The Growth Inhibition of Polyethylene Nanoplastics on the Bait-Microalgae Isochrysis galbana Based on the Transcriptome Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microalgae Strain and Growth Analysis
2.2. Determination of Microalgae Biomass, Pigment, and Soluble Protein Content
2.3. Measurement of MDA Levels and Antioxidant Enzyme Activity
2.4. Transcriptome Sequencing and Analysis
2.5. Analysis of Microbial Community Structure and Diversity
2.6. Co-Culture Experiments
2.7. Statistical Analysis
3. Results
3.1. Microalgal Growth and Physiology Analysis
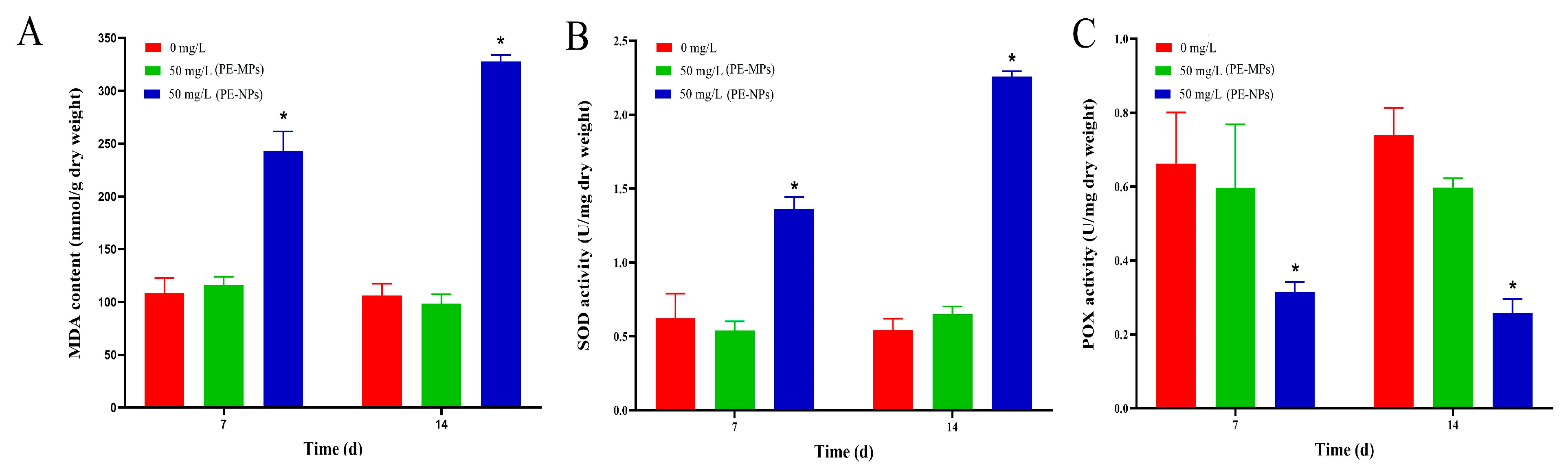
3.2. Test of Antioxidant Enzymes Activity
3.3. Overall Transcriptional Response Profile of I. galbana to MPs Exposure
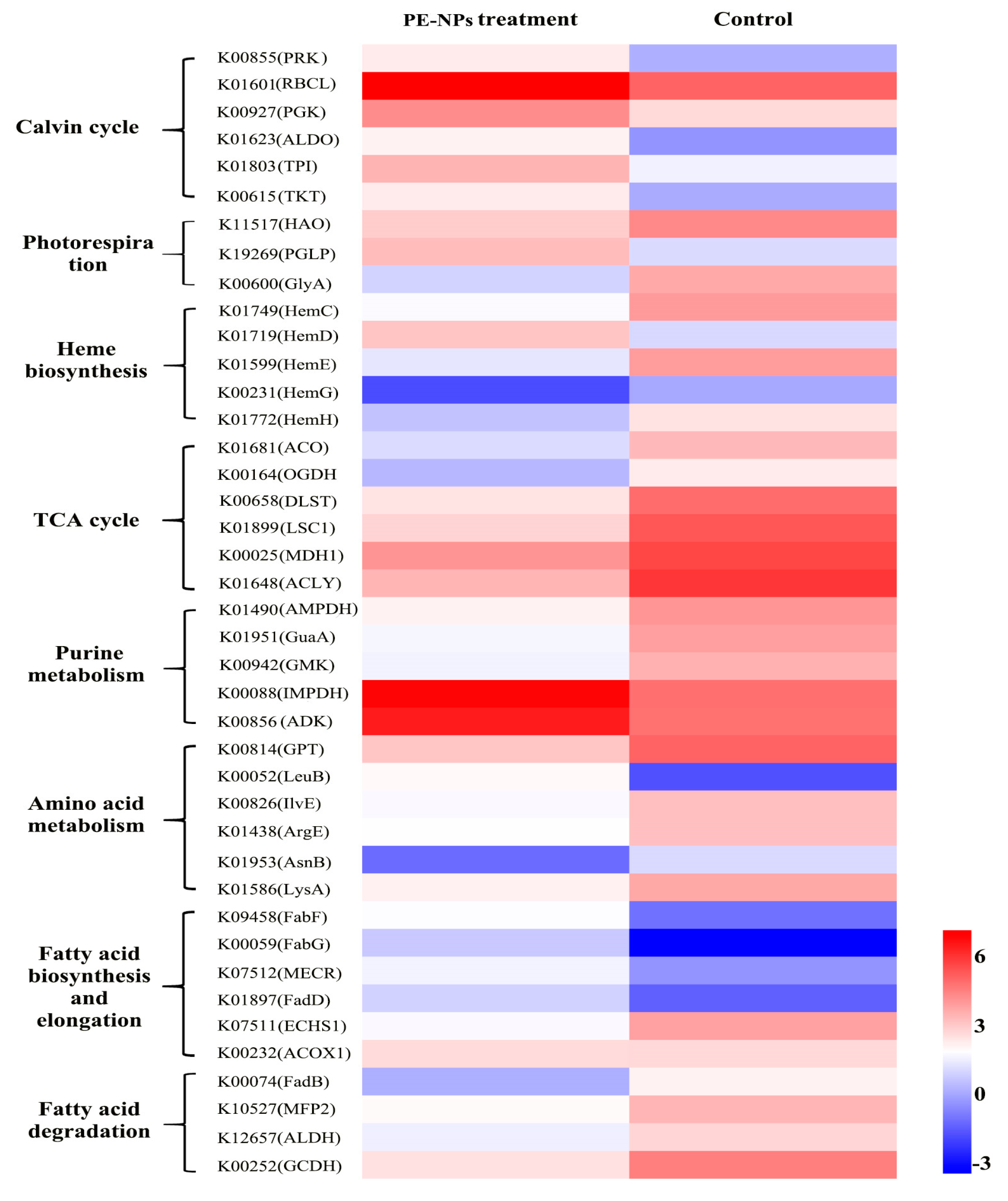
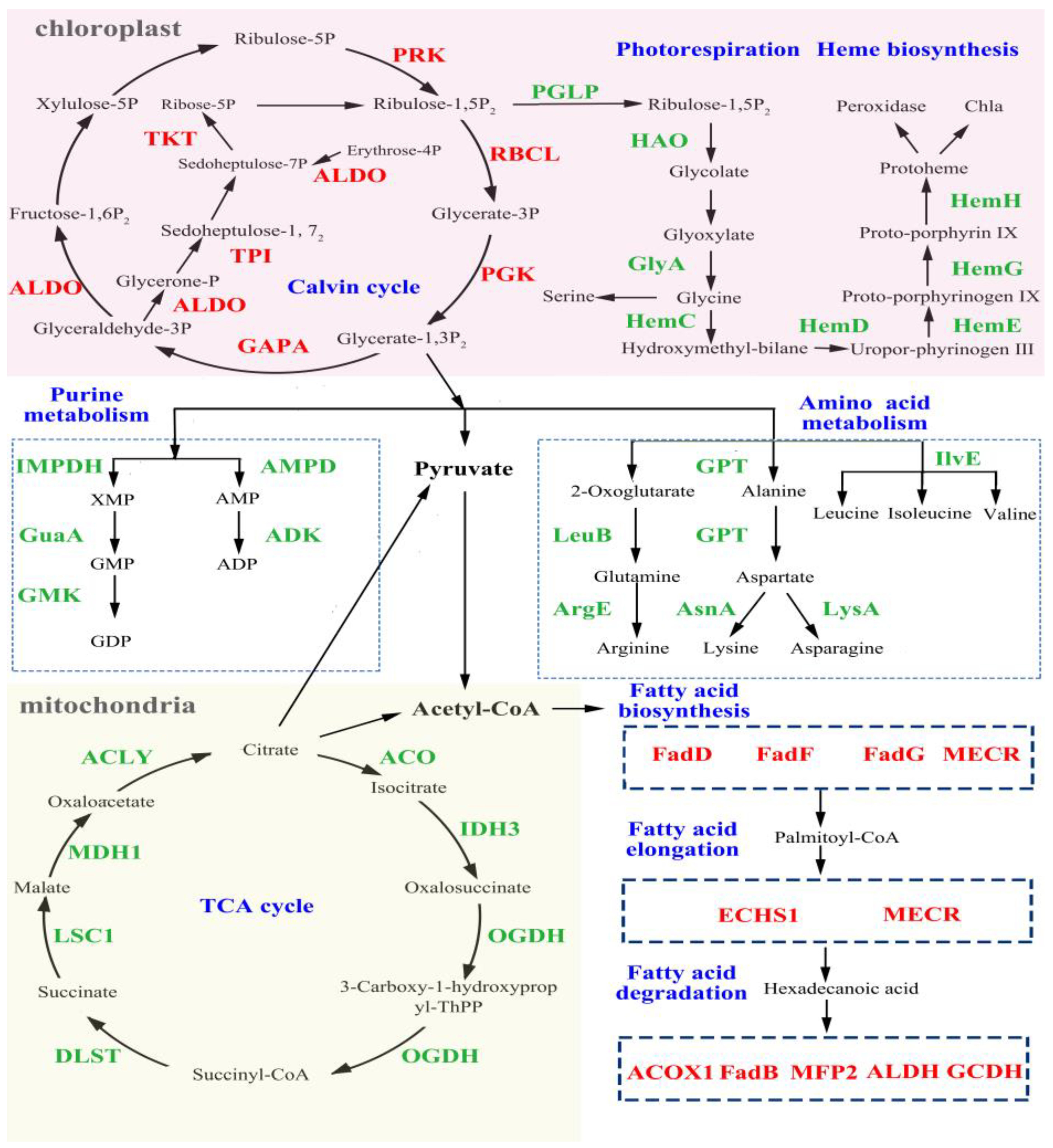
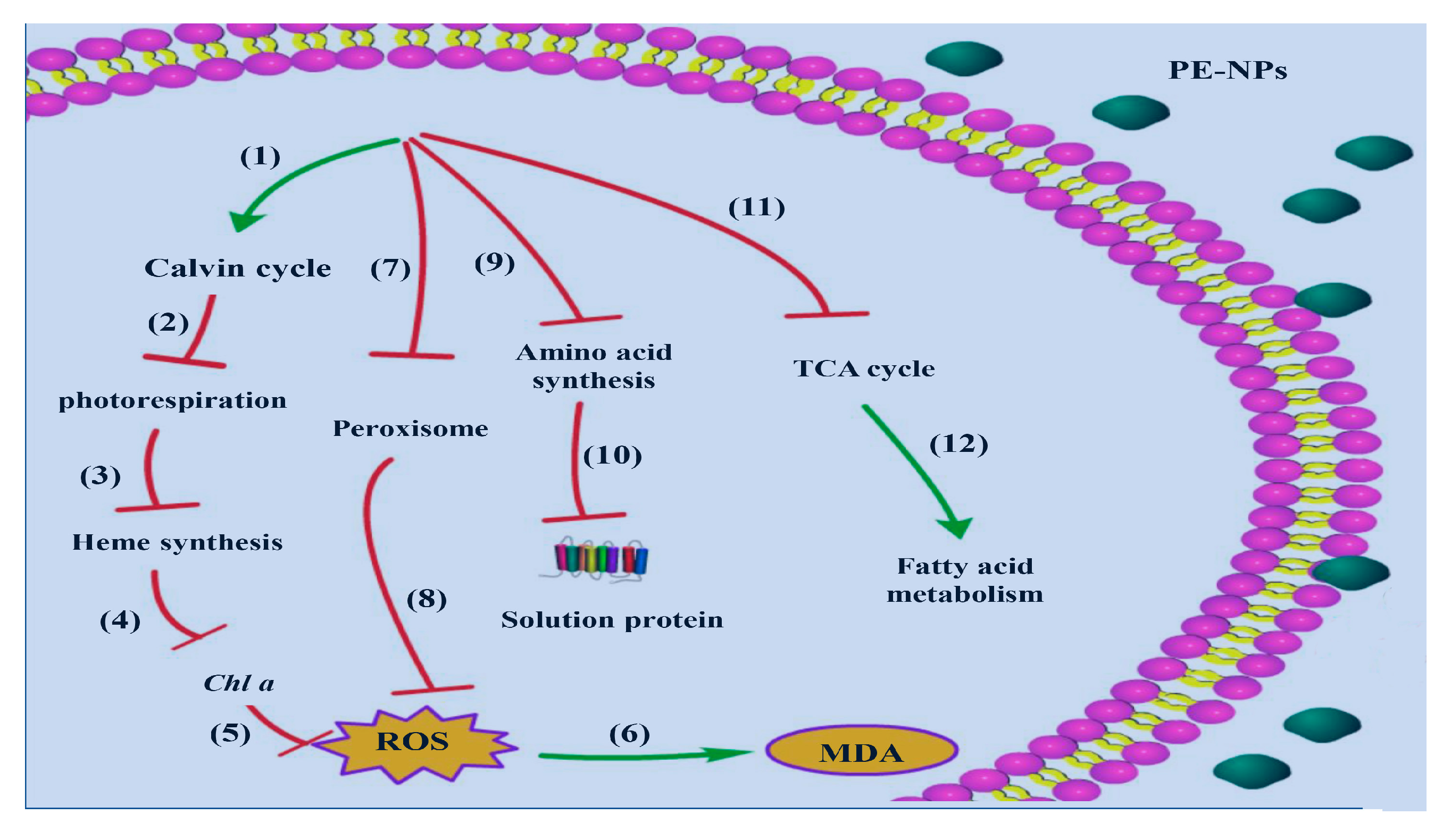
3.4. Analysis of DEGs Related to Cell Metabolism and Photosynthesis
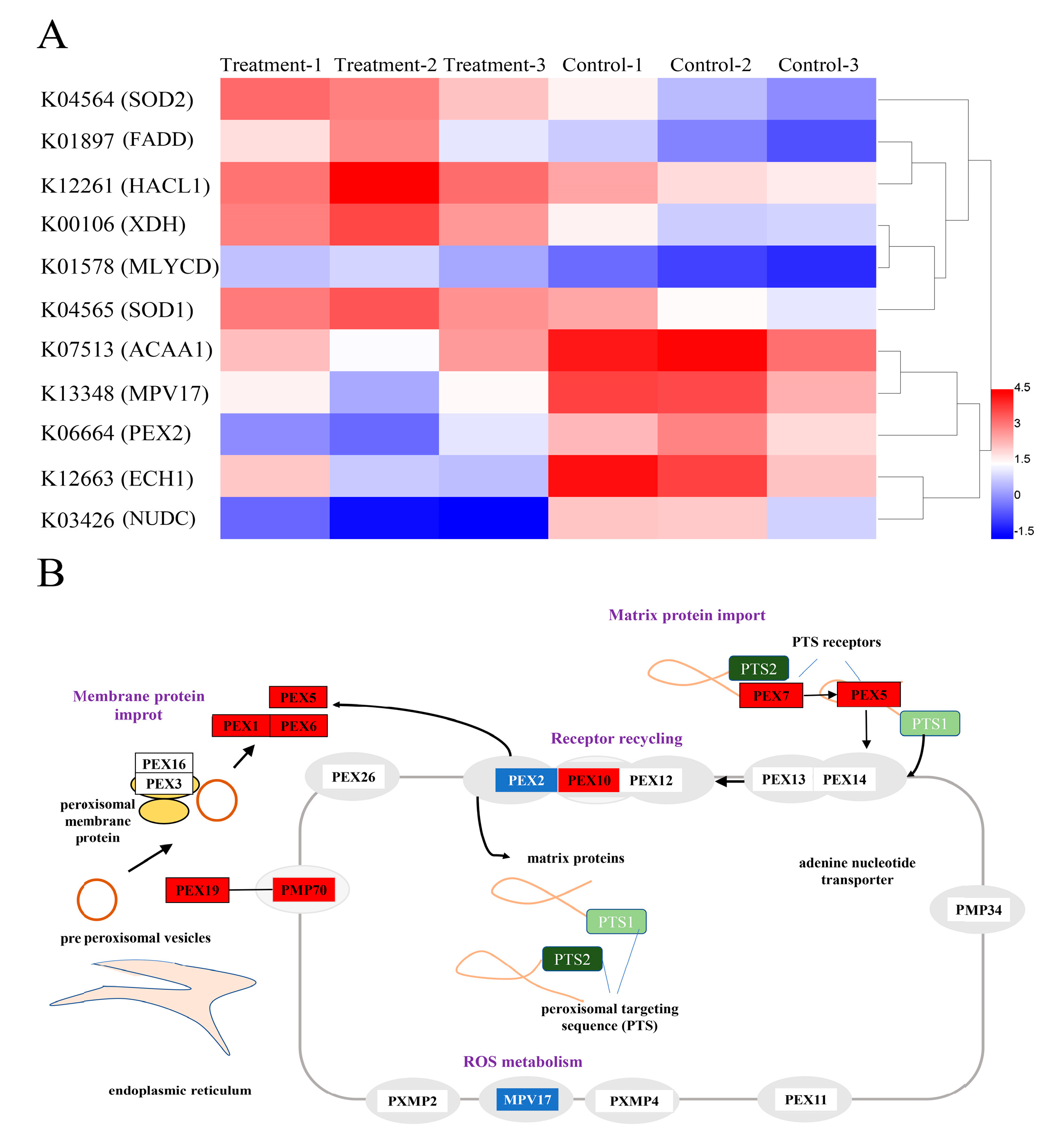
3.5. Analysis of DEGs Related to Antioxidant Defense
3.6. Bacterial Community Composition Analysis
3.7. The Effect of Symbiotic Bacteria on the Microalgal Cell Growth
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Qu, H.; Ma, R.X.; Barrett, H.; Wang, B.; Han, J.J.; Wang, F.; Chen, P.; Wang, W.; Peng, G.L.; Yu, G. How microplastics affect chiral illicit drug methamphetamine in aquatic food chain? From green alga (Chlorella pyrenoidosa) to freshwater snail (Cipangopaludian cathayensis). Environ. Int. 2020, 136, 105480. [Google Scholar] [CrossRef]
- Fu, D.D.; Zhang, Q.J.; Fan, Z.Q.; Qi, H.Y.; Wang, Z.Z.; Peng, L.C. Aged microplastics polyvinyl chloride interact with copper and cause oxidative stress towards microalgae Chlorella vulgaris. Aquat. Toxicol. 2019, 216, 105319. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Li, C.; Ma, L.Y.; Xin, P.; Li, F.J.; Xu, Z.J. Quantitative analysis of microplastics in coastal tidal-flat reclamation in Dongtai, China. Front. Environ. Sci. Eng. 2022, 16, 107. [Google Scholar] [CrossRef]
- Gardon, T.; Reisser, C.; Soyez, C.; Quillien, V.; Moullac, G.L. Microplastics affect energy balance and gametogenesis in the pearl oyster Pinctada margaritifera. Environ. Sci. Technol. 2018, 52, 5277–5286. [Google Scholar] [CrossRef] [PubMed]
- Maroušek, J.; Strunecký, O.; Maroušková, A. Insect rearing on biowaste represents a competitive advantage for fish farming. Rev. Aquac. 2022. Early View. [Google Scholar] [CrossRef]
- Kliestik, T.; Zvarikova, K.; Lăzăroiu, G. Data-driven machine learning and neural network algorithms in the retailing environment: Consumer engagement, experience, and purchase behaviors. Econ. Manag. Financ. Mark. 2022, 17, 57–69. [Google Scholar]
- Razminienė, K.; Vinogradova, I.; Tvaronavičienė, M. Clusters in transition to circular economy: Evaluation of relation. Acta Montan. Slovaca 2021, 26, 3. [Google Scholar] [CrossRef]
- Maroušek, J.; Maroušková, A.; Gavurová, B.; Tuček, D.; Strunecký, O. Competitive algae biodiesel depends on advances in mass algae cultivation. Bioresour. Technol. 2023, 374, 128802. [Google Scholar] [CrossRef]
- Hazeem, L.J.; Yesilay, G.; Bououdina, M.; Perna, S.; Cetin, D.; Suludere, Z.; Barras, A.; Boukherroub, R. Investigation of the toxic effects of different polystyrene micro-and nanoplastics on microalgae Chlorella vulgaris by analysis of cell viability, pigment content, oxidative stress and ultrastructural changes. Mar. Pollut. Bull. 2020, 156, 111278. [Google Scholar] [CrossRef]
- Xiao, Y.; Jiang, X.F.; Liao, Y.C.; Zhao, W.G.; Zhao, P.; Li, M. Adverse physiological and molecular level effects of polystyrene microplastics on freshwater microalgae. Chemosphere 2020, 255, 126914. [Google Scholar] [CrossRef]
- Campa-Córdova, A.; Luna-Gonzalez, A.; Ascencio, F.; Cortés-Jacinto, E.; Cáceres-Martínez, C. Effects of chloramphenicol, erythromycin, and furazolidone on growth of Isochrysis galbana and Chaetoceros gracilis. Aquaculture 2006, 260, 145–150. [Google Scholar] [CrossRef]
- Guschina, I.A.; Hayes, A.J.; Ormerod, S.J. Polystyrene microplastics decrease accumulation of essential fatty acids in common freshwater algae. Environ. Pollut. 2020, 263, 114425. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, N.; Ahmad, F.; Lu, Y.D. Synergy between microalgae and microbiome in polluted waters. Trends. Microbiol. 2022, 31, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.Y.; Kong, Z.Y.; Zhang, Y.F.; Ling, T.; Xu, J.L.; Liao, K.; Zhou, C.X.; Yan, X.J. Bacterial community diversity and screening of growth-affecting bacteria from Isochrysis galbana following antibiotic treatment. Front. Microbiol. 2019, 10, 994. [Google Scholar] [CrossRef]
- Jin, M.; Xiao, X.F.; Qin, L.G.; Geng, W.W.; Gao, Y.; Li, L.; Xue, J.L. Physiological and morphological responses and tolerance mechanisms of Isochrysis galbana to Cr(VI) stress. Bioresour. Technol. 2020, 302, 122860. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Qin, L.G.; Jin, M.; Zhang, L.L.; Geng, W.W.; Xiao, X.F. Extracellular adsorption, intracellular accumulation and tolerance mechanisms of Cyclotella sp. to Cr(VI) stress. Chemosphere 2021, 270, 128662. [Google Scholar] [CrossRef]
- Xia, J.H.; Xin, L.L.; Zhu, W.L.; Li, L.; Li, C.X.; Wang, Y.F.; Mu, Y.L.; Yang, S.L.; Li, K. Characterization of long non-coding RNA transcriptome in high-energy diet induced nonalcoholic steatohepatitis minipigs. Sci. Rep. 2016, 6, 30709. [Google Scholar] [CrossRef] [PubMed]
- Ling, T.; Zhang, Y.-F.; Cao, J.-Y.; Xu, J.-L.; Kong, Z.-Y.; Zhang, L.; Liao, K.; Zhou, C.-X.; Yan, X.-J. Analysis of bacterial community diversity within seven bait-microalgae. Algal. Res. 2020, 51, 102033. [Google Scholar] [CrossRef]
- Zhang, L.L.; Sun, C.M. Fengycins, cyclic lipopeptides from marine Bacillus subtilis strains, kill the plant-pathogenic fungus Magnaporthe grisea by inducing reactive oxygen species production and chromatin condensation. Appl. Environ. Microbiol. 2018, 84, e00445-00418. [Google Scholar] [CrossRef]
- Maroušek, J.; Strunecký, O.; Bartoš, V.; Vochozka, M. Revisiting competitiveness of hydrogen and algae biodiesel. Fuel 2022, 328, 125317. [Google Scholar] [CrossRef]
- Khan, A.A.; Quigley, J.G. Control of intracellular heme levels: Heme transporters and heme oxygenases. Biochim. Biophys. Acta 2011, 1813, 668–682. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ma, W.; Li, J.; Liu, W.; Qi, P.; Ye, Y.; Guo, B.; Zhang, J.; Qu, C. Effects of microplastics on growth, phenanthrene stress, and lipid accumulation in a diatom, Phaeodactylum tricornutum. Environ. Pollut. 2020, 257, 113628. [Google Scholar] [CrossRef] [PubMed]
- Sjollema, S.B.; Redondo-Hasselerharm, P.; Leslie, H.A.; Kraak, M.; Vethaak, A.D. Do plastic particles affect microalgal photosynthesis and growth? Aquat. Toxicol. 2015, 170, 259–261. [Google Scholar] [CrossRef]
- Nava, V.; Leoni, B. A critical review of interactions between microplastics, microalgae and aquatic ecosystem function. Water Res. 2021, 188, 116476. [Google Scholar] [CrossRef]
- Prata, J.C.; da Costa, J.P.; Lopes, I.; Duarte, A.C.; Rocha-Santos, T. Effects of microplastics on microalgae populations: A critical review. Sci. Total Environ. 2019, 665, 400–405. [Google Scholar] [CrossRef]
- Chae, Y.; Kim, D.; An, Y.J. Effects of micro-sized polyethylene spheres on the marine microalga Dunaliella salina: Focusing on the algal cell to plastic particle size ratio. Aquat. Toxicol. 2019, 216, 105296. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.; Fanning, K.; Netzel, M.; Turner, W.; Li, Y.; Schenk, P.M. Profiling of carotenoids and antioxidant capacity of microalgae from subtropical coastal and brackish waters. Food Chem. 2014, 165, 300–306. [Google Scholar] [CrossRef]
- Ansari, F.A.; Ratha, S.K.; Renuka, N.; Ramanna, L.; Bux, F. Effect of microplastics on growth and biochemical composition of microalga Acutodesmus obliquus. Algal. Res. 2021, 56, 102296. [Google Scholar] [CrossRef]
- Yang, W.F.; Gao, X.X.; Wu, Y.X.; Wan, L.; Tan, L.; Yuan, S.M.; Ding, H.J.; Zhang, W.H. The combined toxicity influence of microplastics and nonylphenol on microalgae Chlorella pyrenoidosa. Ecotoxicol. Environ. Saf. 2020, 195, 110484. [Google Scholar] [CrossRef]
- Song, C.; Liu, Z.; Wang, C.; Li, S.; Kitamura, Y. Different interaction performance between microplastics and microalgae: The bio-elimination potential of Chlorella sp. L38 and Phaeodactylum tricornutum MASCC-0025. Sci. Total Environ. 2020, 723, 138146. [Google Scholar] [CrossRef]
- Lin, W.; Su, F.; Lin, M.Z.; Jin, M.; Li, Y.H.; Ding, K.W.; Chen, Q.H.; Qian, Q.R.; Sun, X.L. Effect of microplastics PAN polymer and/or Cu(2+) pollution on the growth of Chlorella pyrenoidosa. Environ. Pollut. 2020, 265, 114985. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.L.; Zhang, K.D.; Zhou, Z.; Wang, J.R.; Yang, X.H.; Tang, J.; Li, H.F.; Lin, S. Microplastic exposure represses the growth of endosymbiotic dinoflagellate Cladocopium goreaui in culture through affecting its apoptosis and metabolism. Chemosphere 2020, 244, 125485. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Lv, L.; Liu, H.; Wang, M.; Sui, Y.; Wang, Y. Effects of ocean acidification and microplastics on microflora community composition in the digestive tract of the thick shell mussel Mytilus coruscus through 16s RNA gene sequencing. Bull. Environ. Contam. Toxicol. 2020, 107, 616–625. [Google Scholar] [CrossRef]
- Lupette, J.; Lami, R.; Krasovec, M.; Grimsley, N.; Moreau, H.; Piganeau, G.; Sanchez-Ferandin, S. Marinobacter dominates the bacterial community of the Ostreococcus tauri phycosphere in culture. Front. Microbiol. 2016, 7, 1414. [Google Scholar] [CrossRef]
- Cho, D.H.; Ramanan, R.; Heo, J.; Lee, J.; Kim, B.H.; Oh, H.M.; Kim, H.S. Enhancing microalgal biomass productivity by engineering a microalgal-bacterial community. Bioresour. Technol. 2015, 175, 578–585. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, X.; Li, W.; Li, S.; Zuo, X.; Liu, J.; Guo, L.; Lu, X.; Zhang, L. The Growth Inhibition of Polyethylene Nanoplastics on the Bait-Microalgae Isochrysis galbana Based on the Transcriptome Analysis. Microorganisms 2023, 11, 1108. https://doi.org/10.3390/microorganisms11051108
Xiao X, Li W, Li S, Zuo X, Liu J, Guo L, Lu X, Zhang L. The Growth Inhibition of Polyethylene Nanoplastics on the Bait-Microalgae Isochrysis galbana Based on the Transcriptome Analysis. Microorganisms. 2023; 11(5):1108. https://doi.org/10.3390/microorganisms11051108
Chicago/Turabian StyleXiao, Xinfeng, Wenfang Li, Shuangwei Li, Xingsheng Zuo, Jie Liu, Linke Guo, Xiao Lu, and Linlin Zhang. 2023. "The Growth Inhibition of Polyethylene Nanoplastics on the Bait-Microalgae Isochrysis galbana Based on the Transcriptome Analysis" Microorganisms 11, no. 5: 1108. https://doi.org/10.3390/microorganisms11051108
APA StyleXiao, X., Li, W., Li, S., Zuo, X., Liu, J., Guo, L., Lu, X., & Zhang, L. (2023). The Growth Inhibition of Polyethylene Nanoplastics on the Bait-Microalgae Isochrysis galbana Based on the Transcriptome Analysis. Microorganisms, 11(5), 1108. https://doi.org/10.3390/microorganisms11051108




