In Vitro Probiotic Properties and In Vivo Anti-Ageing Effects of Lactoplantibacillus plantarum PFA2018AU Strain Isolated from Carrots on Caenorhabditis elegans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of L. plantarum PFA2018AU and Growth Conditions
2.2. Resistance to Lysozyme, Acid pH, and Bile Salts
2.3. Evaluation of Hydrophobicity, Auto-Aggregation and Co-Aggregation Ability
2.4. Antibiotic Resistance
2.5. Antimicrobial Activity
2.6. C. elegans Lifespan and Fertility Assay
2.7. Colonisation Analysis
2.8. Ageing Markers Analysis
2.9. Real Time qPCR
2.10. Statistical Analysis
3. Results
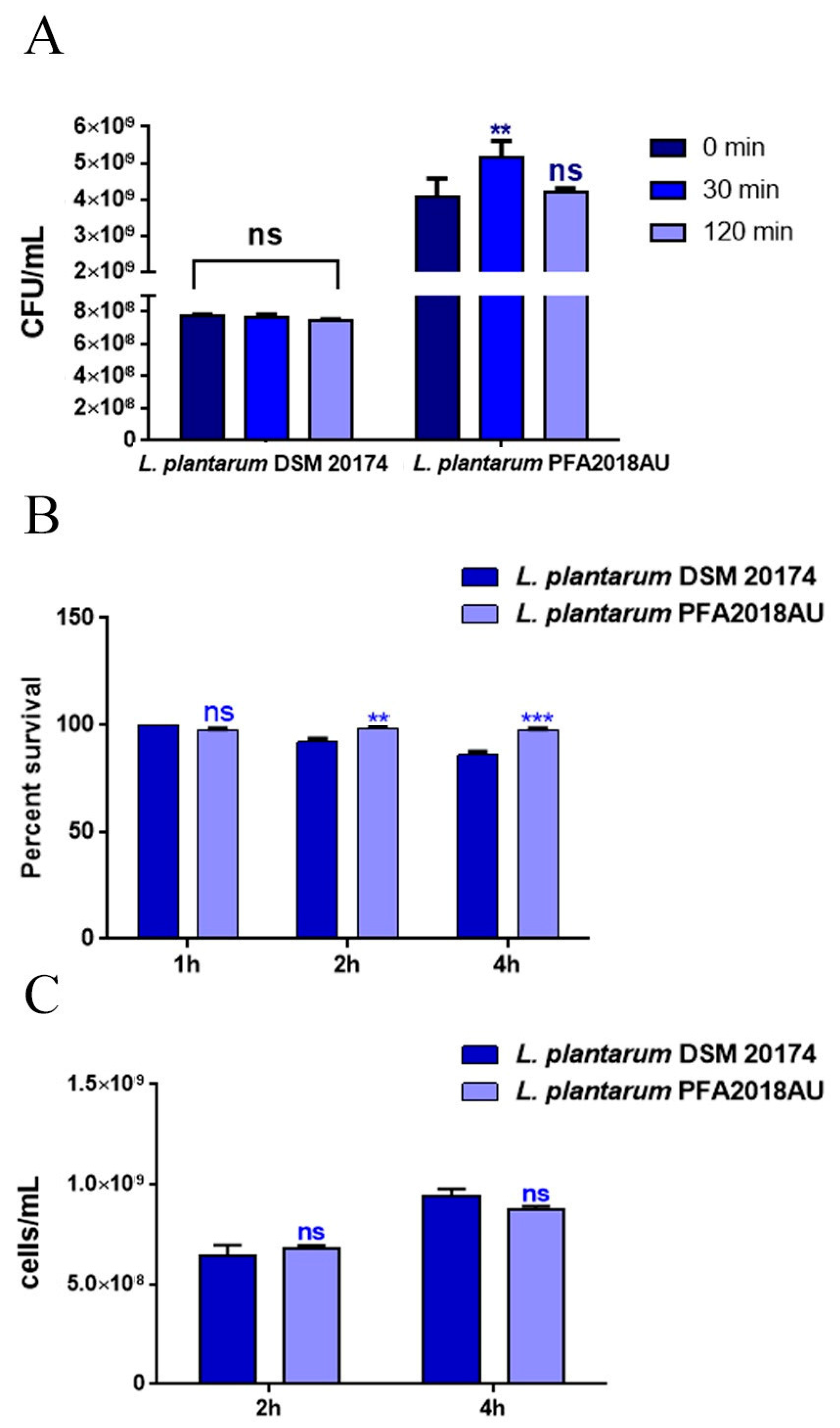
3.1. Resistance to Lysozyme, Low pH, and Bile Salts
3.2. Hydrophobicity, Aggregation, and Co-Aggregation Properties
3.3. Resistance to Antibiotics and Anti-Pathogenicity
3.4. Impact on C. elegans Lifespan and Colonisation Capability
3.5. Evaluation of Ageing Processes and Innate Immunity Stimulation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ranjha, M.M.A.N.; Shafique, B.; Batool, M.; Kowalczewski, P.Ł.; Shehzad, Q.; Usman, M.; Manzoor, M.F.; Zahra, S.M.; Yaqub, S.; Aadil, R.M. Nutritional and Health Potential of Probiotics: A Review. Appl. Sci. 2021, 11, 11204. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, P.; Zhang, X. Probiotics Regulate Gut Microbiota: An Effective Method to Improve Immunity. Molecules 2021, 26, 6076. [Google Scholar] [CrossRef]
- George Kerry, R.; Patra, J.K.; Gouda, S.; Park, Y.; Shin, H.-S.; Das, G. Benefaction of Probiotics for Human Health: A Review. J. Food Drug Anal. 2018, 26, 927–939. [Google Scholar] [CrossRef] [PubMed]
- FAO/WHO Probiotics in Food. Health and Nutritional Properties and Guidelines for Evaluation 2006. Available online: http://www.fao.org/food/food-safety-quality/a-z-index/probiotics/en/ (accessed on 16 September 2021).
- Han, S.; Lu, Y.; Xie, J.; Fei, Y.; Zheng, G.; Wang, Z.; Liu, J.; Lv, L.; Ling, Z.; Berglund, B.; et al. Probiotic Gastrointestinal Transit and Colonization After Oral Administration: A Long Journey. Front. Cell Infect. Microbiol. 2021, 11, 609722. [Google Scholar] [CrossRef]
- Tegegne, B.A.; Kebede, B. Probiotics, Their Prophylactic and Therapeutic Applications in Human Health Development: A Review of the Literature. Heliyon 2022, 8, e09725. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.E.; Akkermans, L.M.A.; Haller, D.; Hammerman, C.; Heimbach, J.T.; Hörmannsperger, G.; Huys, G. Safety Assessment of Probiotics for Human Use. Gut Microbes 2010, 1, 164–185. [Google Scholar] [CrossRef]
- Kang, H.-J.; Im, S.-H. Probiotics as an Immune Modulator. J. Nutr. Sci. Vitaminol. 2015, 61, S103–S105. [Google Scholar] [CrossRef] [PubMed]
- Küçükgöz, K.; Trząskowska, M. Nondairy Probiotic Products: Functional Foods That Require More Attention. Nutrients 2022, 14, 753. [Google Scholar] [CrossRef]
- Morelli, L.; Calleagri, M.; Vogensen, F. Genetics of Lactic Acid Bacteria. In Lactic Acid Bacteria; Von Wright, A., Ed.; CRC Press: Boca Raton, FL, USA, 2011; pp. 17–37. ISBN 978-1-4398-3677-4. [Google Scholar]
- Soltan Dallal, M.M.; Zamaniahari, S.; Davoodabadi, A.; Hosseini, M.; Rajabi, Z. Identification and Characterization of Probiotic Lactic Acid Bacteria Isolated from Traditional Persian Pickled Vegetables. GMS Hyg. Infect. Control 2017, 12, Doc15. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, B.; Bangar, S.P.; Echegaray, N.; Suri, S.; Tomasevic, I.; Manuel Lorenzo, J.; Melekoglu, E.; Rocha, J.M.; Ozogul, F. The Impacts of Lactiplantibacillus Plantarum on the Functional Properties of Fermented Foods: A Review of Current Knowledge. Microorganisms 2022, 10, 826. [Google Scholar] [CrossRef]
- Zhang, F.; Li, Y.; Wang, X.; Wang, S.; Bi, D. The Impact of Lactobacillus plantarum on the Gut Microbiota of Mice with DSS-Induced Colitis. BioMed. Res. Int. 2019, 2019, 1–10. [Google Scholar] [CrossRef]
- Arena, M.P.; Silvain, A.; Normanno, G.; Grieco, F.; Drider, D.; Spano, G.; Fiocco, D. Use of Lactobacillus plantarum Strains as a Bio-Control Strategy against Food-Borne Pathogenic Microorganisms. Front. Microbiol. 2016, 7, 464. [Google Scholar] [CrossRef]
- Jiang, H.; Wang, D. The Microbial Zoo in the C. elegans Intestine: Bacteria, Fungi and Viruses. Viruses 2018, 10, 85. [Google Scholar] [CrossRef]
- Poupet, C.; Chassard, C.; Nivoliez, A.; Bornes, S. Caenorhabditis elegans, a Host to Investigate the Probiotic Properties of Beneficial Microorganisms. Front. Nutr. 2020, 7, 135. [Google Scholar] [CrossRef]
- Roselli, M.; Schifano, E.; Guantario, B.; Zinno, P.; Uccelletti, D.; Devirgiliis, C. Caenorhabditis elegans and Probiotics Interactions from a Prolongevity Perspective. Int. J. Mol. Sci. 2019, 20, 5020. [Google Scholar] [CrossRef]
- Schifano, E.; Conta, G.; Preziosi, A.; Ferrante, C.; Batignani, G.; Mancini, P.; Tomassini, A.; Sciubba, F.; Scopigno, T.; Uccelletti, D.; et al. 2-Hydroxyisobutyric Acid (2-HIBA) Modulates Ageing and Fat Deposition in Caenorhabditis elegans. Front. Mol. Biosci. 2022, 9, 986022. [Google Scholar] [CrossRef] [PubMed]
- Guantario, B.; Zinno, P.; Schifano, E.; Roselli, M.; Perozzi, G.; Palleschi, C.; Uccelletti, D.; Devirgiliis, C. In Vitro and in Vivo Selection of Potentially Probiotic Lactobacilli from Nocellara Del Belice Table Olives. Front. Microbiol. 2018, 9, 595. [Google Scholar] [CrossRef] [PubMed]
- Veisseire, P.; Bonnet, M.; Saraoui, T.; Poupet, C.; Camarès, O.; Gachinat, M.; Callon, C.; Febvre, G.; Chassard, C.; Bornes, S. Investigation into In Vitro and In Vivo Caenorhabditis elegans Models to Select Cheese Yeasts as Probiotic Candidates for Their Preventive Effects against Salmonella Typhimurium. Microorganisms 2020, 8, 922. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Gao, L.; Huang, W.; Ma, Y.; Muhammad, I.; Hanif, A.; Ding, Z.; Guo, X. Antioxidant Properties of Lactic Acid Bacteria Isolated from Traditional Fermented Yak Milk and Their Probiotic Effects on the Oxidative Senescence of Caenorhabditis elegans. Food Funct. 2022, 13, 3690–3703. [Google Scholar] [CrossRef]
- Schifano, E.; Tomassini, A.; Preziosi, A.; Montes, J.; Aureli, W.; Mancini, P.; Miccheli, A.; Uccelletti, D. Leuconostoc Mesenteroides Strains Isolated from Carrots Show Probiotic Features. Microorganisms 2021, 9, 2290. [Google Scholar] [CrossRef]
- Vadrucci, M.; De Bellis, G.; Mazzuca, C.; Mercuri, F.; Borgognoni, F.; Schifano, E.; Uccelletti, D.; Cicero, C. Effects of the Ionizing Radiation Disinfection Treatment on Historical Leather. Front. Mater. 2020, 7, 21. [Google Scholar] [CrossRef]
- Schifano, E.; Zinno, P.; Guantario, B.; Roselli, M.; Marcoccia, S.; Devirgiliis, C.; Uccelletti, D. The Foodborne Strain Lactobacillus fermentum MBC2 Triggers Pept-1-Dependent Pro-Longevity Effects in Caenorhabditis elegans. Microorganisms 2019, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Zanni, E.; Schifano, E.; Motta, S.; Sciubba, F.; Palleschi, C.; Mauri, P.; Perozzi, G.; Uccelletti, D.; Devirgiliis, C.; Miccheli, A. Combination of Metabolomic and Proteomic Analysis Revealed Different Features among Lactobacillus delbrueckii Subspecies bulgaricus and lactis Strains While In Vivo Testing in the Model Organism Caenorhabditis elegans Highlighted Probiotic Properties. Front. Microbiol. 2017, 8, 1206. [Google Scholar] [CrossRef] [PubMed]
- Schifano, E.; Marazzato, M.; Ammendolia, M.G.; Zanni, E.; Ricci, M.; Comanducci, A.; Goldoni, P.; Conte, M.P.; Uccelletti, D.; Longhi, C. Virulence Behavior of Uropathogenic Escherichia coli Strains in the Host Model Caenorhabditis elegans. MicrobiologyOpen 2019, 8, e00756. [Google Scholar] [CrossRef] [PubMed]
- Yun, B.; Ryu, S.; Kang, M.; Lee, J.; Yoo, J.; Kim, Y.; Oh, S. Probiotic Lacticaseibacillus rhamnosus GG Increased Longevity and Resistance Against Foodborne Pathogens in Caenorhabditis elegans by Regulating MicroRNA MiR-34. Front. Cell Infect. Microbiol. 2022, 11, 819328. [Google Scholar] [CrossRef]
- Park, M.R.; Ryu, S.; Maburutse, B.E.; Oh, N.S.; Kim, S.H.; Oh, S.; Jeong, S.-Y.; Jeong, D.-Y.; Oh, S.; Kim, Y. Probiotic Lactobacillus fermentum Strain JDFM216 Stimulates the Longevity and Immune Response of Caenorhabditis elegans through a Nuclear Hormone Receptor. Sci. Rep. 2018, 8, 7441. [Google Scholar] [CrossRef]
- Trindade, D.P.d.A.; Barbosa, J.P.; Martins, E.M.F.; Tette, P.A.S. Isolation and Identification of Lactic Acid Bacteria in Fruit Processing Residues from the Brazilian Cerrado and Its Probiotic Potential. Food Biosci. 2022, 48, 101739. [Google Scholar] [CrossRef]
- Xu, X.; Luo, D.; Bao, Y.; Liao, X.; Wu, J. Characterization of Diversity and Probiotic Efficiency of the Autochthonous Lactic Acid Bacteria in the Fermentation of Selected Raw Fruit and Vegetable Juices. Front. Microbiol. 2018, 9, 2539. [Google Scholar] [CrossRef]
- Sornplang, P.; Piyadeatsoontorn, S. Probiotic Isolates from Unconventional Sources: A Review. J. Anim. Sci. Technol. 2016, 58, 26. [Google Scholar] [CrossRef]
- Tomassini, A.; Sciubba, F.; Di Cocco, M.E.; Capuani, G.; Delfini, M.; Aureli, W.; Miccheli, A. 1H NMR-Based Metabolomics Reveals a Pedoclimatic Metabolic Imprinting in Ready-to-Drink Carrot Juices. J. Agric. Food Chem. 2016, 64, 5284–5291. [Google Scholar] [CrossRef]
- Di Cagno, R.; Surico, R.F.; Siragusa, S.; De Angelis, M.; Paradiso, A.; Minervini, F.; De Gara, L.; Gobbetti, M. Selection and Use of Autochthonous Mixed Starter for Lactic Acid Fermentation of Carrots, French Beans or Marrows. Int. J. Food Microbiol. 2008, 127, 220–228. [Google Scholar] [CrossRef]
- Leuzzi, A.; Grossi, M.; Di Martino, M.L.; Pasqua, M.; Micheli, G.; Colonna, B.; Prosseda, G. Role of the SRRz/Rz1 lambdoid lysis cassette in the pathoadaptive evolution of Shigella. Int. J. Med. Microbiol. 2017, 307, 268–275. [Google Scholar] [CrossRef]
- Chaffanel, F.; Charron-Bourgoin, F.; Soligot, C.; Kebouchi, M.; Bertin, S.; Payot, S.; Le Roux, Y.; Leblond-Bourget, N. Surface Proteins Involved in the Adhesion of Streptococcus salivarius to Human Intestinal Epithelial Cells. Appl. Microbiol. Biotechnol. 2018, 102, 2851–2865. [Google Scholar] [CrossRef] [PubMed]
- Falah, F.; Vasiee, A.; Behbahani, B.A.; Yazdi, F.T.; Moradi, S.; Mortazavi, S.A.; Roshanak, S. Evaluation of Adherence and Anti-Infective Properties of Probiotic Lactobacillus fermentum Strain 4–17 against Escherichia coli Causing Urinary Tract Infection in Humans. Microb. Pathog. 2019, 131, 246–253. [Google Scholar] [CrossRef] [PubMed]
- de Souza, B.M.S.; Borgonovi, T.F.; Casarotti, S.N.; Todorov, S.D.; Penna, A.L.B. Lactobacillus casei and Lactobacillus fermentum Strains Isolated from Mozzarella Cheese: Probiotic Potential, Safety, Acidifying Kinetic Parameters and Viability under Gastrointestinal Tract Conditions. Probiotics Antimicrob. Proteins 2019, 11, 382–396. [Google Scholar] [CrossRef]
- Lim, S.-M. Factors Affecting Adhesion of Lactic Acid Bacteria to Caco-2 Cells and Inhibitory Effect on Infection of Salmonella Typhimurium. J. Microbiol. Biotechnol. 2012, 22, 1731–1739. [Google Scholar] [CrossRef] [PubMed]
- Hutt, P.; Shchepetova, J.; Loivukene, K.; Kullisaar, T.; Mikelsaar, M. Antagonistic Activity of Probiotic Lactobacilli and Bifidobacteria against Entero- and Uropathogens. J. Appl. Microbiol. 2006, 100, 1324–1332. [Google Scholar] [CrossRef]
- Ramos, A.N.; Sesto Cabral, M.E.; Noseda, D.; Bosch, A.; Yantorno, O.M.; Valdez, J.C. Antipathogenic Properties of Lactobacillus plantarum on Pseudomonas aeruginosa: The Potential Use of Its Supernatants in the Treatment of Infected Chronic Wounds: Antipathogenic Properties of L. plantarum on P. aeruginosa. Wound Repair Regen. 2012, 20, 552–562. [Google Scholar] [CrossRef]
- Seddik, H.A.; Bendali, F.; Gancel, F.; Fliss, I.; Spano, G.; Drider, D. Lactobacillus plantarum and Its Probiotic and Food Potentialities. Probiotics Antimicro. Prot. 2017, 9, 111–122. [Google Scholar] [CrossRef]
- Kumar, A.; Joishy, T.; Das, S.; Kalita, M.C.; Mukherjee, A.K.; Khan, M.R. A Potential Probiotic Lactobacillus plantarum JBC5 Improves Longevity and Healthy Ageing by Modulating Antioxidative, Innate Immunity and Serotonin-Signaling Pathways in Caenorhabditis elegans. Antioxidants 2022, 11, 268. [Google Scholar] [CrossRef]
- Sharma, K.; Pooranachithra, M.; Balamurugan, K.; Goel, G. Multivariate Analysis of Increase in Life Span of Caenorhabditis elegans Through Intestinal Colonization by Indigenous Probiotic Strains. Probiotics Antimicrob. Proteins 2019, 11, 865–873. [Google Scholar] [CrossRef]
- Oh, A.; Daliri, E.B.-M.; Oh, D.H. Screening for Potential Probiotic Bacteria from Korean Fermented Soybean Paste: In Vitro and Caenorhabditis elegans Model Testing. LWT 2018, 88, 132–138. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Bou-Teen, D.; Kaludercic, N.; Weissman, D.; Turan, B.; Maack, C.; Di Lisa, F.; Ruiz-Meana, M. Mitochondrial ROS and Mitochondria-Targeted Antioxidants in the Aged Heart. Free Radic. Biol. Med. 2021, 167, 109–124. [Google Scholar] [CrossRef]
- Zuo, L.; Sypert, D.C.; Clark, A.D.; Xu, Z.; Garrison, D.E.; He, F. Redox Mechanism of Reactive Oxygen Species in Gastrointestinal Tract Diseases. In Gastrointestinal Tissue; Elsevier: Amsterdam, The Netherlands, 2017; pp. 21–27. ISBN 978-0-12-805377-5. [Google Scholar]
- Brenneisen, P.; Steinbrenner, H.; Sies, H. Selenium, Oxidative Stress, and Health Aspects. Mol. Asp. Med. 2005, 26, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Marques, T.M.; Cryan, J.F.; Shanahan, F.; Fitzgerald, G.F.; Ross, R.P.; Dinan, T.G.; Stanton, C. Gut Microbiota Modulation and Implications for Host Health: Dietary Strategies to Influence the Gut–Brain Axis. Innov. Food Sci. Emerg. Technol. 2014, 22, 239–247. [Google Scholar] [CrossRef]
- Aquilano, K.; Baldelli, S.; Ciriolo, M.R. Glutathione: New Roles in Redox Signaling for an Old Antioxidant. Front. Pharmacol. 2014, 5, 196. [Google Scholar] [CrossRef] [PubMed]
- Venz, R.; Pekec, T.; Katic, I.; Ciosk, R.; Ewald, C.Y. End-of-Life Targeted Degradation of DAF-2 Insulin/IGF-1 Receptor Promotes Longevity Free from Growth-Related Pathologies. eLife 2021, 10, e71335. [Google Scholar] [CrossRef]
- Qu, Y.; Shi, L.; Liu, Y.; Huang, L.; Luo, H.-R.; Wu, G.-S. Orientin Prolongs the Longevity of Caenorhabditis elegans and Postpones the Development of Neurodegenerative Diseases via Nutrition Sensing and Cellular Protective Pathways. Oxidative Med. Cell Longev. 2022, 2022, 8878923. [Google Scholar] [CrossRef]
- Deng, Y.; Liu, H.; Huang, Q.; Tu, L.; Hu, L.; Zheng, B.; Sun, H.; Lu, D.; Guo, C.; Zhou, L. Mechanism of Longevity Extension of Caenorhabditis elegans Induced by Schizophyllum commune Fermented Supernatant with Added Radix Puerariae. Front. Nutr. 2022, 9, 847064. [Google Scholar] [CrossRef]
- Dinić, M.; Jakovljević, S.; Đokić, J.; Popović, N.; Radojević, D.; Strahinić, I.; Golić, N. Probiotic-Mediated P38 MAPK Immune Signaling Prolongs the Survival of Caenorhabditis elegans Exposed to Pathogenic Bacteria. Sci. Rep. 2021, 11, 21258. [Google Scholar] [CrossRef] [PubMed]
- Yavorov-Dayliev, D.; Milagro, F.I.; Ayo, J.; Oneca, M.; Aranaz, P. Pediococcus acidilactici CECT9879 (PA1c) Counteracts the Effect of a High-Glucose Exposure in C. elegans by Affecting the Insulin Signaling Pathway (IIS). Int. J. Mol. Sci. 2022, 23, 2689. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, J.; Gong, J.; Yu, H.; Pacan, J.C.; Niu, Z.; Si, W.; Sabour, P.M. Use of Caenorhabditis elegans for Preselecting Lactobacillus Isolates to Control Salmonella Typhimurium. J. Food Prot. 2011, 74, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Xiao, K.; Zhang, L.; Liu, M.; Li, L.; Zhu, H.; Wang, W.; Yi, C.; Yu, F.; Li, Q.; et al. The Use of Caenorhabditis elegans Model to Screen Lactobacilli for the Control of Patulin. Food Control 2022, 137, 108963. [Google Scholar] [CrossRef]




| Antibiotic | L. plantarum DSM 20174 | L. plantarum PFA2018AU | p-Value |
|---|---|---|---|
| Vancomycin | 0 | 0 | ns |
| Clindamycin | 0.9 cm ± 0.35 | 0.5 cm ± 0.20 | p < 0.001 |
| Cefalotin | 0.1 cm ± 0.12 | 0.5 cm ± 0.53 | p < 0.001 |
| Cefuroxime | 1 cm ± 0.23 | 0 | p < 0.001 |
| Tobramycin | 0 | 0.6 cm ± 0.05 | p < 0.001 |
| Ampicillin | 0.5 cm ± 0.20 | 0.8 cm ± 0.08 | p < 0.001 |
| Cefotaxime | 0.6 cm ± 0.81 | 0 | p < 0.001 |
| Chloramphenicol | 1 cm ± 0.50 | 0.9 cm ± 0.83 | p < 0.05 |
| Tetracycline | 0.6 cm ± 0.24 | 0.8 cm ± 0.23 | p < 0.01 |
| Erythromycin | 0.7 cm ± 0.20 | 0.6 cm ± 0.15 | p < 0.05 |
| Amikacin | 0 | 0 | ns |
| Oxacillin | 1 cm ± 1.10 | 1 cm ± 1.2 | ns |
| Fosfomycin | 0 | 0 | ns |
| Rifampicin | 1 cm ± 0.08 | 0.9 cm ± 0.10 | p < 0.05 |
| Gentamicin | 0 | 0 | ns |
| Penicillin | 1 cm ± 0.72 | 0.6 cm ± 0.31 | p < 0.001 |
| Aztreonam | 1 cm ± 0.60 | 0.1 cm ± 0.50 | p < 0.001 |
| Carbenicillin | 0.3 cm ± 0.58 | 1 cm ± 0.92 | p < 0.001 |
| Mezlocillin | 1 cm ± 0.3 | 0.9 cm ± 0.45 | p < 0.05 |
| Streptomycin | 0.5 cm ± 0.9 | 0 | p < 0.001 |
| S. enterica | L. monocytogenes | S. aureus | P. aeruginosa | p-Value | |
|---|---|---|---|---|---|
| L. plantarum DSM 20174 | 2.2 ± 0.36 | 2.3 ± 0.32 | 2.2 ± 0.34 | 2.3 ± 0.5 | ns |
| L. plantarum PFA2018AU | 2.4 ± 0.67 | 2.5 ± 0.46 | 2.5 ± 0.45 | 2.7 ± 0.6 | ns |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pompa, L.; Montanari, A.; Tomassini, A.; Bianchi, M.M.; Aureli, W.; Miccheli, A.; Uccelletti, D.; Schifano, E. In Vitro Probiotic Properties and In Vivo Anti-Ageing Effects of Lactoplantibacillus plantarum PFA2018AU Strain Isolated from Carrots on Caenorhabditis elegans. Microorganisms 2023, 11, 1087. https://doi.org/10.3390/microorganisms11041087
Pompa L, Montanari A, Tomassini A, Bianchi MM, Aureli W, Miccheli A, Uccelletti D, Schifano E. In Vitro Probiotic Properties and In Vivo Anti-Ageing Effects of Lactoplantibacillus plantarum PFA2018AU Strain Isolated from Carrots on Caenorhabditis elegans. Microorganisms. 2023; 11(4):1087. https://doi.org/10.3390/microorganisms11041087
Chicago/Turabian StylePompa, Laura, Arianna Montanari, Alberta Tomassini, Michele Maria Bianchi, Walter Aureli, Alfredo Miccheli, Daniela Uccelletti, and Emily Schifano. 2023. "In Vitro Probiotic Properties and In Vivo Anti-Ageing Effects of Lactoplantibacillus plantarum PFA2018AU Strain Isolated from Carrots on Caenorhabditis elegans" Microorganisms 11, no. 4: 1087. https://doi.org/10.3390/microorganisms11041087
APA StylePompa, L., Montanari, A., Tomassini, A., Bianchi, M. M., Aureli, W., Miccheli, A., Uccelletti, D., & Schifano, E. (2023). In Vitro Probiotic Properties and In Vivo Anti-Ageing Effects of Lactoplantibacillus plantarum PFA2018AU Strain Isolated from Carrots on Caenorhabditis elegans. Microorganisms, 11(4), 1087. https://doi.org/10.3390/microorganisms11041087






