Neotropical Frog Foam Nest’s Microbiomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Biochemical Characterization of the Foam Nests
2.2.1. Protein and Carbohydrate Determination
2.2.2. Surface Tension
2.2.3. SDS-PAGE
2.3. DNA Extraction and Sequencing
2.4. Data Processing
2.5. Statistical Analysis
3. Results
3.1. Biochemical Characterization of the Foam Nests
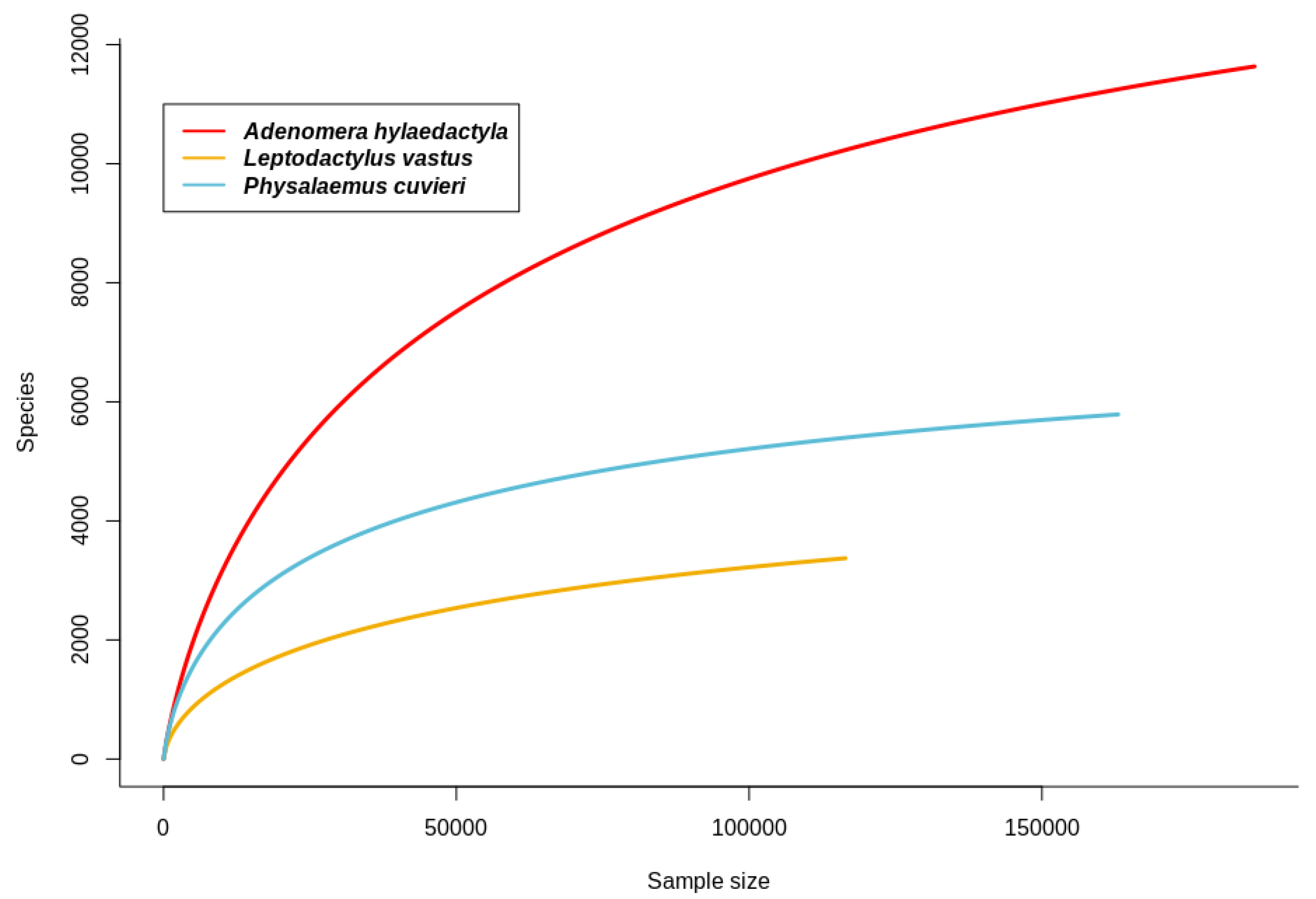
3.2. Estimation of Bacterial Richness and Diversity in Foam Nests
3.3. Microbial Community Structure and Composition
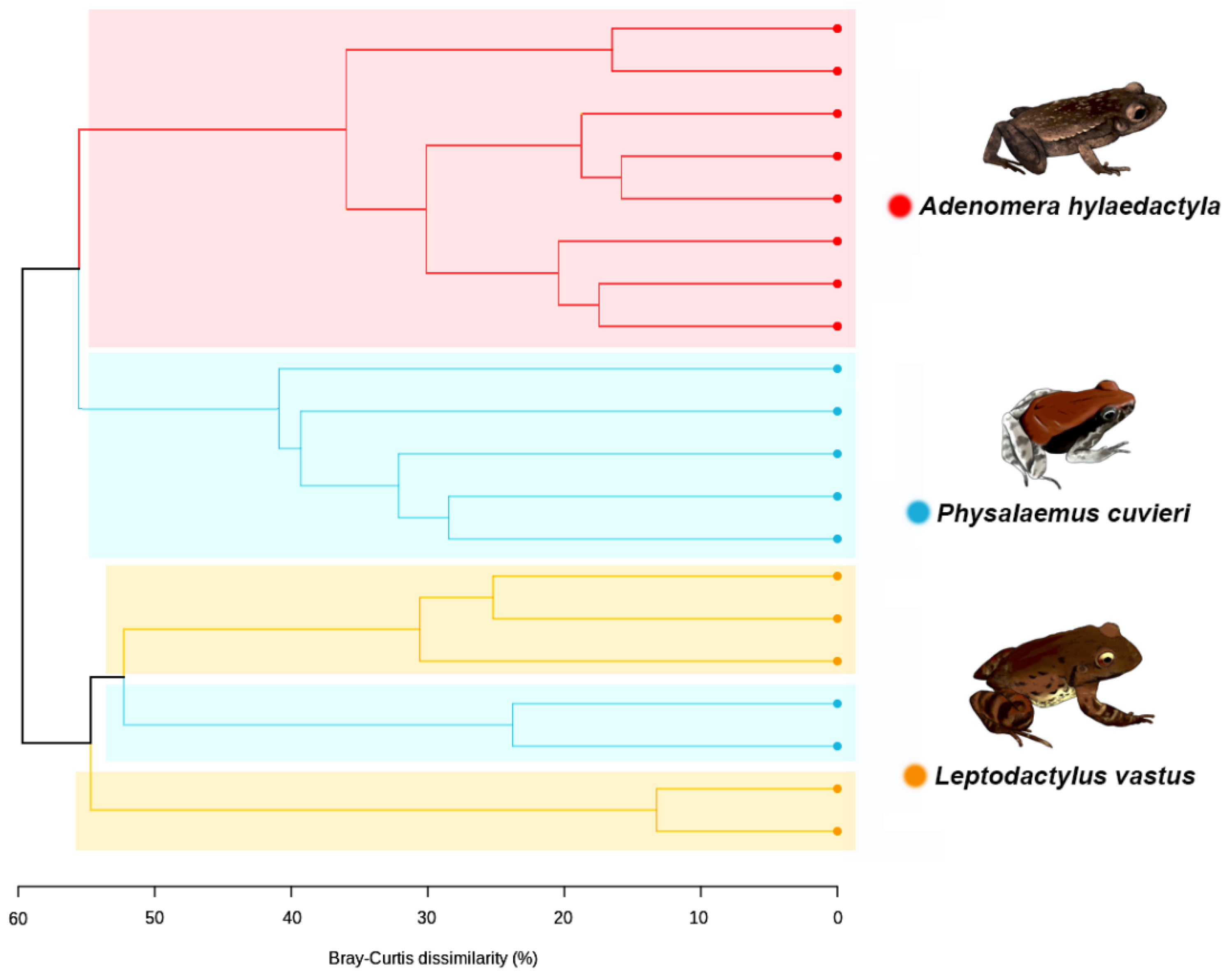
3.4. Beta Diversity of Foam Nest Bacterial Community
4. Discussion
4.1. Biochemical Characterization of the Foam Nests
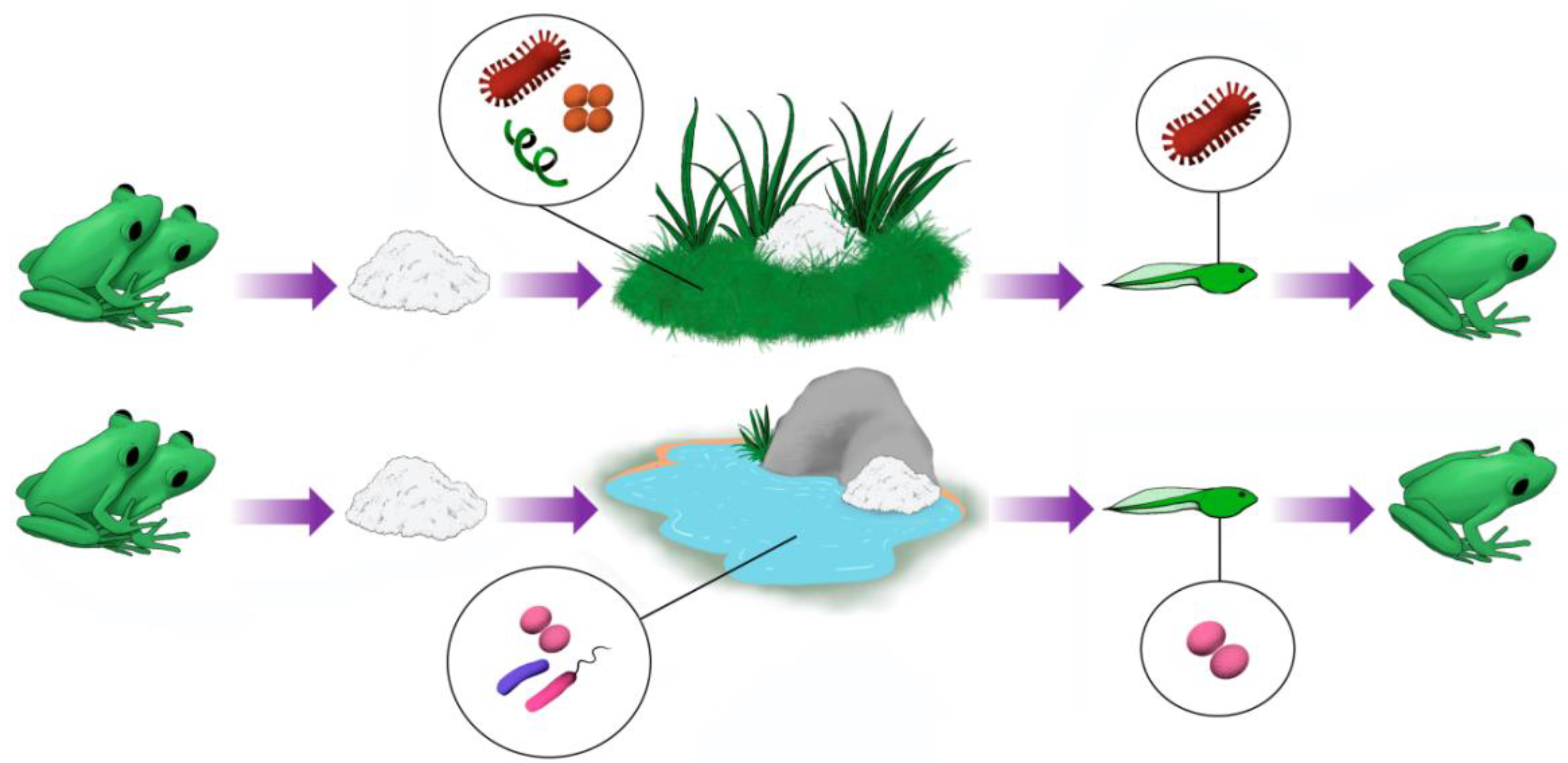
4.2. A distinctive Frog foam Nest Community
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wells, K.D. Chapter 10: The natural history of amphibian reproduction. In The Ecology and Behavior of Amphibians; Wells, K.D., Ed.; University of Chicago Press: Chicago, IL, USA, 2007; pp. 451–515. [Google Scholar]
- Vitt, L.J.; Caldwell, J.P. Herpetology: An Introductory Biology of Ampibians and Reptiles, 4th ed.; Academic Press: San Diego, CA, USA, 2014. [Google Scholar]
- Hissa, D.C.; Vasconcelos, I.M.; Carvalho, A.F.U.; Nogueira, V.L.R.; Cascon, P.; Antunes, A.S.L.; de Macedo, G.R.; Melo, V.M.M. Novel surfactant proteins are involved in the structure and stability of foam nests from the frog Leptodactylus vastus. J. Exp. Biol. 2008, 211, 2707–2711. [Google Scholar] [CrossRef] [PubMed]
- Heyer, R.W. The adaptative ecology of the species groups the genus Leptodactylus (Amphibians, Leptodactylidae). Evolution 1969, 23, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.; Kennedy, M.W.; Fleming, R.I.; Wilson, E.H.; Videler, H.; Wokosin, D.L.; Su, T.-J.; Green, R.J.; Lu, J.R. Adsorption of frog foam nest proteins at the air–water interface. Biophys. J. 2004, 88, 2114–2125. [Google Scholar] [CrossRef] [PubMed]
- Fleming, R.I.; Mackenzie, C.D.; Cooper, A.; Kennedy, M.W. Foam nest components of the tungara frog: A cocktail of proteins conferring physical and biological resilience. Proc. Royal Soc. B Biol. Sci. 2009, 276, 1787–1795. [Google Scholar] [CrossRef] [PubMed]
- Haddad, C.F.B.; Prado, C.P.A. Reproductive modes in frogs and their unexpected diversity in the Atlantic Forest of Brazil. BioScience 2005, 55, 207–217. [Google Scholar] [CrossRef]
- Pombal, J.P., Jr.; Haddad, C.F.B. Estratégias e modos reprodutivos em anuros. In Herpetologia no Brasil II; Nascimento, L.B., Oliveira, M.E., Eds.; Sociedade Brasileira de Herpetologia: Belo Horizonte, Brazil, 2007; pp. 101–116. [Google Scholar]
- Méndez-Narváez, J.; Flechas, S.V.; Amézquita, A. Foam nests provide context-dependent thermal insulation to embryos of three leptodactylid frogs. Physiol. Biochem. Zool. 2015, 88, 246–253. [Google Scholar] [CrossRef]
- Arzabe, C. Reproductive activity patterns of anurans in two different altitudinal sites within the Brazilian Caatinga. Rev. Bras. Zool. 1999, 16, 851–864. [Google Scholar] [CrossRef]
- Pereira, E.B.; Pinto-Ledezma, J.N.; de Freitas, C.G.; Villalobos, F.; Collevatti, R.G.; Maciel, N.M. Evolution of the anuran foam nest: Trait conservatism and lineage diversification. Biol. J. Linn. Soc. 2017, 122, 814–823. [Google Scholar] [CrossRef]
- McGrath-Blaser, S.; Steffen, M.; Grafe, T.U.; Torres-Sánchez, M.; McLeod, D.S.; Muletz-Wolz, C.R. Early life skin microbial trajectory as a function of vertical and environmental transmission in Bornean foam-nesting frogs. Anim. Microbiome 2021, 3, 83. [Google Scholar] [CrossRef]
- Hopkins, W.A. Amphibians as models for studying environmental change. ILAR J. 2007, 48, 270–277. [Google Scholar] [CrossRef]
- Relyea, R.A.; Schoeppner, N.M.; Hoverman, J.T. Pesticides and amphibians: The importance of community context. Ecol. Appl. 2005, 15, 1125–1134. [Google Scholar] [CrossRef]
- Scheele, B.C.; Rebouças, R.; Toledo, L.F. Amphibian fungal panzootic causes catastrophic and ongoing loss of biodiversity. Science 2019, 363, 1459–1463. [Google Scholar] [CrossRef] [PubMed]
- Knutie, S.A.; Wilkinson, C.L.; Kohl, K.D.; Rohr, J.R. Early-life disruption of amphibian microbiota decreases later-life resistance to parasites. Nat. Commun. 2017, 8, 86. [Google Scholar] [CrossRef] [PubMed]
- Lam, B.A. Proportion of individuals with anti–Batrachochytrium dendrobatidis skin bacteria is associated with population persistence in the frog Rana Muscosa. Biol. Conserv. 2010, 143, 529–531. [Google Scholar] [CrossRef]
- Leary, S.; Underwood, W.; Anthony, R.; Cartner, S.; Corey, D.; Grandin, T.; Greenacre, C.; Gwaltney-Brant, S.; McCrackin, M.A.; Meyer, R.; et al. AVMA Guidelines for the Euthanasia of Animals; American Veterinary Medical Association: Schaumburg, IL, USA, 2013. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Albalasmeh, A.A.; Berhe, A.A.; Ghezzehei, T.A. A new method for rapid determination of carbohydrate and total carbon concentrations using UV spectrophotometry. Carbohydr. Polym. 2013, 97, 253–261. [Google Scholar] [CrossRef]
- Du Noüy, P.L. An interfacial tensiometer for universal use. J. Gen. Physiol. 1925, 7, 625. [Google Scholar] [CrossRef] [PubMed]
- Swift, M.L. GraphPad prism, data analysis, and scientific graphing. J. Chem. Inf. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]
- Schägger, H.; Von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]
- Neuhoff, V.; Arold, N.; Taube, D.; Ehrhardt, W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 1988, 9, 255–262. [Google Scholar] [CrossRef]
- Warner, S.A.J. Genomic DNA isolation and lambda library construction. In Plant Gene Isolation. Principles and Practice; Foster, G.D., Twell, D., Eds.; John Wiley and Sons: Chichester, NH, USA, 1996; pp. 51–74. [Google Scholar]
- Caporaso, J.G.; Lauber, C.L.; Costello, E.K.; Berg-Lyons, D.; Gonzalez, A.; Stombaugh, J.; Knights, D.; Gajer, P.; Ravel, J.; Fierer, N.; et al. Moving pictures of the human microbiome. Genome Biol. 2011, 12, R50. [Google Scholar] [CrossRef]
- Andrews, S.; Krueger, F.; Segonds-Pichon, A.; Biggins, L.; Krueger, C.; Wingett, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics: Cambridge, UK, 2010; Available online: www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 16 February 2023).
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mah, E.F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical: Vienna, Austria, 2020; Available online: https://www.r-project.org/> (accessed on 16 February 2023).
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [PubMed]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acid. Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-7. 2013. Available online: https://CRAN.R-project.org/package=vegan (accessed on 9 May 2022).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Hissa, D.C.; Bezerra, G.A.; Birner-Gruenberger, R.; Silva, L.P.; Usón, I.; Gruber, K.; Melo, V.M.M. Unique Crystal Structure of a Novel Surfactant Protein from the Foam Nest of the Frog Leptodactylus vastus. ChemBioChem 2014, 15, 393–398. [Google Scholar] [CrossRef]
- Hissa, D.C.; Bezerra, W.M.; Freitas CD, T.D.; Ramos, M.V.; Lopes JL, D.S.; Beltramini, L.M.; Cascon, I.J.R.P.; Melo, V.M.M. Frog foam Nest protein diversity and synthesis. J. Exp. Zool. A Ecol. Genet. Physiol. 2016, 325, 425–433. [Google Scholar] [CrossRef]
- Cooper, A.; Kennedy, M.W. Biofoams and natural protein surfactants. Biophys. Chem. 2010, 151, 96–104. [Google Scholar] [CrossRef]
- Shigeri, Y.; Nakata, M.; Kubota, H.Y.; Tomari, N.; Yamamoto, Y.; Uegaki, K.; Haramoto, Y.; Bumb, C.; Tanaka, Y.; Kinumi, T.; et al. Identification of novel proteins in foam nests of the Japanese Forest Green Tree Frog, Rhacophorus arboreus. Zool. Sci. 2020, 38, 8–19. [Google Scholar] [CrossRef]
- Mackenzie, C.D.; Smith, B.O.; Meister, A.; Blume, A.; Zhao, X.; Lu, J.R.; Kennedy, M.W.; Cooper, A. Ranaspumin-2: Structure and function of a surfactant protein from the foam nests of a tropical frog. Biophys. J. 2009, 96, 4984–4992. [Google Scholar] [CrossRef]
- Pyron, R.A.; Wiens, J.J. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenet. Evol. 2011, 61, 543–583. [Google Scholar] [CrossRef]
- Frost, D.R. Amphibian Species of the World: An Online Reference; Version 6.0; American Museum of Natural History: New York, NY, USA, 2019; Available online: http://research.amnh.org/herpetology/amphibia/index.html/ (accessed on 10 August 2019).
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Bletz, M.C.; Archer, H.; Harris, R.N.; McKenzie, V.J.; Rabemananjara, F.C.; Rakotoarison, A.; Vences, M. Host ecology rather than host phylogeny drives amphibian skin microbial community structure in the biodiversity hotspot of Madagascar. Front. Microbiol. 2017, 8, 1530. [Google Scholar] [CrossRef]
- Kueneman, J.G.; Bletz, M.C.; McKenzie, V.J.; Becker, C.G.; Joseph, M.B.; Abarca, J.G.; Archer, H.; Arellano, A.L.; Bataille, A.; Becker, M.; et al. Community richness of amphibian skin bacteria correlates with bioclimate at the global scale. Nat. Ecol. Evol. 2019, 3, 381–389. [Google Scholar] [CrossRef]
- Brunetti, A.E.; Lyra, M.L.; Melo, W.G.; Andrade, L.E.; Palacios-Rodríguez, P.; Prado, B.M.; Haddad, C.F.B.; Pupo, M.T.; Lopes, N.P. Symbiotic skin bacteria as a source for sex-specific scents in frogs. Proc. Natl. Acad. Sci. USA 2019, 116, 2124–2129. [Google Scholar] [CrossRef]
- Becker, M.H.; Walke, J.B.; Murrill, L.; Woodhams, D.C.; Reinert, L.K.; Rollins-Smith, L.A.; Burzynski, E.A.; Umile, T.P.; Minbiole, K.P.C.; Belden, L.K. Phylogenetic distribution of symbiotic bacteria from Panamanian amphibians that inhibit growth of the lethal fungal pathogen Batrachochytrium dendrobatidis. Mol. Ecol. 2015, 24, 1628–1641. [Google Scholar] [CrossRef]
- Martin, H.C.; Ibáñez, R.; Nothias, L.F.; Boya, P.C.A.; Reinert, L.K.; Rollins-Smith, L.A.; Dorrestein, P.C.; Gutiérrez, M. Viscosin-like lipopeptides from frog skin bacteria inhibit Aspergillus fumigatus and Batrachochytrium dendrobatidis detected by imaging mass spectrometry and molecular networking. Sci. Rep. 2019, 9, 3091. [Google Scholar] [CrossRef] [PubMed]
- Abarca, J.G.; Vargas, G.; Zuniga, I.; Whitfield, S.M.; Woodhams, D.C.; Kerby, J.; McKenzie, V.J.; Murillo-Cruz, C.; Pinto-Tomas, A.A. Assessment of bacterial communities associated with the skin of costa rican amphibians at la selva biological station. Front. Microbiol. 2018, 9, 2001. [Google Scholar] [CrossRef]
- Kueneman, J.; Woodhams, D.; Van Treuren, W.; Archer, H.M.; Knight, R.; McKenzie, V.J. Inhibitory bacteria reduce fungi on early life stages of endangered Colorado boreal toads (Anaxyrus boreas). ISME J. 2016, 10, 934–944. [Google Scholar] [CrossRef]
- Kohl, K.D.; Yahn, J. Effects of environmental temperature on the gut microbial communities of tadpoles. Environ. Microbiol. 2016, 18, 1561–1565. [Google Scholar] [CrossRef]
- Harris, R.N.; James, T.Y.; Lauer, A.; Simon, M.A.; Patel, A. Amphibian Pathogen Batrachochytrium dendrobatidis Is Inhibited by the Cutaneous Bacteria of Amphibian Species. EcoHealth 2006, 3, 53. [Google Scholar] [CrossRef]
- Kueneman, J.G.; Parfrey, L.W.; Woodhams, D.C.; Archer, H.M.; Knight, R.; McKenzie, V.J. The amphibian skin-associated microbiome across species, space and life history stages. Mol. Ecol. 2014, 23, 1238–1250. [Google Scholar] [CrossRef]
- Loudon, A.H.; Woodhams, D.C.; Parfrey, L.W.; Archer, H.; Knight, R.; McKenzie, V.; Harris, R.N. Microbial community dynamics and effect of environmental microbial reservoirs on red-backed salamanders (Plethodon cinereus). ISME J. 2014, 8, 830–840. [Google Scholar] [CrossRef]
- Walke, J.B.; Becker, M.H.; Hughey, M.C.; Swartwout, M.C.; Jensen, R.V.; Belden, L.K. Most of the dominant members of amphibian skin bacterial communities can be readily cultured. Appl. Environ. Microbiol. 2015, 81, 6589–6600. [Google Scholar] [CrossRef]
- De Assis, A.B.; Barreto, C.C.; Navas, C.A. Skin microbiota in frogs from the Brazilian Atlantic Forest: Species, forest type, and potential against pathogens. PLoS ONE 2017, 12, e0179628. [Google Scholar] [CrossRef]
- Catenazzi, A.; Flechas, S.V.; Burkart, D.; Hooven, N.D.; Townsend, J.; Vredenburg, V.T. Widespread elevational occurrence of antifungal bacteria in Andean amphibians decimated by disease: A complex role for skin symbionts in defense against chytridiomycosis. Front. Microbiol. 2018, 9, 465. [Google Scholar] [CrossRef]
- Fontaine, S.S.; Novarro, A.J.; Kohl, K.D. Environmental temperature alters the digestive performance and gut microbiota of a terrestrial amphibian. J. Exp. Biol. 2018, 221, jeb187559. [Google Scholar] [CrossRef]
- Griffiths, S.M.; Harrison, X.A.; Weldon, C.; Wood, M.D.; Pretorius, A.; Hopkins, K.; Fox, G.; Preziosi, R.F.; Antwis, R.E. Genetic variability and ontogeny predict microbiome structure in a disease-challenged montane amphibian. ISME J. 2018, 12, 2506–2517. [Google Scholar] [CrossRef]
- Passos, L.F.; Garcia, G.; Young, R.J. Comparing the bacterial communities of wild and captive golden mantella frogs: Implications for amphibian conservation. PLoS ONE 2018, 13, e0205652. [Google Scholar] [CrossRef]
- Brunetti, A.E.; Bunk, B.; Lyra, M.L.; Fuzo, C.A.; Marani, M.M.; Spröer, C.; Haddad, C.F.B.; Lopez, N.P.; Overmann, J. Molecular basis of a bacterial-amphibian symbiosis revealed by comparative genomics, modeling, and functional testing. ISME J. 2022, 16, 788–800. [Google Scholar] [CrossRef]
- Grimes, D.J.; Woese, C.R.; MacDonell, M.T.; Colwell, R.R. Systematic study of the genus Vogesella gen. nov. and its type species, Vogesella indigofera comb. nov. Int. J. Syst. Evol. Microbiol. 1997, 47, 19–27. [Google Scholar] [CrossRef]
- Jørgensen, N.O.; Brandt, K.K.; Nybroe, O.; Hansen, M. Vogesella mureinivorans sp. nov., a peptidoglycan-degrading bacterium from lake water. Int. J. Syst. Evol. Microbiol. 2010, 60, 2467–2472. [Google Scholar] [CrossRef] [PubMed]
- Sheu, S.Y.; Chen, J.C.; Young, C.C.; Chen, W.M. Vogesella fluminis sp. nov., isolated from a freshwater river, and emended description of the genus Vogesella. Int. J. Syst. Evol. Microbiol. 2013, 63, 3043–3049. [Google Scholar] [CrossRef] [PubMed]
- Rapala, J.; Berg, K.A.; Lyra, C.; Niemi, R.M.; Manz, W.; Suomalainen, S.; Paulin, L.; Lahti, K. Paucibacter toxinivorans gen. nov., sp. nov., a bacterium that degrades cyclic cyanobacterial hepatotoxins microcystins and nodularin. Int. J. Syst. Evol. Microbiol. 2005, 55, 1563–1568. [Google Scholar] [CrossRef] [PubMed]
- Pheng, S.; Lee, J.J.; Eom, M.K.; Lee, K.H.; Kim, S.G. Paucibacter oligotrophus sp. nov., isolated from fresh water, and emended description of the genus Paucibacter. Int. J. Syst. Evol. Microbiol. 2017, 67, 2231–2235. [Google Scholar] [CrossRef]
- Nam, Y.H.; Choi, A.; Hwang, J.M.; Yim, K.J.; Kim, J.H.; Choi, G.G.; Chung, E.J. Paucibacter aquatile sp. nov. isolated from freshwater of the Nakdong River, Republic of Korea. Arch. Microbiol. 2018, 200, 877–882. [Google Scholar] [CrossRef]
- Thomas, F.; Hehemann, J.H.; Rebuffet, E.; Czjzek, M.; Michel, G. Environmental and gut bacteroidetes: The food connection. Front. Microbiol. 2011, 2, 93. [Google Scholar] [CrossRef]
- Proença, D.N.; Nobre, M.F.; Morais, P.V. Chitinophaga costaii sp. nov., an endophyte of Pinus pinaster, and emended description of Chitinophaga niabensis. Int. J. Syst. Evol. Microbiol. 2014, 64, 1237–1243. [Google Scholar] [CrossRef]
- Kämpfer, P.; Busse, H.J.; Kleinhagauer, T.; McInroy, J.A.; Glaeser, S.P. Sphingobacterium zeae sp. nov., an endophyte of maize. Int. J. Syst. Evol. Microbiol. 2016, 66, 2643–2649. [Google Scholar] [CrossRef]
- Lee, Y.; Jin, H.M.; Jung, H.S.; Jeon, C.O. Sphingobacterium humi sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 4632–4638. [Google Scholar] [CrossRef]
- Shaffer, J.P.; U’Ren, J.M.; Gallery, R.E.; Baltrus, D.A.; Arnold, A.E. An endohyphal bacterium (Chitinophaga, Bacteroidetes) alters carbon source use by Fusarium keratoplasticum (F. solani species complex, Nectriaceae). Front. Microbiol. 2017, 8, 350. [Google Scholar] [CrossRef]
- Xu, L.; Sun, J.Q.; Wang, L.J.; Gao, Z.W.; Sun, L.Z.; Wu, X.L. Sphingobacterium alkalisoli sp. nov., isolated from a saline-alkaline soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 1943–1948. [Google Scholar] [CrossRef] [PubMed]
- Kohl, K.D.; Cary, T.L.; Karasov, W.H.; Dearing, M.D. Restructuring of the amphibian gut microbiota through metamorphosis. Environ. Microbiol. Rep. 2013, 5, 899–903. [Google Scholar] [CrossRef] [PubMed]
- Pereira, E.B.; Collevatti, R.G.; Kokubum, M.N.D.C.; Miranda, N.E.D.O.; Maciel, N.M. Ancestral reconstruction of reproductive traits shows no tendency toward terrestriality in leptodactyline frogs. BMC Evol. Biol. 2015, 15, 91. [Google Scholar] [CrossRef] [PubMed]
- Duellman, W.E.; Trueb, L. Chapter 2: Reproductive Strategies. In Biology of Amphibians; The Johns Hopkins University Press: Baltimore, MD, USA, 1994. [Google Scholar]
- Oke, M.; Ching, R.T.Y.; Carter, L.G.; Johnson, K.A.; Liu, H.; McMahon, S.A.; White, M.F.; Bloch, C., Jr.; Botting, C.H.; Walsh, M.A.; et al. Unusual chromophore and cross-links in ranasmurfin: A blue protein from the foam nests of a tropical frog. Angew. Chem. 2008, 47, 7853–7856. [Google Scholar] [CrossRef]





| Species | Protein (mg/mL) | Carbohydrate (mg/mL) | Surface Tension (mN/m) | Reproductive Mode † |
|---|---|---|---|---|
| P. cuvieri | 2.27 ± 0.48 a | 1.16 ± 0.06 a | 39.66 ± 0.50 a | RM 11 |
| L. vastus | 1.37 ± 0.24 b | 0.23 ± 0.01 b | 45.34 ± 0.70 b | RM 13 |
| A. hylaedactyla | 0.75 ± 0.15 b | N/A | 50.73 ± 0.35 c | RM 32 |
| Richness/α-Diversity Index | Leptodactylidae Species | ||
|---|---|---|---|
| A. hylaedactyla | P. cuvieri | L. vastus | |
| ASVs | 3555.38 ± 167.89 | 1706.43 ± 307.93 | 1198.60 ± 159.07 |
| Chao1 | 4631.60 ± 316.39 | 1977.25 ± 355.38 | 1544.21 ± 219.95 |
| Shannon | 6.21 ± 0.15 | 5.93 ± 0.31 | 4.95 ± 0.20 |
| Inverse Simpson | 57.83 ± 11.48 | 165.86 ± 56.58 | 40.89 6.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monteiro, F.A.C.; Bezerra, S.G.d.S.; Castro, L.G.Z.d.; Oliveira, F.A.d.S.; Normando, L.R.O.; Melo, V.M.M.; Hissa, D.C. Neotropical Frog Foam Nest’s Microbiomes. Microorganisms 2023, 11, 900. https://doi.org/10.3390/microorganisms11040900
Monteiro FAC, Bezerra SGdS, Castro LGZd, Oliveira FAdS, Normando LRO, Melo VMM, Hissa DC. Neotropical Frog Foam Nest’s Microbiomes. Microorganisms. 2023; 11(4):900. https://doi.org/10.3390/microorganisms11040900
Chicago/Turabian StyleMonteiro, Felipe Augusto Correia, Saulo Gonçalves de Santiago Bezerra, Luzia Gabrielle Zeferino de Castro, Francisca Andrea da Silva Oliveira, Leonardo Ribeiro Oliveira Normando, Vânia Maria Maciel Melo, and Denise Cavalcante Hissa. 2023. "Neotropical Frog Foam Nest’s Microbiomes" Microorganisms 11, no. 4: 900. https://doi.org/10.3390/microorganisms11040900
APA StyleMonteiro, F. A. C., Bezerra, S. G. d. S., Castro, L. G. Z. d., Oliveira, F. A. d. S., Normando, L. R. O., Melo, V. M. M., & Hissa, D. C. (2023). Neotropical Frog Foam Nest’s Microbiomes. Microorganisms, 11(4), 900. https://doi.org/10.3390/microorganisms11040900






