Insights on Novel Effectors and Characterization of Metacaspase (RS107_6) as a Potential Cell Death-Inducing Protein in Rhizoctonia solani
Abstract
1. Introduction
2. Material and Methods
2.1. Fungal Strain, Plant Material, and Rice Inoculation
2.2. In Silico Identification and Characterization of Putative Cell Death Effectors in R. solani
2.3. Phylogenetic Analysis of Cell Death Effectors
2.4. Molecular Docking Analysis and Molecular Dynamic (MD) Simulation Studies
2.5. RNA Extraction, cDNA Synthesis and Cloning of R. solani Genes Encoding Putative Cell Death Effectors
2.6. Expression and Purification of Recombinant Protein RS107_6
2.7. MS Analysis of Recombinant RS107_6 Protein Using TripleTOF 5600+
2.8. Gene Expression Analysis (qPCR) of Predicted Effector Genes in Planta
2.9. Statistical Analysis
3. Results
3.1. Molecular Identification of Fungal Isolate
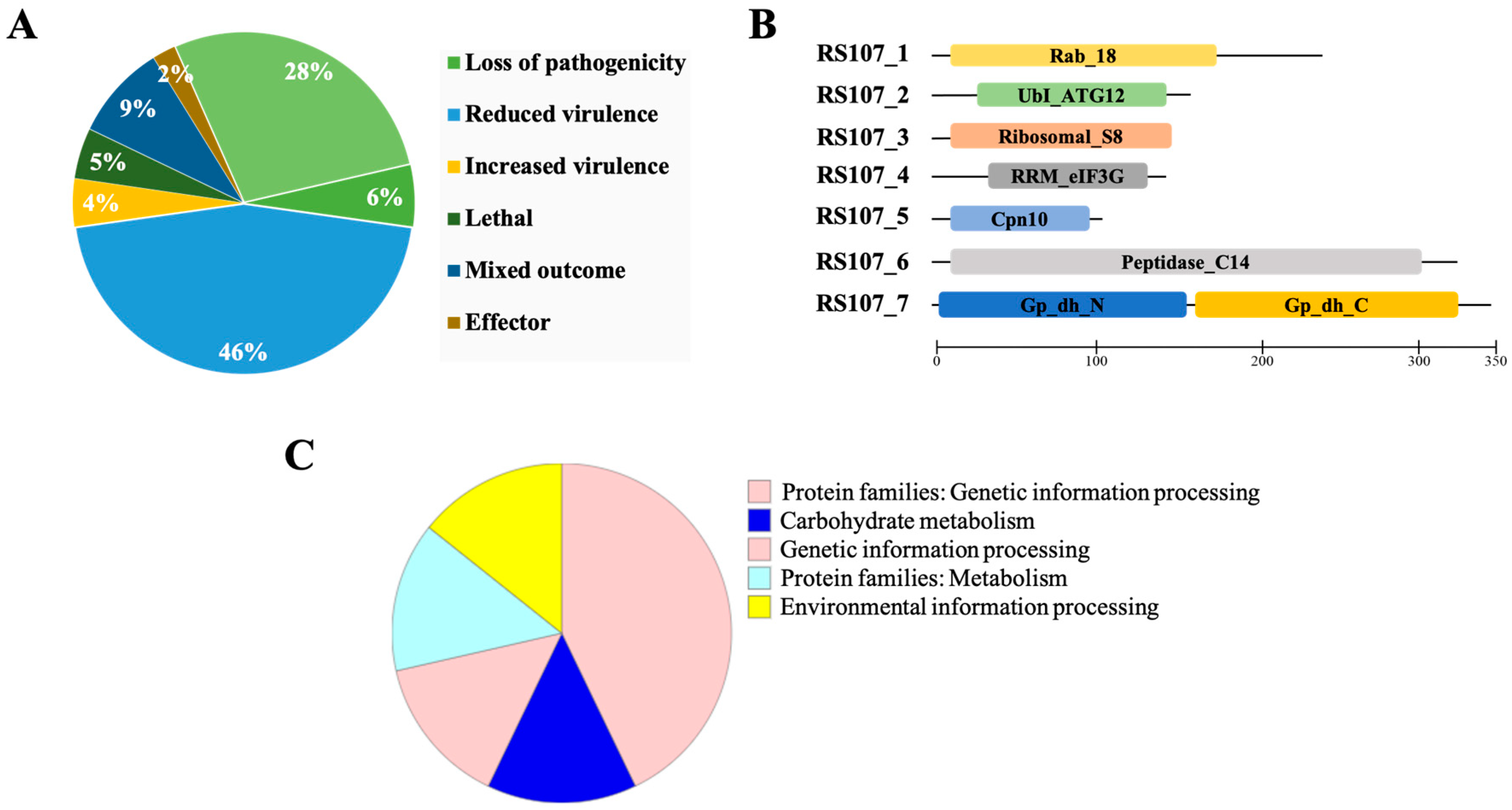
3.2. Genome-Wide Identification of Putative Cell Death Effectors in R. solani
3.3. In Silico Characterization of Putative Effector Proteins
3.4. Physiochemical Characterization
3.5. Cell Death Effectors Are Highly Conserved among R. solani Isolates
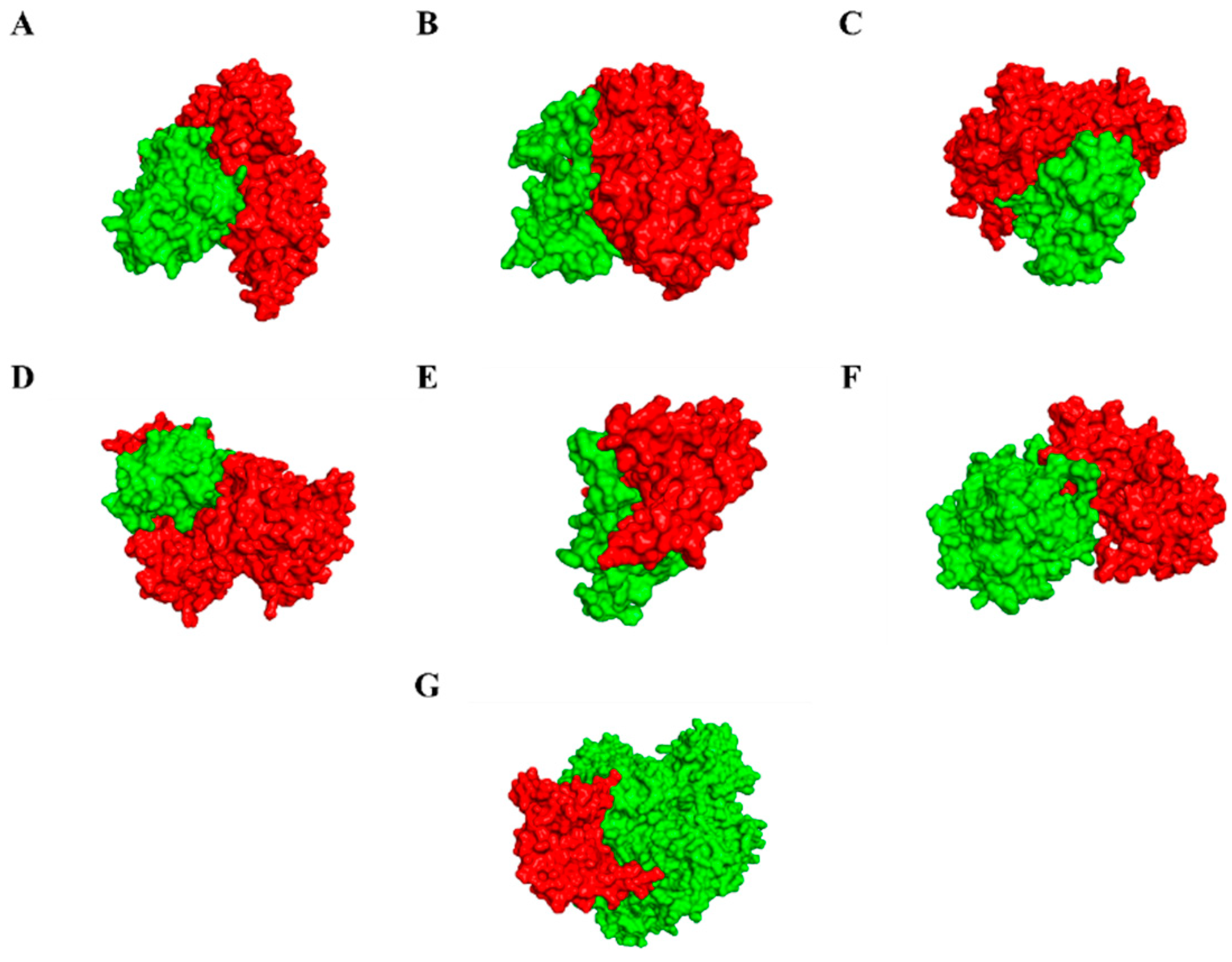
3.6. Modelling of Effectors and Their Interacting Partners in Rice System
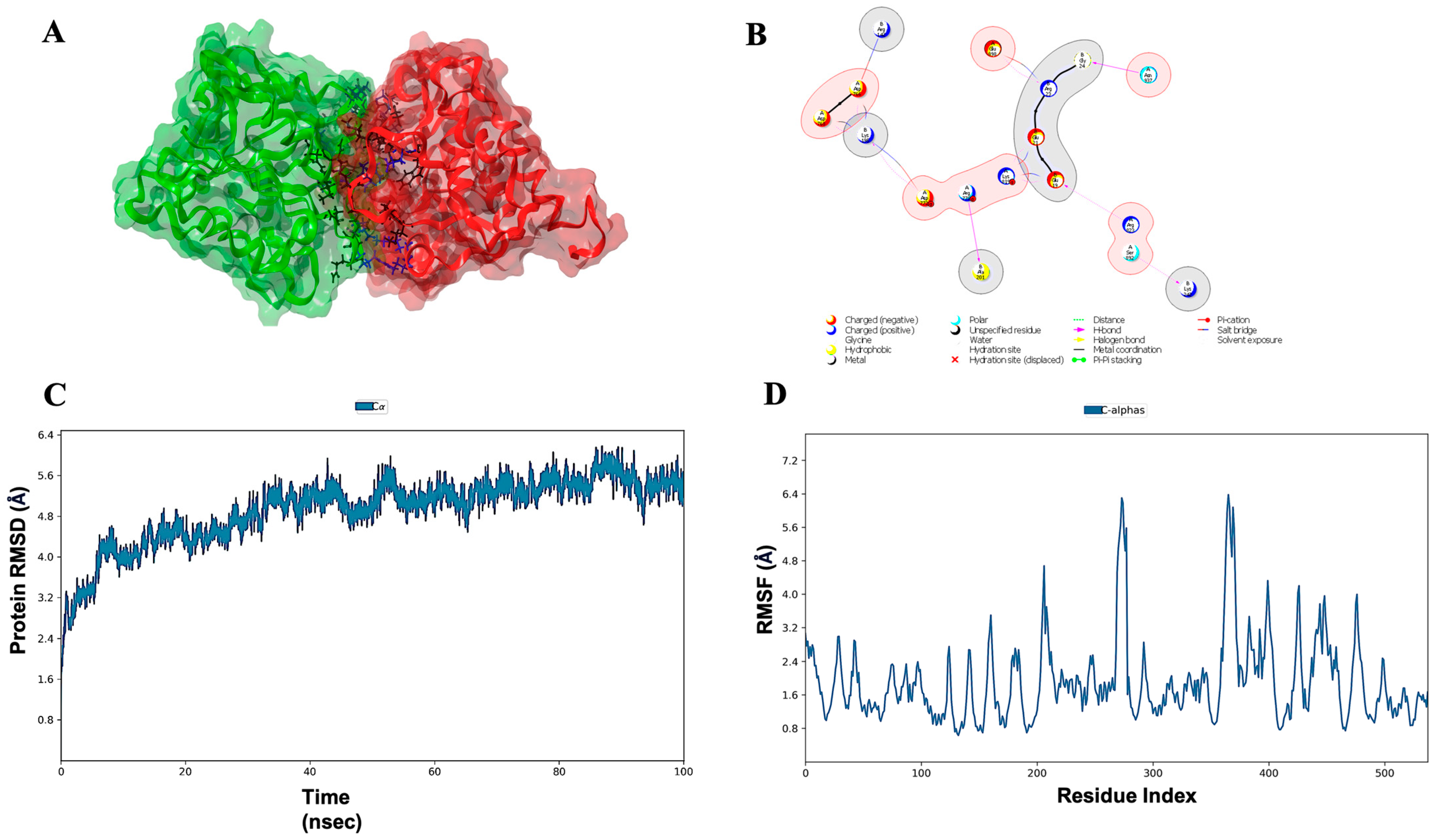
3.7. Molecular Docking, Simulations, and Analysis of Docked Structures
3.8. Cloning, Expression, and Purification of the Recombinant RS107_6 Protein
3.9. RS107_6 Protein Characterization
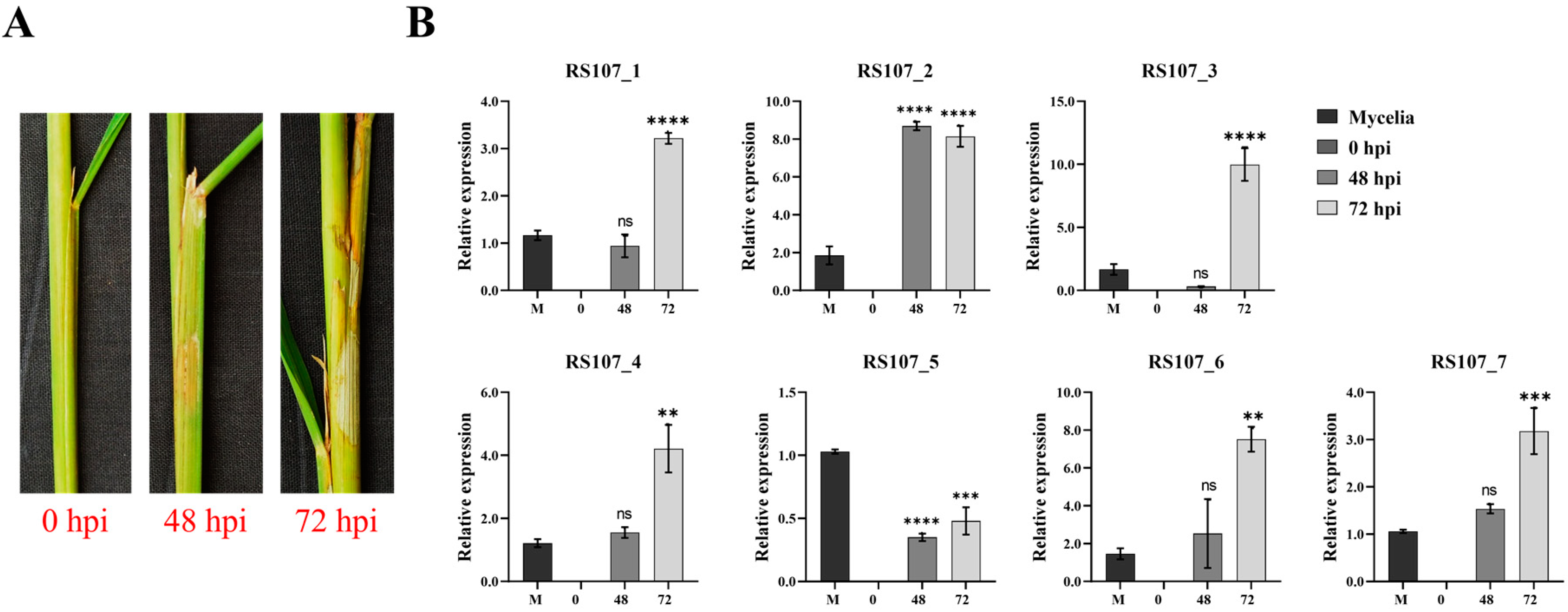
3.10. Relative Expression Patterns of Effector Genes during Rice Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anderson, N.A. The genetics and pathology of Rhizoctonia solani. Annu. Rev. Phytopathol. 1982, 20, 329–347. [Google Scholar] [CrossRef]
- Singh, P.; Mazumdar, P.; Harikrishna, J.A.; Babu, S. Sheath blight of rice: A review and identification of priorities for future research. Planta 2019, 250, 1387–1407. [Google Scholar] [CrossRef] [PubMed]
- Yellareddygari, S.K.R.; Reddy, M.S.; Kloepper, J.W.; Lawrence, K.S.; Fadamiro, H. Rice sheath blight: A review of disease and pathogen management approaches. J. Plant Pathol. Microb. 2014, 5, 1. [Google Scholar]
- Zheng, A.; Lin, R.; Zhang, D.; Qin, P.; Xu, L.; Ai, P.; Wang, Y.; Chen, Y.; Liu, Y.; Sun, Z.; et al. The evolution and pathogenic mechanisms of the rice sheath blight pathogen. Nat. Commun. 2013, 4, 1424. [Google Scholar] [CrossRef]
- Wibberg, D.; Jelonek, L.; Rupp, O.; Hennig, M.; Eikmeyer, F.; Goesmann, A.; Hartmann, A.; Borriss, R.; Grosch, R.; Pühler, A.; et al. Establishment and interpretation of the genome sequence of the phytopathogenic fungus Rhizoctonia solani AG1-IB isolate 7/3/14. J. Biotechnol. 2013, 167, 142–155. [Google Scholar] [CrossRef]
- Cubeta, M.A.; Thomas, E.; Dean, R.A.; Jabaji, S.; Neate, S.M.; Tavantzis, S.; Toda, T.; Vilgalys, R.; Bharathan, N.; Fedorova-Abrams, N.; et al. Draft genome sequence of the plant-pathogenic soil fungus Rhizoctonia solani anastomosis group 3 strain Rhs1AP. Genome Announc. 2014, 2, e01072-14. [Google Scholar] [CrossRef]
- Hane, J.K.; Anderson, J.P.; Williams, A.H.; Sperschneider, J.; Singh, K.B. Genome sequencing and comparative genomics of the broad host-range pathogen Rhizoctonia solani AG8. PLoS Genet. 2014, 10, e1004281. [Google Scholar] [CrossRef] [PubMed]
- Wibberg, D.; Andersson, L.; Tzelepis, G.; Rupp, O.; Blom, J.; Jelonek, L.; Pühler, A.; Fogelqvist, J.; Varrelmann, M.; Schlüter, A.; et al. Genome analysis of the sugar beet pathogen Rhizoctonia solani AG2-2IIIB revealed high numbers in secreted proteins and cell wall degrading enzymes. BMC Genom. 2016, 17, 245. [Google Scholar] [CrossRef]
- Singh, S.K.; Shree, A.; Verma, S.; Singh, K.; Kumar, K.; Srivastava, V.; Singh, R.; Saxena, S.; Singh, A.P.; Pandey, A.; et al. The nuclear effector ArPEC25 from the necrotrophic fungus Ascochyta rabiei targets the chickpea transcription factor CaβLIM1a and negatively modulates lignin biosynthesis, increasing host susceptibility. Plant Cell 2023, 35, 1134–1159. [Google Scholar] [CrossRef]
- Zhang, S.; Li, C.; Si, J.; Han, Z.; Chen, D. Action mechanisms of effectors in plant-pathogen interaction. Int. J. Mol. Sci. 2022, 23, 6758. [Google Scholar] [CrossRef]
- Anderson, J.P.; Sperschneider, J.; Win, J.; Kidd, B.; Yoshida, K.; Hane, J.; Saunders, D.G.O.; Singh, K.B. Comparative secretome analysis of Rhizoctonia solani isolates with different host ranges reveals unique secretomes and cell death inducing effectors. Sci. Rep. 2017, 7, 10410. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, L.; He, Z.; Zhang, J. Molecular cloning and functional analysis of two novel polygalacturonase genes in Rhizoctonia solani. Can. J. Plant Pathol. 2017, 40, 39–47. [Google Scholar] [CrossRef]
- Li, S.; Peng, X.; Wang, Y.; Hua, K.; Xing, F.; Zheng, Y.; Liu, W.; Sun, W.; Wei, S. The effector AGLIP1 in Rhizoctonia solani AG1 IA triggers cell death in plants and promotes disease development through inhibiting PAMP-triggered immunity in Arabidopsis thaliana. Front. Microbiol. 2019, 10, 2228. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Wang, A.; Liu, Y.; Ma, L.; Niu, X.; Zheng, A. Identification of the novel effector RsIA_NP8 in Rhizoctonia solani AG1 IA that induces cell death and triggers defense responses in non-host plants. Front. Microbiol. 2020, 11, 1115. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Yamamoto, N.; Yang, G.; Lin, H.; Jiang, L.; Liu, Y.; Zheng, A. A small secreted protein, RsMf8HN, in Rhizoctonia solani triggers plant immune response, which interacts with rice OsHIPP28. Microbiol. Res. 2023, 266, 127219. [Google Scholar] [CrossRef] [PubMed]
- Dolfors, F.; Holmquist, L.; Dixelius, C.; Tzelepis, G. A LysM effector protein from the basidiomycete Rhizoctonia solani contributes to virulence through suppression of chitin-triggered immunity. Mol. Genet. Genom. 2019, 294, 1211–1218. [Google Scholar] [CrossRef]
- Tzelepis, G.; Dölfors, F.; Holmquist, L.; Dixelius, C. Plant mitochondria and chloroplasts are targeted by the Rhizoctonia solani RsCRP1 effector. Biochem. Biophys. Res. Commun. 2021, 544, 86–90. [Google Scholar] [CrossRef]
- Charova, S.N.; Dölfors, F.; Holmquist, L.; Moschou, P.N.; Dixelius, C.; Tzelepis, G. The RsRlpA effector is a protease inhibitor promoting Rhizoctonia solani virulence through suppression of the hypersensitive response. Int. J. Mol. Sci. 2020, 21, 8070. [Google Scholar] [CrossRef]
- Barnett, H.L.; Hunter, B.B. Illustrated Genera of Imperfect Fungi, 4th ed.; APS Press: St. Paul, MN, USA, 1998; p. 218. [Google Scholar]
- White, T.J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; Volume 31, pp. 315–322. [Google Scholar]
- Park, D.S.; Sayler, R.J.; Hong, Y.G.; Nam, M.H.; Yang, Y. A method for inoculation and evaluation of rice sheath blight disease. Plant Dis. 2008, 92, 25–29. [Google Scholar]
- Sperschneider, J.; Dodds, P.N. EffectorP 3.0: Prediction of apoplastic and cytoplasmic effectors in fungi and oomycetes. Mol. Plant Microbe Interact. 2022, 35, 146–156. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kallberg, M.; Margaryan, G.; Wang, S.; Ma, J.; Xu, J. RaptorX server: A resource for template-based protein structure modeling. Methods Mol. Biol. 2012, 1137, 17–27. [Google Scholar]
- Ramachandran, G.N.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Macindoe, G.; Mavridis, L.; Venkatraman, V.; Devignes, M.D.; Ritchie, D.W. Hex Server: An FFT-based protein docking server powered by graphics processors. Nucleic Acids Res. 2010, 38, 445–449. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the ACM/IEEE Conference on Supercomputing (SC’06), Tampa, FL, USA, 11–17 November 2006. [Google Scholar]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Price, D.J.; Brooks, C.L. A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 2004, 121, 10096–10103. [Google Scholar] [CrossRef]
- Bradford, M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Guruprasad, K.; Reddy, B.V.; Pandit, M.W. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1980, 4, 155–161. [Google Scholar] [CrossRef]
- Ikai, A. Thermostability and aliphatic index of globular proteins. J. Biochem. 1980, 88, 1895–1898. [Google Scholar] [PubMed]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [PubMed]
- Reddy, C.H.S.; Vijayasarathy, K.; Srinivas, E.; Sastry, G.M.; Sastry, G.N. Homology modeling of membrane proteins: A critical assessment. Comput. Biol. Chem. 2006, 30, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Tan, S.; Zhao, Q.; Lin, D.L.; Xu, Z.H.; Friml, J.; Xue, H.W. mRNA surveillance complex PELOTA-HBS1 regulates phosphoinositide-dependent protein kinase1 and plant growth. Plant Physiol. 2021, 186, 2003–2020. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.B.; Feng, B.H.; Wang, H.M.; Xu, X.; Shi, Y.F.; He, Y.; Chen, Z.; Sathe, A.P.; Shi, L.; Wu, J.L. A substitution mutation in OsPELOTA confers bacterial blight resistance by activating the salicylic acid pathway. J. Integr. Plant Biol. 2018, 60, 160–172. [Google Scholar] [CrossRef]
- Yang, Z.; Xing, J.; Wang, L.; Liu, Y.; Qu, J.; Tan, Y.; Fu, X.; Lin, Q.; Deng, H.; Yu, F. Mutations of two FERONIA-like receptor genes enhance rice blast resistance without growth penalty. J. Exp. Bot. 2020, 71, 2112–2126. [Google Scholar] [CrossRef]
- Juszkiewicz, S.; Speldewinde, S.H.; Wan, L.; Svejstrup, J.Q.; Hegde, R.S. The ASC-1 complex disassembles collided ribosomes. Mol. Cell 2020, 79, 603–614. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Y.; Ma, X.; Meng, L.; Jing, R.; Wang, F.; Wang, S.; Cheng, Z.; Zhang, X.; Jiang, L.; et al. Disruption of gene SPL35, encoding a novel CUE domain-containing protein, leads to cell death and enhanced disease response in rice. Plant Biotechnol. J. 2019, 17, 1679–1693. [Google Scholar] [CrossRef]
- Nawaz, Z.; Kakar, K.U.; Saand, M.A.; Shu, Q.Y. Cyclic nucleotide-gated ion channel gene family in rice, identification, characterization and experimental analysis of expression response to plant hormones, biotic and abiotic stresses. BMC Genom. 2014, 15, 853. [Google Scholar] [CrossRef]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant-Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef]
- Shen, X.; Liu, H.; Yuan, B.; Li, X.; Xu, C.; Wang, S. OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant Cell Environ. 2011, 34, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Manosalva, P.M.; Bruce, M.; Leach, J.E. Rice 14-3-3 protein (GF14e) negatively affects cell death and disease resistance. Plant J. 2011, 68, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Sonah, H.; Deshmukh, R.K.; Bélanger, R.R. Computational prediction of effector proteins in fungi: Opportunities and challenges. Front. Plant Sci. 2016, 7, 126. [Google Scholar] [CrossRef]
- Sperschneider, J.; Gardiner, D.M.; Dodds, P.N.; Tini, F.; Covarelli, L.; Singh, K.B.; Manners, J.M.; Taylor, J.M. EffectorP: Predicting fungal effector proteins from secretomes using machine learning. New Phytol. 2015, 210, 743–761. [Google Scholar] [CrossRef]
- Liu, T.; Song, T.; Zhang, X.; Yuan, H.; Su, L.; Li, W.; Xu, J.; Liu, S.; Chen, L.; Chen, T.; et al. Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 2014, 5, 4686. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ceron, D.; Lowe, R.G.T.; McKenna, J.A.; Brain, L.M.; Dawson, C.S.; Clark, B.; Berkowitz, O.; Faou, P.; Whelan, J.; Bleackley, M.R.; et al. Extracellular vesicles from Fusarium graminearum contain protein effectors expressed during infection of corn. J. Fungi 2021, 7, 977. [Google Scholar] [CrossRef] [PubMed]
- Selin, C.; de Kievit, T.R.; Belmonte, M.F.; Fernando, W.G. Elucidating the role of effectors in plant-fungal interactions: Progress and challenges. Front. Microbiol. 2016, 7, 600. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Djavaheri, M.; Wang, H.; Larkan, N.J.; Haddadi, P.; Beynon, E.; Gropp, G.; Borhan, M.H. Leptosphaeria maculans effector protein AvrLm1 modulates plant immunity by enhancing MAP Kinase 9 phosphorylation. iScience 2018, 3, 177–191. [Google Scholar] [CrossRef]
- Wang, B.; Song, N.; Tang, C.; Ma, J.; Wang, N.; Sun, Y.; Kang, Z. PsRPs26, a 40S ribosomal protein subunit, regulates the growth and pathogenicity of Puccinia striiformis f. sp.tritici. Front. Microbiol. 2019, 10, 968. [Google Scholar] [CrossRef]
- Verwaaijen, B.; Wibberg, D.; Krober, M.; Winkler, A.; Zrenner, R.; Bednarz, H.; Niehaus, K.; Grosch, R.; Puhler, A.; Schluter, A. The Rhizoctonia solani AG1-IB (isolate 7/3/14) transcriptome during interaction with the host plant lettuce (Lactuca sativa L.). PLoS ONE 2017, 12, e0177278. [Google Scholar] [CrossRef]
- Liu, T.B.; Liu, X.H.; Lu, J.P.; Zhang, L.; Min, H.; Lin, F.C. The cysteine protease MoAtg4 interacts with MoAtg8 and is required for differentiation and pathogenesis in Magnaporthe oryzae. Autophagy 2010, 6, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhao, J.; Guo, W.; Zhang, T. Functional analysis of autophagy genes via Agrobacterium-mediated transformation in the vascular wilt fungus Verticillium dahliae. J. Genet. Genom. 2013, 40, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Khalid, A.R.; Zhang, S.; Luo, X.; Shaheen, H.; Majeed, A.; Maqbool, M.; Zahid, N.; Rahim, J.; Ren, M.; Qiu, D. Functional analysis of autophagy-related gene ATG12 in potato dry rot fungus Fusarium oxysporum. Int. J. Mol. Sci. 2021, 22, 4932. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Qin, G.; Li, B.; Tian, S. Knocking out Bcsas1 in Botrytis cinerea impacts growth, development, and secretion of extracellular proteins, which decreases virulence. Mol. Plant-Microbe Interact. 2014, 27, 590–600. [Google Scholar] [CrossRef]
- Yang, C.D.; Dang, X.; Zheng, H.W.; Chen, X.F.; Lin, X.L.; Zhang, D.M.; Abubakar, Y.S.; Chen, X.; Lu, G.; Wang, Z.; et al. Rab5 homologs are essential for the development and pathogenicity of the rice blast fungus Magnaporthe oryzae. Front. Plant Sci. 2017, 8, 620. [Google Scholar] [CrossRef]
- Jia, H.; Halilao, A.I.; Hu, L.; Cai, W.; Liu, J.; Huang, B. Heat shock protein 10 (HSP10) in immune-related diseases: One coin, two sides. Int. J. Biochem. Mol. Biol. 2011, 2, 47–57. [Google Scholar]
- Fernandez, J.; Lopez, V.; Kinch, L.; Pfeifer, M.A.; Gray, H.; Garcia, N.; Grishin, N.V.; Khang, C.H.; Orth, K. Role of two metacaspases in development and pathogenicity of the rice blast fungus Magnaporthe oryzae. mBio 2021, 12, e03471-20. [Google Scholar] [CrossRef]
- Mukherjee, D.; Gupta, S.; Saran, N.; Datta, R.; Ghosh, A. Induction of apoptosis-like cell death and clearance of stress-induced intracellular protein aggregates: Dual roles for Ustilago maydis metacaspase Mca1. Mol. Microbiol. 2017, 106, 815–831. [Google Scholar] [CrossRef]
- Lee, R.E.; Brunette, S.; Puente, L.G.; Megeney, L.A. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc. Natl. Acad. Sci. USA 2010, 107, 13348–13353. [Google Scholar] [CrossRef]
- Conchou, L.; Doumèche, B.; Galisson, F.; Violot, S.; Dugelay, C.; Diesis, E.; Page, A.; Bienvenu, A.L.; Picot, S.; Aghajari, N.; et al. Structural and molecular determinants of Candida glabrata metacaspase maturation and activation by calcium. Commun. Biol. 2022, 5, 1158. [Google Scholar] [CrossRef]
- Leang, L.; Mcdonald, M.C.; Mineo, C.R.; Jones, B.; Barker, T.; Gagliardi, C.; Fox, K.M. Identification and characterization of Schizophyllum commune type I metacaspases. Biochem. Biophys. Rep. 2019, 20, 100706. [Google Scholar] [CrossRef] [PubMed]
- Strobel, I.; Osiewacz, H.D. Poly(ADP-ribose) polymerase is a substrate recognized by two metacaspases of Podospora anserina. Eukaryot. Cell 2013, 12, 900–912. [Google Scholar] [CrossRef] [PubMed]
- Coll, N.S.; Vercammen, D.; Smidler, A.; Clover, C.; Van Breusegem, F.; Dangl, J.L.; Epple, P. Arabidopsis type I metacaspases control cell death. Science 2010, 330, 1393–1397. [Google Scholar] [CrossRef] [PubMed]


| Cell Death Effectors | Conserved Domain | Subcellular Localization | Complete Coding Sequence (bp) | Amino Acid Residues | Molecular Weight (kDa) | Cysteine Content | Isoelectric Point(pI) | Instability Index | Aliphatic Index | GRAVY Index |
|---|---|---|---|---|---|---|---|---|---|---|
| RS107_1 | Rab_18 domain | Cytoplasmic | 642 | 213 | 23.15 | 3 | 5.41 | 27.99 | 78.17 | -0.268 |
| RS107_2 | UbI-like ATG12 domain | Cytoplasmic-nuclear | 423 | 140 | 15.75 | 2 | 9.23 | 34.36 | 81.57 | -0.064 |
| RS107_3 | Ribosomal_S8 | Cytoplasmic/apoplastic | 393 | 130 | 14.66 | 1 | 10.33 | 37.32 | 113.77 | -0.071 |
| RS107_4 | RRM_eIF3G | Cytoplasmic-nuclear | 396 | 131 | 14.39 | 0 | 6.60 | 38.97 | 49.92 | -0.886 |
| RS107_5 | Cpn10 domain | Cytoplasmic | 297 | 98 | 10.12 | 1 | 9.8 | 32.07 | 98.47 | 0.118 |
| RS107_6 | Peptidase_C14 | Cytoplasmic | 906 | 301 | 33.10 | 7 | 5.39 | 44.73 | 75.22 | -0.437 |
| RS107_7 | GAPDH | Cytoplasmic | 1032 | 343 | 36.85 | 2 | 6.34 | 21.79 | 90.35 | -0.103 |
| Rice Target Genes | Protein Name | Role in Plants | References |
|---|---|---|---|
| LOC_Os04g56480 (OsPELOTA) | PELOTA protein | mRNA surveillance and quality control; enhance resistance against rice bacterial blight pathogen by activating salicylic acid pathway | Kong et al. [37]; Zhang et al. [38] |
| LOC_Os01g56330 (FLR2) | Feronia protein | Receptor-like kinases; recognize the PAMPs and trigger oxidative bursts | Yang et al. [39] |
| LOC_Os03g10750 (SPL35) | ASC-1 complex subunit | Ribosome-associated quality control; regulates cell death and defense response through its ubiquitination and vesicular trafficking pathways | Juszkiewicz et al. [40]; Ma et al. [41] |
| LOC_Os01g02720 (SPL33) | HBS-1-like protein | mRNA surveillance and quality control | Kong et al. [37] |
| LOC_Os09g38580 (OsCNGC9) | Cyclic nucleotide-gated ion channel protein | Passages for conducting calcium ions into the cytosol in response to pathogen invasion as an early event in defense signaling | Nawaz et al. [42] |
| LOC_Os03g06410 (OsEDR1) | Serine–threonine protein kinases/mitogen- activated protein kinases | Signaling and plant defense | Afzal et al. [43]; Shen et al. [44] |
| LOC_Os02g36974 (OsGF14e) | 14-3-3-like protein GF14E | Negatively regulates the induction of plant defense response genes, cell death, and contributes to broad-spectrum resistance in rice | Manosalva et al. [45] |
| Purification Step | Protein Concentration (mg/mL) | Total Protein (mg) | Yield (%) | Purification (Fold) |
|---|---|---|---|---|
| Crude lysate | 1.85 | 27.7 | 100 | 1 |
| Affinity purification | 1.16 | 11.6 | 41.8 | 0.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kavya, N.; Prasannakumar, M.K.; Venkateshbabu, G.; Niranjan, V.; Uttarkar, A.; Buela Parivallal, P.; Banakar, S.N.; Mahesh, H.B.; Devanna, P.; Manasa, K.G.; et al. Insights on Novel Effectors and Characterization of Metacaspase (RS107_6) as a Potential Cell Death-Inducing Protein in Rhizoctonia solani. Microorganisms 2023, 11, 920. https://doi.org/10.3390/microorganisms11040920
Kavya N, Prasannakumar MK, Venkateshbabu G, Niranjan V, Uttarkar A, Buela Parivallal P, Banakar SN, Mahesh HB, Devanna P, Manasa KG, et al. Insights on Novel Effectors and Characterization of Metacaspase (RS107_6) as a Potential Cell Death-Inducing Protein in Rhizoctonia solani. Microorganisms. 2023; 11(4):920. https://doi.org/10.3390/microorganisms11040920
Chicago/Turabian StyleKavya, N., M. K. Prasannakumar, Gopal Venkateshbabu, Vidya Niranjan, Akshay Uttarkar, P. Buela Parivallal, Sahana N. Banakar, H. B. Mahesh, Pramesh Devanna, K. G. Manasa, and et al. 2023. "Insights on Novel Effectors and Characterization of Metacaspase (RS107_6) as a Potential Cell Death-Inducing Protein in Rhizoctonia solani" Microorganisms 11, no. 4: 920. https://doi.org/10.3390/microorganisms11040920
APA StyleKavya, N., Prasannakumar, M. K., Venkateshbabu, G., Niranjan, V., Uttarkar, A., Buela Parivallal, P., Banakar, S. N., Mahesh, H. B., Devanna, P., Manasa, K. G., & Shivakumara, T. N. (2023). Insights on Novel Effectors and Characterization of Metacaspase (RS107_6) as a Potential Cell Death-Inducing Protein in Rhizoctonia solani. Microorganisms, 11(4), 920. https://doi.org/10.3390/microorganisms11040920






