseudomonas species. Environ. Microbiol. Rep. 2010, 2, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Leeman, M.; Van Oorschot, M.M.; Van der Sluis, I.; Schippers, B.; Bakker, P. Dose-response relationships in biological control of Fusarium wilt of radish by Pseudomonas spp. Phytopathology 1995, 85, 1075–1080. [Google Scholar] [CrossRef]
- Visca, P.; Imperi, F.; Lamont, I.L. Pyoverdine siderophores: From biogenesis to biosignificance. Trends Microbiol. 2007, 15, 22–30. [Google Scholar] [CrossRef]
- Picard, C.; Bosco, M. Maize heterosis affects the structure and dynamics of indigenous rhizospheric auxins-producing Pseudomonas populations. FEMS Microbiol. Ecol. 2005, 53, 349–357. [Google Scholar] [CrossRef] [Green Version]
- Blaha, D.; Prigent-Combaret, C.; Mirza, M.S.; Moënne-Loccoz, Y. Phylogeny of the 1-aminocyclopropane-1-carboxylic acid deaminase-encoding gene acdS in phytobeneficial and pathogenic Proteobacteria and relation with strain biogeography. FEMS Microbiol. Ecol. 2006, 56, 455–470. [Google Scholar] [CrossRef] [Green Version]
- Couillerot, O.; Combes-Meynet, E.; Pothier, J.F.; Bellvert, F.; Challita, E.; Poirier, M.-A.; Rohr, R.; Comte, G.; Moënne-Loccoz, Y.; Prigent-Combaret, C. The role of the antimicrobial compound 2,4-diacetylphloroglucinol in the impact of biocontrol Pseudomonas fluorescens F113 on Azospirillum brasilense phytostimulators. Microbiology 2011, 157, 1694–1705. [Google Scholar] [CrossRef] [Green Version]
- Nandi, M.; Selin, C.; Brawerman, G.; Fernando, W.D.; de Kievit, T. Hydrogen cyanide, which contributes to Pseudomonas chlororaphis strain PA23 biocontrol, is upregulated in the presence of glycine. Biol. Control 2017, 108, 47–54. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Redondo-Nieto, M.; Martin, M.; Rivilla, R. Comparative genomics of the Pseudomonas corrugata subgroup reveals high species diversity and allows the description of Pseudomonas ogarae sp. nov. Microb. Genom. 2021, 7, 000593. [Google Scholar] [CrossRef]
- Shanahan, P.; O’Sullivan, D.J.; Simpson, P.; Glennon, J.D.; O’Gara, F. Isolation of 2, 4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl. Environ. Microbiol. 1992, 58, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Simons, M.; Van Der Bij, A.; Brand, I.; De Weger, L.; Wijffelman, C.; Lugtenberg, B. Gnotobiotic system for studying rhizosphere colonization by plant growth-promoting Pseudomonas bacteria. Mol. Plant-Microbe Interact. MPMI 1996, 9, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Cronin, D.; Moënne-Loccoz, Y.; Fenton, A.; Dunne, C.; Dowling, D.N.; O’Gara, F. Ecological interaction of a biocontrol Pseudomonas fluorescens strain producing 2, 4-diacetylphloroglucinol with the soft rot potato pathogen Erwinia carotovora subsp. atroseptica. FEMS Microbiol. Ecol. 1997, 23, 95–106. [Google Scholar] [CrossRef]
- Naseby, D.; Lynch, J. Effects of Pseudomonas fluorescens F113 on ecological functions in the pea rhizosphere are dependent on pH. Microb. Ecol. 1999, 37, 248–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dekkers, L.C.; Mulders, I.H.; Phoelich, C.C.; Chin-A-Woeng, T.F.; Wijfjes, A.H.; Lugtenberg, B.J. The sss colonization gene of the tomato-Fusarium oxysporum f. sp. radicis-lycopersici biocontrol strain Pseudomonas fluorescens WCS365 can improve root colonization of other wild-type Pseudomonas spp. bacteria. Mol. Plant-Microbe Interact. 2000, 13, 1177–1183. [Google Scholar] [PubMed] [Green Version]
- Villacieros, M.; Power, B.; Sánchez-Contreras, M.; Lloret, J.; Oruezabal, R.I.; Martín, M.; Fernández-Piñas, F.; Bonilla, I.; Whelan, C.; Dowling, D.N.; et al. Colonization behaviour of Pseudomonas fluorescens and Sinorhizobium meliloti in the alfalfa (Medicago sativa) rhizosphere. Plant Soil 2003, 251, 47–54. [Google Scholar] [CrossRef]
- Villacieros, M.; Whelan, C.; Mackova, M.; Molgaard, J.; Sánchez-Contreras, M.; Lloret, J.; de Cárcer, D.A.; Oruezábal, R.I.; Bolaños, L.; Macek, T.; et al. Polychlorinated biphenyl rhizoremediation by Pseudomonas fluorescens F113 derivatives, using a Sinorhizobium meliloti nod system to drive bph gene expression. Appl. Environ. Microbiol. 2005, 71, 2687–2694. [Google Scholar] [CrossRef] [Green Version]
- De La Fuente, L.; Landa, B.B.; Weller, D.M. Host crop affects rhizosphere colonization and competitiveness of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens. Phytopathology 2006, 96, 751–762. [Google Scholar] [CrossRef] [Green Version]
- De Cárcer, D.A.; Martín, M.; Karlson, U.; Rivilla, R. Changes in bacterial populations and in biphenyl dioxygenase gene diversity in a polychlorinated biphenyl-polluted soil after introduction of willow trees for rhizoremediation. Appl. Environ. Microbiol. 2007, 73, 6224–6232. [Google Scholar] [CrossRef] [Green Version]
- Rein, A.; Fernqvist, M.M.; Mayer, P.; Trapp, S.; Bittens, M.; Karlson, U.G. Degradation of PCB congeners by bacterial strains. Appl. Microbiol. Biotechnol. 2007, 77, 469–481. [Google Scholar] [CrossRef] [Green Version]
- Barahona, E.; Navazo, A.; Yousef-Coronado, F.; Aguirre de Cárcer, D.; Martínez-Granero, F.; Espinosa-Urgel, M.; Martín, M.; Rivilla, R. Efficient rhizosphere colonization by Pseudomonas fluorescens F113 mutants unable to form biofilms on abiotic surfaces. Environ. Microbiol. 2010, 12, 3185–3195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barahona, E.; Navazo, A.; Martínez-Granero, F.; Zea-Bonilla, T.; Pérez-Jiménez, R.M.; Martín, M.; Rivilla, R. Pseudomonas fluorescens F113 mutant with enhanced competitive colonization ability and improved biocontrol activity against fungal root pathogens. Appl. Environ. Microbiol. 2011, 77, 5412–5419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vacheron, J.; Combes-Meynet, E.; Walker, V.; Gouesnard, B.; Muller, D.; Moënne-Loccoz, Y.; Prigent-Combaret, C. Expression on roots and contribution to maize phytostimulation of 1-aminocyclopropane-1-decarboxylate deaminase gene acdS in Pseudomonas fluorescens F113. Plant Soil 2016, 407, 187–202. [Google Scholar] [CrossRef]
- Vacheron, J.; Desbrosses, G.; Renoud, S.; Padilla, R.; Walker, V.; Muller, D.; Prigent-Combaret, C. Differential contribution of plant-beneficial functions from Pseudomonas kilonensis F113 to root system architecture alterations in Arabidopsis thaliana and Zea mays. Mol. Plant-Microbe Interact. 2018, 31, 212–223. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Granero, F.; Rivilla, R.; Martín, M. Rhizosphere selection of highly motile phenotypic variants of Pseudomonas fluorescens with enhanced competitive colonization ability. Appl. Environ. Microbiol. 2006, 72, 3429–3434. [Google Scholar] [CrossRef] [Green Version]
- Redondo-Nieto, M.; Barret, M.; Morrisey, J.P.; Germaine, K.; Martínez-Granero, F.; Barahona, E.; Navazo, A.; Sánchez-Contreras, M.; Moynihan, J.A.; Giddens, S.R.; et al. Genome sequence of the biocontrol strain Pseudomonas fluorescens F113. J. Bacteriol. 2012, 194, 1273–1274. [Google Scholar] [CrossRef] [Green Version]
- Redondo-Nieto, M.; Barret, M.; Morrissey, J.; Germaine, K.; Martínez-Granero, F.; Barahona, E.; Navazo, A.; Sánchez-Contreras, M.; Moynihan, J.A.; Muriel, C.; et al. Genome sequence reveals that Pseudomonas fluorescens F113 possesses a large and diverse array of systems for rhizosphere function and host interaction. BMC Genom. 2013, 14, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rezzonico, F.; Zala, M.; Keel, C.; Duffy, B.; Moënne-Loccoz, Y.; Défago, G. Is the ability of biocontrol fluorescent pseudomonads to produce the antifungal metabolite 2, 4-diacetylphloroglucinol really synonymous with higher plant protection? New Phytol. 2007, 173, 861–872. [Google Scholar] [CrossRef]
- Muriel, C.; Jalvo, B.; Redondo-Nieto, M.; Rivilla, R.; Martín, M. Chemotactic motility of Pseudomonas fluorescens F113 under aerobic and denitrification conditions. PLoS ONE 2015, 10, e0132242. [Google Scholar] [CrossRef] [Green Version]
- Ghirardi, S.; Dessaint, F.; Mazurier, S.; Corberand, T.; Raaijmakers, J.M.; Meyer, J.-M.; Dessaux, Y.; Lemanceau, P. Identification of traits shared by rhizosphere-competent strains of fluorescent pseudomonads. Microb. Ecol. 2012, 64, 725–737. [Google Scholar] [CrossRef]
- Fenton, A.; Stephens, P.; Crowley, J.; O’Callaghan, M.; O’Gara, F. Exploitation of gene (s) involved in 2, 4-diacetylphloroglucinol biosynthesis to confer a new biocontrol capability to a Pseudomonas strain. Appl. Environ. Microbiol. 1992, 58, 3873–3878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moënne-Loccoz, Y.; Tichy, H.-V.; O’Donnell, A.; Simon, R.; O’Gara, F. Impact of 2, 4-Diacetylphloroglucinol-Producing Biocontrol Strain Pseudomonas fluorescens F113 on Intraspecific Diversity of Resident Culturable Fluorescent Pseudomonads Associated with the Roots of Field-Grown Sugar Beet Seedlings. Appl. Environ. Microbiol. 2001, 67, 3418–3425. [Google Scholar] [CrossRef] [Green Version]
- Marshall, K.C. Planktonic versus sessile life of prokaryotes. Prokaryotes 2006, 2, 3–15. [Google Scholar]
- Sauer, K.; Camper, A.K.; Ehrlich, G.D.; Costerton, J.W.; Davies, D.G. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 2002, 184, 1140–1154. [Google Scholar] [CrossRef] [Green Version]
- Wan, N.; Wang, H.; Ng, C.K.; Mukherjee, M.; Ren, D.; Cao, B.; Tang, Y.J. Bacterial metabolism during biofilm growth investigated by 13C tracing. Front. Microbiol. 2018, 9, 2657. [Google Scholar] [CrossRef] [Green Version]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef] [PubMed]
- Lugtenberg, B.J.; Dekkers, L.; Bloemberg, G.V. Molecular determinants of rhizosphere colonization by Pseudomonas. Annu. Rev. Phytopathol. 2001, 39, 461–490. [Google Scholar] [CrossRef]
- Chin-A-Woeng, T.F.; Bloemberg, G.V.; Mulders, I.H.; Dekkers, L.C.; Lugtenberg, B.J. Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol. Plant-Microbe Interact. 2000, 13, 1340–1345. [Google Scholar] [CrossRef] [Green Version]
- Pliego, C.; De Weert, S.; Lamers, G.; De Vicente, A.; Bloemberg, G.; Cazorla, F.M.; Ramos, C. Two similar enhanced root-colonizing Pseudomonas strains differ largely in their colonization strategies of avocado roots and Rosellinia necatrix hyphae. Environ. Microbiol. 2008, 10, 3295–3304. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [Green Version]
- Capdevila, S.; Martínez-Granero, F.M.; Sánchez-Contreras, M.; Rivilla, R.; Martín, M. Analysis of Pseudomonas fluorescens F113 genes implicated in flagellar filament synthesis and their role in competitive root colonization. Microbiology 2004, 150, 3889–3897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saunders, N.J.; Moxon, E.R.; Gravenor, M.B. Mutation rates: Estimating phase variation rates when fitness differences are present and their impact on population structure. Microbiology 2003, 149, 485–495. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Contreras, M.; Martín, M.; Villacieros, M.; O’Gara, F.; Bonilla, I.; Rivilla, R. Phenotypic selection and phase variation occur during alfalfa root colonization by Pseudomonas fluorescens F113. J. Bacteriol. 2002, 184, 1587–1596. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Granero, F.; Capdevila, S.; Sanchez-Contreras, M.; Martin, M.; Rivilla, R. Two site-specific recombinases are implicated in phenotypic variation and competitive rhizosphere colonization in Pseudomonas fluorescens. Microbiology 2005, 151, 975–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos-González, M.I.; Matilla, M.A.; Quesada, J.M.; Ramos, J.L.; Espinosa-Urgel, M. Using genomics to unveil bacterial determinants of rhizosphere life style. Mol. Microb. Ecol. Rhizosphere 2013, 1, 5–16. [Google Scholar]
- Levy, A.; Conway, J.M.; Dangl, J.L.; Woyke, T. Elucidating bacterial gene functions in the plant microbiome. Cell Host Microbe 2018, 24, 475–485. [Google Scholar] [CrossRef] [Green Version]
- Levy, A.; González, I.S.; Mittelviefhaus, M.; Clingenpeel, S.; Paredes, S.H.; Miao, J.; Wang, K.; Devescovi, G.; Stillman, K.; Monteiro, F. Genomic features of bacterial adaptation to plants. Nat. Genet. 2018, 50, 138–150. [Google Scholar] [CrossRef] [Green Version]
- Ofek-Lalzar, M.; Sela, N.; Goldman-Voronov, M.; Green, S.J.; Hadar, Y.; Minz, D. Niche and host-associated functional signatures of the root surface microbiome. Nat. Commun. 2014, 5, 4950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, B.J.; Feltcher, M.E.; Waters, R.J.; Wetmore, K.M.; Mucyn, T.S.; Ryan, E.M.; Wang, G.; Ul-Hasan, S.; McDonald, M.; Yoshikuni, Y. Genome-wide identification of bacterial plant colonization genes. PLoS Biol. 2017, 15, e2002860. [Google Scholar] [CrossRef] [PubMed]
- Helmann, T.C.; Deutschbauer, A.M.; Lindow, S.E. Genome-wide identification of Pseudomonas syringae genes required for fitness during colonization of the leaf surface and apoplast. Proc. Natl. Acad. Sci. USA 2019, 116, 18900–18910. [Google Scholar] [CrossRef] [Green Version]
- Armanhi, J.S.L.; de Souza, R.S.C.; Damasceno, N.d.B.; de Araújo, L.M.; Imperial, J.; Arruda, P. A community-based culture collection for targeting novel plant growth-promoting bacteria from the sugarcane microbiome. Front. Plant Sci. 2018, 8, 2191. [Google Scholar] [CrossRef] [Green Version]
- Arruda, B.; Rodrigues, M.; Gumiere, T.; Richardson, A.E.; Andreote, F.D.; Soltangheisi, A.; Gatiboni, L.C.; Pavinato, P.S. The impact of sugarcane filter cake on the availability of P in the rhizosphere and associated microbial community structure. Soil Use Manag. 2019, 35, 334–345. [Google Scholar] [CrossRef]
- Wei, Y.; Wang, X.; Liu, J.; Nememan, I.; Singh, A.H.; Weiss, H.; Levin, B.R. The population dynamics of bacteria in physically structured habitats and the adaptive virtue of random motility. Proc. Natl. Acad. Sci. USA 2011, 108, 4047–4052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henrichsen, J. Bacterial surface translocation: A survey and a classification. Bacteriol. Rev. 1972, 36, 478. [Google Scholar] [CrossRef]
- Mattingly, A.E.; Weaver, A.A.; Dimkovikj, A.; Shrout, J.D. Assessing travel conditions: Environmental and host influences on bacterial surface motility. J. Bacteriol. 2018, 200, e00014-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shrout, J.D.; Chopp, D.L.; Just, C.L.; Hentzer, M.; Givskov, M.; Parsek, M.R. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 2006, 62, 1264–1277. [Google Scholar] [CrossRef] [PubMed]
- Burrows, L.L. Pseudomonas aeruginosa twitching motility: Type IV pili in action. Annu. Rev. Microbiol. 2012, 66, 493–520. [Google Scholar] [CrossRef] [Green Version]
- Nan, B. Bacterial gliding motility: Rolling out a consensus model. Curr. Biol. 2017, 27, R154–R156. [Google Scholar] [CrossRef] [Green Version]
- Nogales, J.; Vargas, P.; Farias, G.A.; Olmedilla, A.; Sanjuán, J.; Gallegos, M.-T. FleQ coordinates flagellum-dependent and-independent motilities in Pseudomonas syringae pv. tomato DC3000. Appl. Environ. Microbiol. 2015, 81, 7533–7545. [Google Scholar] [CrossRef] [Green Version]
- Sampedro, I.; Parales, R.E.; Krell, T.; Hill, J.E. Pseudomonas chemotaxis. FEMS Microbiol. Rev. 2015, 39, 17–46. [Google Scholar]
- Bashan, Y. Migration of the rhizosphere bacteria Azospirillum brasilense and Pseudomonas fluorescens towards wheat roots in the soil. Microbiology 1986, 132, 3407–3414. [Google Scholar] [CrossRef] [Green Version]
- Espinosa-Urgel, M.; Kolter, R.; Ramos, J.L. Root colonization by Pseudomonas putida: Love at first sight. Microbiology 2002, 148, 341–343. [Google Scholar] [CrossRef] [Green Version]
- De Weert, S.; Vermeiren, H.; Mulders, I.H.; Kuiper, I.; Hendrickx, N.; Bloemberg, G.V.; Vanderleyden, J.; De Mot, R.; Lugtenberg, B.J. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol. Plant-Microbe Interact. 2002, 15, 1173–1180. [Google Scholar] [CrossRef] [Green Version]
- Harshey, R.M. Bacterial motility on a surface: Many ways to a common goal. Annu. Rev. Microbiol. 2003, 57, 249–273. [Google Scholar] [CrossRef]
- Kearns, D.B. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 2010, 8, 634–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armitage, J.P. Bacterial tactic responses. In Advances in Microbial Physiology; Elsevier: Amsterdam, The Netherlands, 1999; Volume 41, pp. 229–289. [Google Scholar]
- Thormann, K.M.; Paulick, A. Tuning the flagellar motor. Microbiology 2010, 156, 1275–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sowa, Y.; Berry, R.M. Bacterial flagellar motor. Q. Rev. Biophys. 2008, 41, 103–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arora, S.K.; Ritchings, B.W.; Almira, E.C.; Lory, S.; Ramphal, R. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 1997, 179, 5574–5581. [Google Scholar] [CrossRef] [Green Version]
- Dasgupta, N.; Wolfgang, M.C.; Goodman, A.L.; Arora, S.K.; Jyot, J.; Lory, S.; Ramphal, R. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 2003, 50, 809–824. [Google Scholar] [CrossRef] [Green Version]
- Barahona, E.; Navazo, A.; Garrido-Sanz, D.; Muriel, C.; Martínez-Granero, F.; Redondo-Nieto, M.; Martín, M.; Rivilla, R. Pseudomonas fluorescens F113 can produce a second flagellar apparatus, which is important for plant root colonization. Front. Microbiol. 2016, 7, 1471. [Google Scholar] [CrossRef] [Green Version]
- Navazo, A.; Barahona, E.; Redondo-Nieto, M.; Martínez-Granero, F.; Rivilla, R.; Martín, M. Three independent signalling pathways repress motility in Pseudomonas fluorescens F113. Microb. Biotechnol. 2009, 2, 489–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muriel, C.; Blanco-Romero, E.; Trampari, E.; Arrebola, E.; Durán, D.; Redondo-Nieto, M.; Malone, J.G.; Martín, M.; Rivilla, R. The diguanylate cyclase AdrA regulates flagellar biosynthesis in Pseudomonas fluorescens F113 through SadB. Sci. Rep. 2019, 9, 8096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Römling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Povolotsky, T.L.; Hengge, R. ‘Life-style’control networks in Escherichia coli: Signaling by the second messenger c-di-GMP. J. Biotechnol. 2012, 160, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Granero, F.; Navazo, A.; Barahona, E.; Redondo-Nieto, M.; Rivilla, R.; Martín, M. The Gac-Rsm and SadB signal transduction pathways converge on AlgU to downregulate motility in Pseudomonas fluorescens. PLoS ONE 2012, 7, e31765. [Google Scholar] [CrossRef] [Green Version]
- Schubert, M.; Lapouge, K.; Duss, O.; Oberstrass, F.C.; Jelesarov, I.; Haas, D.; Allain, F.H. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat. Struct. Mol. Biol. 2007, 14, 807–813. [Google Scholar] [CrossRef]
- Martínez-Granero, F.; Redondo-Nieto, M.; Vesga, P.; Martín, M.; Rivilla, R. AmrZ is a global transcriptional regulator implicated in iron uptake and environmental adaption in P. fluorescens F113. BMC Genom. 2014, 15, 237. [Google Scholar] [CrossRef] [Green Version]
- Muriel, C.; Arrebola, E.; Redondo-Nieto, M.; Martínez-Granero, F.; Jalvo, B.; Pfeilmeier, S.; Blanco-Romero, E.; Baena, I.; Malone, J.G.; Rivilla, R.; et al. AmrZ is a major determinant of c-di-GMP levels in Pseudomonas fluorescens F113. Sci. Rep. 2018, 8, 1979. [Google Scholar] [CrossRef] [Green Version]
- Dasgupta, N.; Ferrell, E.P.; Kanack, K.J.; West, S.E.; Ramphal, R. fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is σ70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J. Bacteriol. 2002, 184, 5240–5250. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Granero, F.; Navazo, A.; Barahona, E.; Redondo-Nieto, M.; De Heredia, E.G.; Baena, I.; Martín-Martín, I.; Rivilla, R.; Martín, M. Identification of flgZ as a flagellar gene encoding a PilZ domain protein that regulates swimming motility and biofilm formation in Pseudomonas. PLoS ONE 2014, 9, e87608. [Google Scholar] [CrossRef] [Green Version]
- Matilla, M.A.; Krell, T. The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol. Rev. 2018, 42, fux052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hess, J.F.; Oosawa, K.; Matsumura, P.; Simon, M.I. Protein phosphorylation is involved in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 1987, 84, 7609–7613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scharf, B.E.; Hynes, M.F.; Alexandre, G.M. Chemotaxis signaling systems in model beneficial plant–bacteria associations. Plant Mol. Biol. 2016, 90, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Oku, S.; Komatsu, A.; Tajima, T.; Nakashimada, Y.; Kato, J. Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0-1 and their involvement in chemotaxis to tomato root exudate and root colonization. Microbes Environ. 2012, 27, 462–469. [Google Scholar] [CrossRef] [Green Version]
- Oku, S.; Komatsu, A.; Nakashimada, Y.; Tajima, T.; Kato, J. Identification of Pseudomonas fluorescens chemotaxis sensory proteins for malate, succinate, and fumarate, and their involvement in root colonization. Microbes Environ. 2014, 29, 413–419. [Google Scholar] [CrossRef] [Green Version]
- Schweinitzer, T.; Josenhans, C. Bacterial energy taxis: A global strategy? Arch. Microbiol. 2010, 192, 507–520. [Google Scholar] [CrossRef] [Green Version]
- Costerton, J. The role of bacterial exopolysaccharides in nature and disease. J. Ind. Microbiol. Biotechnol. 1999, 22, 551–563. [Google Scholar] [CrossRef]
- Høiby, N. A personal history of research on microbial biofilms and biofilm infections. Pathog. Dis. 2014, 70, 205–211. [Google Scholar] [CrossRef] [Green Version]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- Rudrappa, T.; Biedrzycki, M.L.; Bais, H.P. Causes and consequences of plant-associated biofilms. FEMS Microbiol. Ecol. 2008, 64, 153–166. [Google Scholar] [CrossRef]
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm Matrixome: Extracellular Components in Structured Microbial Communities. Trends Microbiol. 2020, 28, 668–681. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Dragoš, A.; Kovács, Á.T. The peculiar functions of the bacterial extracellular matrix. Trends Microbiol. 2017, 25, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Mann, E.E.; Wozniak, D.J. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 2012, 36, 893–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanco-Romero, E.; Garrido-Sanz, D.; Rivilla, R.; Redondo-Nieto, M.; Martin, M. In Silico Characterization and Phylogenetic Distribution of Extracellular Matrix Components in the Model Rhizobacteria Pseudomonas fluorescens F113 and Other Pseudomonads. Microorganisms 2020, 8, 1740. [Google Scholar] [CrossRef]
- Ha, D.-G.; O’Toole, G.A. c-di-GMP and its effects on biofilm formation and dispersion: A Pseudomonas aeruginosa review. Microb. Biofilms 2015, 3, 301–317. [Google Scholar] [CrossRef] [Green Version]
- Fazli, M.; Almblad, H.; Rybtke, M.L.; Givskov, M.; Eberl, L.; Tolker-Nielsen, T. Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ. Microbiol. 2014, 16, 1961–1981. [Google Scholar] [CrossRef]
- Davies, J.A.; Harrison, J.J.; Marques, L.L.; Foglia, G.R.; Stremick, C.A.; Storey, D.G.; Turner, R.J.; Olson, M.E.; Ceri, H. The GacS sensor kinase controls phenotypic reversion of small colony variants isolated from biofilms of Pseudomonas aeruginosa PA14. FEMS Microbiol. Ecol. 2007, 59, 32–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Toole, G.A.; Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef]
- Hickman, J.W.; Harwood, C.S. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 2008, 69, 376–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Gil, M.; Ramos-González, M.I.; Espinosa-Urgel, M. Roles of cyclic di-GMP and the Gac system in transcriptional control of the genes coding for the Pseudomonas putida adhesins LapA and LapF. J. Bacteriol. 2014, 196, 1484–1495. [Google Scholar] [CrossRef] [Green Version]
- Matsuyama, B.Y.; Krasteva, P.V.; Baraquet, C.; Harwood, C.S.; Sondermann, H.; Navarro, M.V. Mechanistic insights into c-di-GMP–dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2016, 113, E209–E218. [Google Scholar] [CrossRef] [Green Version]
- Baraquet, C.; Murakami, K.; Parsek, M.R.; Harwood, C.S. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 2012, 40, 7207–7218. [Google Scholar] [CrossRef] [PubMed]
- Navarro, M.V.; Newell, P.D.; Krasteva, P.V.; Chatterjee, D.; Madden, D.R.; O’Toole, G.A.; Sondermann, H. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLoS Biol. 2011, 9, e1000588. [Google Scholar] [CrossRef]
- Newell, P.D.; Yoshioka, S.; Hvorecny, K.L.; Monds, R.D.; O’Toole, G.A. Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0-1. J. Bacteriol. 2011, 193, 4685–4698. [Google Scholar] [CrossRef] [Green Version]
- Monds, R.D.; Newell, P.D.; Gross, R.H.; O’Toole, G.A. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 2007, 63, 656–679. [Google Scholar] [CrossRef]
- López-Sánchez, A.; Jiménez-Fernández, A.; Calero, P.; Gallego, L.D.; Govantes, F. New methods for the isolation and characterization of biofilm-persistent mutants in Pseudomonas putida. Environ. Microbiol. Rep. 2013, 5, 679–685. [Google Scholar]
- Valentini, M.; Filloux, A. Biofilms and cyclic di-GMP (c-di-GMP) signaling: Lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 2016, 291, 12547–12555. [Google Scholar] [CrossRef] [Green Version]
- Liang, F.; Zhang, B.; Yang, Q.; Zhang, Y.; Zheng, D.; Zhang, L.-q.; Yan, Q.; Wu, X. Cyclic-di-GMP Regulates the Quorum-Sensing System and Biocontrol Activity of Pseudomonas fluorescens 2P24 through the RsmA and RsmE Proteins. Appl. Environ. Microbiol. 2020, 86, e02016–e02020. [Google Scholar] [CrossRef] [PubMed]
- Flores-Bautista, E.; Hernández-Guerrero, R.; Huerta-Saquero, A.; Tenorio-Salgado, S.; Rivera-Gómez, N.; Romero, A.; Ibarra, J.A.; Pérez-Rueda, E. Deciphering the functional diversity of DNA-binding transcription factors in Bacteria and Archaea organisms. PLoS ONE 2020, 15, e0237135. [Google Scholar] [CrossRef]
- Pérez-Rueda, E.; Hernández-Guerrero, R.; Martínez-Núñez, M.A.; Armenta-Medina, D.; Sánchez, I.; Ibarra, J.A. Abundance, diversity and domain architecture variability in prokaryotic DNA-binding transcription factors. PLoS ONE 2018, 13, e0195332. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, I.; Hernández-Guerrero, R.; Méndez-Monroy, P.E.; Martínez-Núñez, M.A.; Ibarra, J.A.; Pérez-Rueda, E. Evaluation of the abundance of DNA-binding transcription factors in prokaryotes. Genes 2020, 11, 52. [Google Scholar] [CrossRef] [Green Version]
- Fan, L.; Wang, T.; Hua, C.; Sun, W.; Li, X.; Grunwald, L.; Liu, J.; Wu, N.; Shao, X.; Yin, Y. A compendium of DNA-binding specificities of transcription factors in Pseudomonas syringae. Nat. Commun. 2020, 11, 4947. [Google Scholar] [CrossRef] [PubMed]
- Baynham, P.J.; Brown, A.L.; Hall, L.L.; Wozniak, D.J. Pseudomonas aeruginosa AlgZ, a ribbon–helix–helix DNA-binding protein, is essential for alginate synthesis and algD transcriptional activation. Mol. Microbiol. 1999, 33, 1069–1080. [Google Scholar] [CrossRef]
- Waligora, E.A.; Ramsey, D.M.; Pryor, E.E.; Lu, H.; Hollis, T.; Sloan, G.P.; Deora, R.; Wozniak, D.J. AmrZ beta-sheet residues are essential for DNA binding and transcriptional control of Pseudomonas aeruginosa virulence genes. J. Bacteriol. 2010, 192, 5390–5401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, C.J.; Newsom, D.; Kelly, B.; Irie, Y.; Jennings, L.K.; Xu, B.; Limoli, D.H.; Harrison, J.J.; Parsek, M.R.; White, P. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 2014, 10, e1003984. [Google Scholar] [CrossRef]
- Wozniak, D.J.; Sprinkle, A.B.; Baynham, P.J. Control of Pseudomonas aeruginosa algZ expression by the alternative sigma factor AlgT. J. Bacteriol. 2003, 185, 7297–7300. [Google Scholar] [CrossRef] [Green Version]
- Baynham, P.J.; Wozniak, D.J. Identification and characterization of AlgZ, an AlgT-dependent DNA-binding protein required for Pseudomonas aeruginosa algD transcription. Mol. Microbiol. 1996, 22, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, D.J.; Ohman, D. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J. Bacteriol. 1994, 176, 6007–6014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramsey, D.M.; Wozniak, D.J. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 2005, 56, 309–322. [Google Scholar] [CrossRef]
- Xu, B.; Ju, Y.; Soukup, R.J.; Ramsey, D.M.; Fishel, R.; Wysocki, V.H.; Wozniak, D.J. The Pseudomonas aeruginosa AmrZ C-terminal domain mediates tetramerization and is required for its activator and repressor functions. Environ. Microbiol. Rep. 2016, 8, 85–90. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.J.; Ryder, C.R.; Mann, E.E.; Wozniak, D.J. AmrZ modulates Pseudomonas aeruginosa biofilm architecture by directly repressing transcription of the psl operon. J. Bacteriol. 2013, 195, 1637–1644. [Google Scholar] [CrossRef] [Green Version]
- Petrova, O.E.; Cherny, K.E.; Sauer, K. The Pseudomonas aeruginosa diguanylate cyclase GcbA, a homolog of P. fluorescens GcbA, promotes initial attachment to surfaces, but not biofilm formation, via regulation of motility. J. Bacteriol. 2014, 196, 2827–2841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, L.; Debru, A.; Chen, Q.; Bao, Q.; Li, K. AmrZ Regulates Swarming Motility through c-di-GMP Dependent Motility Inhibition and Controlling Pel Polysaccharide Production in Pseudomonas aeruginosa PA14. Front. Microbiol. 2019, 10, 1847. [Google Scholar] [CrossRef] [Green Version]
- Garrett, E.S.; Perlegas, D.; Wozniak, D.J. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU). J. Bacteriol. 1999, 181, 7401–7404. [Google Scholar] [CrossRef] [Green Version]
- Tart, A.H.; Blanks, M.J.; Wozniak, D.J. The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J. Bacteriol. 2006, 188, 6483–6489. [Google Scholar] [CrossRef] [Green Version]
- Xu, A.; Zhang, M.; Du, W.; Wang, D.; Ma, L.Z. A molecular mechanism for how sigma factor AlgT and transcriptional regulator AmrZ inhibit twitching motility in Pseudomonas aeruginosa. Environ. Microbiol. 2020, 23, 572–587. [Google Scholar] [CrossRef]
- Baynham, P.J.; Ramsey, D.M.; Gvozdyev, B.V.; Cordonnier, E.M.; Wozniak, D.J. The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J. Bacteriol. 2006, 188, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Pryor, E.E., Jr.; Waligora, E.A.; Xu, B.; Dellos-Nolan, S.; Wozniak, D.J.; Hollis, T. The transcription factor AmrZ utilizes multiple DNA binding modes to recognize activator and repressor sequences of Pseudomonas aeruginosa virulence genes. PLoS Pathog. 2012, 8, e1002648. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Shao, X.; Xie, Y.; Wang, T.; Zhang, Y.; Wang, X.; Deng, X. An integrated genomic regulatory network of virulence-related transcriptional factors in Pseudomonas aeruginosa. Nat. Commun. 2019, 10, 2931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allsopp, L.P.; Wood, T.E.; Howard, S.A.; Maggiorelli, F.; Nolan, L.M.; Wettstadt, S.; Filloux, A. RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2017, 114, 7707–7712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prada-Ramírez, H.A.; Pérez-Mendoza, D.; Felipe, A.; Martínez-Granero, F.; Rivilla, R.; Sanjuán, J.; Gallegos, M.T. AmrZ regulates cellulose production in Pseudomonas syringae pv. tomato DC 3000. Mol. Microbiol. 2016, 99, 960–977. [Google Scholar]
- Pérez-Mendoza, D.; Felipe, A.; Ferreiro, M.D.; Sanjuán, J.; Gallegos, M.T. AmrZ and FleQ Co-regulate cellulose production in Pseudomonas syringae pv. tomato DC3000. Front. Microbiol. 2019, 10, 746. [Google Scholar] [CrossRef] [Green Version]
- Baltrus, D.A.; Dougherty, K.; Díaz, B.; Murillo, R. Evolutionary Plasticity of AmrZ Regulation in Pseudomonas. mSphere 2018, 3, e00132-18. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Yan, H.; Xiao, Y.; Nie, H.; Huang, Q.; Chen, W. The exopolysaccharide gene cluster pea is transcriptionally controlled by RpoS and repressed by AmrZ in Pseudomonas putida KT2440. Microbiol. Res. 2019, 218, 1–11. [Google Scholar] [CrossRef]
- Blanco-Romero, E.; Garrido-Sanz, D.; Duran, D.; Rivilla, R.; Redondo-Nieto, M.; Martin, M. Regulation of extracellular matrix components by AmrZ is mediated by c-di-GMP in Pseudomonas ogarae F113. Sci. Rep. 2022, 12, 11914. [Google Scholar] [CrossRef]
- Xu, B.; Wozniak, D.J. Development of a novel method for analyzing Pseudomonas aeruginosa twitching motility and its application to define the AmrZ regulon. PLoS ONE 2015, 10, e0136426. [Google Scholar] [CrossRef] [Green Version]
- Jyot, J.; Dasgupta, N.; Ramphal, R. FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J. Bacteriol. 2002, 184, 5251–5260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baraquet, C.; Harwood, C.S. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc. Natl. Acad. Sci. USA 2013, 110, 18478–18483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasgupta, N.; Ramphal, R. Interaction of the Antiactivator FleN with the Transcriptional Activator FleQ Regulates Flagellar Number in Pseudomonas aeruginosa. J. Bacteriol. 2001, 183, 6636–6644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, T.; Liu, S.; Wang, K.; Chi, K.; Zhu, D.; Wei, T.; Huang, Y.; Guo, L.; Hu, W.; Xu, S. The REC domain mediated dimerization is critical for FleQ from Pseudomonas aeruginosa to function as a c-di-GMP receptor and flagella gene regulator. J. Struct. Biol. 2015, 192, 1–13. [Google Scholar] [CrossRef]
- Molina-Henares, M.A.; Ramos-González, M.I.; Daddaoua, A.; Fernández-Escamilla, A.M.; Espinosa-Urgel, M. FleQ of Pseudomonas putida KT2440 is a multimeric cyclic diguanylate binding protein that differentially regulates expression of biofilm matrix components. Res. Microbiol. 2017, 168, 36–45. [Google Scholar] [CrossRef]
- Marshall, B.; Robleto, E.A.; Wetzler, R.; Kulle, P.; Casaz, P.; Levy, S.B. The adnA transcriptional factor affects persistence and spread of Pseudomonas fluorescens under natural field conditions. Appl. Environ. Microbiol. 2001, 67, 852–857. [Google Scholar] [CrossRef] [Green Version]
- Casaz, P.; Happel, A.; Keithan, J.; Read, D.L.; Strain, S.R.; Levy, S.B. The Pseudomonas fluorescens transcription activator AdnA is required for adhesion and motility. Microbiology 2001, 147, 355–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mastropaolo, M.D.; Silby, M.W.; Nicoll, J.S.; Levy, S.B. Novel genes involved in Pseudomonas fluorescens Pf0-1 motility and biofilm formation. Appl. Environ. Microbiol. 2012, 78, 4318–4329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baraquet, C.; Harwood, C.S. FleQ DNA binding consensus sequence revealed by studies of FleQ-dependent regulation of biofilm gene expression in Pseudomonas aeruginosa. J. Bacteriol. 2016, 198, 178–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez-Fernández, A.; López-Sánchez, A.; Jiménez-Díaz, L.; Navarrete, B.; Calero, P.; Platero, A.I.; Govantes, F. Complex interplay between FleQ, cyclic diguanylate and multiple σ factors coordinately regulates flagellar motility and biofilm development in Pseudomonas putida. PLoS ONE 2016, 11, e0163142. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Nie, H.; Liu, H.; Luo, X.; Chen, W.; Huang, Q. C-di-GMP regulates the expression of lapA and bcs operons via FleQ in Pseudomonas putida KT2440. Environ. Microbiol. Rep. 2016, 8, 659–666. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Wang, J.; Wang, X. FleQ regulates both the type VI secretion system and flagella in Pseudomonas putida. Biotechnol. Appl. Biochem. 2018, 65, 419–427. [Google Scholar] [CrossRef]
- Navarrete, B.; Leal-Morales, A.; Serrano-Ron, L.; Sarrió, M.; Jiménez-Fernández, A.; Jiménez-Díaz, L.; López-Sánchez, A.; Govantes, F. Transcriptional organization, regulation and functional analysis of flhF and fleN in Pseudomonas putida. PLoS ONE 2019, 14, e0214166. [Google Scholar] [CrossRef] [Green Version]
- Giddens, S.R.; Jackson, R.W.; Moon, C.D.; Jacobs, M.A.; Zhang, X.-X.; Gehrig, S.M.; Rainey, P.B. Mutational activation of niche-specific genes provides insight into regulatory networks and bacterial function in a complex environment. Proc. Natl. Acad. Sci. USA 2007, 104, 18247–18252. [Google Scholar] [CrossRef] [Green Version]
- Nie, H.; Xiao, Y.; Liu, H.; He, J.; Chen, W.; Huang, Q. FleN and FleQ play a synergistic role in regulating lapA and bcs operons in Pseudomonas putida KT2440. Environ. Microbiol. Rep. 2017, 9, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Romero, E.; Duran, D.; Garrido-Sanz, D.; Rivilla, R.; Martin, M.; Redondo-Nieto, M. Transcriptomic analysis of Pseudomonas ogarae F113 reveals the antagonistic roles of AmrZ and FleQ during rhizosphere adaption. Microb. Genom. 2022, 8, 000750. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Romero, E.; Redondo-Nieto, M.; Martinez-Granero, F.; Garrido-Sanz, D.; Ramos-Gonzalez, M.I.; Martin, M.; Rivilla, R. Genome-wide analysis of the FleQ direct regulon in Pseudomonas fluorescens F113 and Pseudomonas putida KT2440. Sci. Rep. 2018, 8, 13145. [Google Scholar] [CrossRef] [PubMed]
- Duran, D.; Bernal, P.; Vazquez-Arias, D.; Blanco-Romero, E.; Garrido-Sanz, D.; Redondo-Nieto, M.; Rivilla, R.; Martin, M. Pseudomonas fluorescens F113 type VI secretion systems mediate bacterial killing and adaption to the rhizosphere microbiome. Sci. Rep. 2021, 11, 5772. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blanco-Romero, E.; Durán, D.; Garrido-Sanz, D.; Redondo-Nieto, M.; Martín, M.; Rivilla, R. Adaption of Pseudomonas ogarae F113 to the Rhizosphere Environment—The AmrZ-FleQ Hub. Microorganisms 2023, 11, 1037. https://doi.org/10.3390/microorganisms11041037
Blanco-Romero E, Durán D, Garrido-Sanz D, Redondo-Nieto M, Martín M, Rivilla R. Adaption of Pseudomonas ogarae F113 to the Rhizosphere Environment—The AmrZ-FleQ Hub. Microorganisms. 2023; 11(4):1037. https://doi.org/10.3390/microorganisms11041037
Chicago/Turabian StyleBlanco-Romero, Esther, David Durán, Daniel Garrido-Sanz, Miguel Redondo-Nieto, Marta Martín, and Rafael Rivilla. 2023. "Adaption of Pseudomonas ogarae F113 to the Rhizosphere Environment—The AmrZ-FleQ Hub" Microorganisms 11, no. 4: 1037. https://doi.org/10.3390/microorganisms11041037





