The Role of the PAA1 Gene on Melatonin Biosynthesis in Saccharomyces cerevisiae: A Search of New Arylalkylamine N-Acetyltransferases
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Media
2.2. Plasmid Construction
2.3. Bioconversion Assays
2.4. Drop Test
2.5. Gene Expression Analysis
2.6. Statistical Analysis
3. Results and Discussion
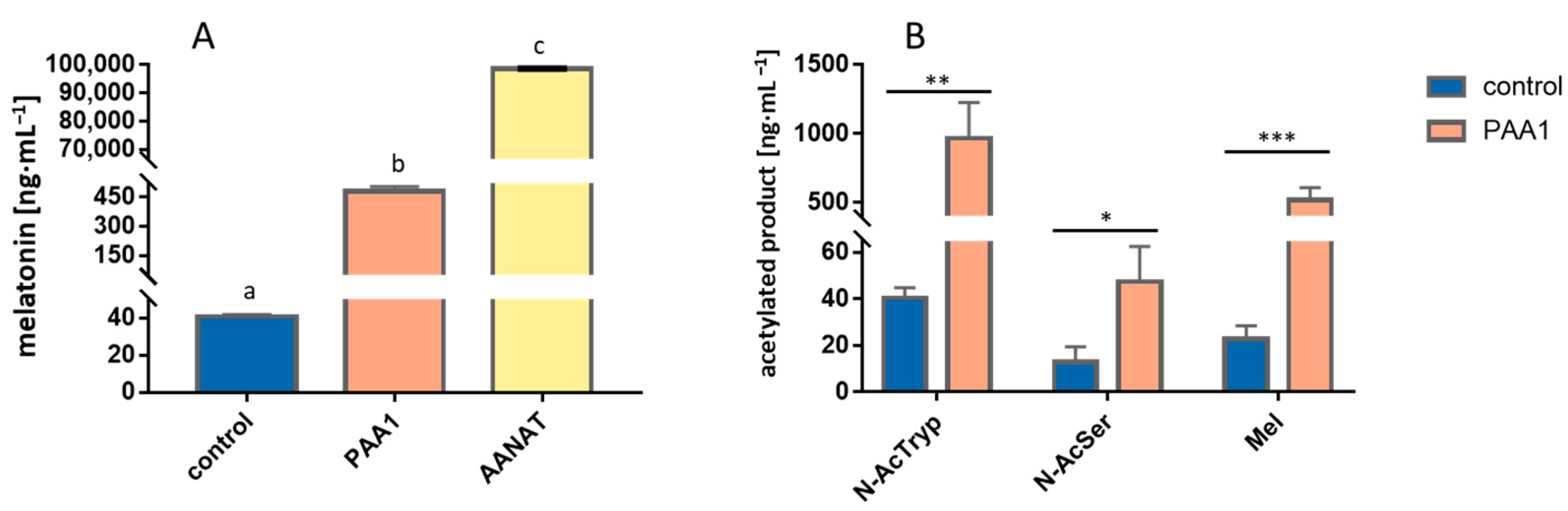
3.1. Overexpression of PAA1 in S. cerevisiae
3.2. Overexpression of PAA1 in E. coli
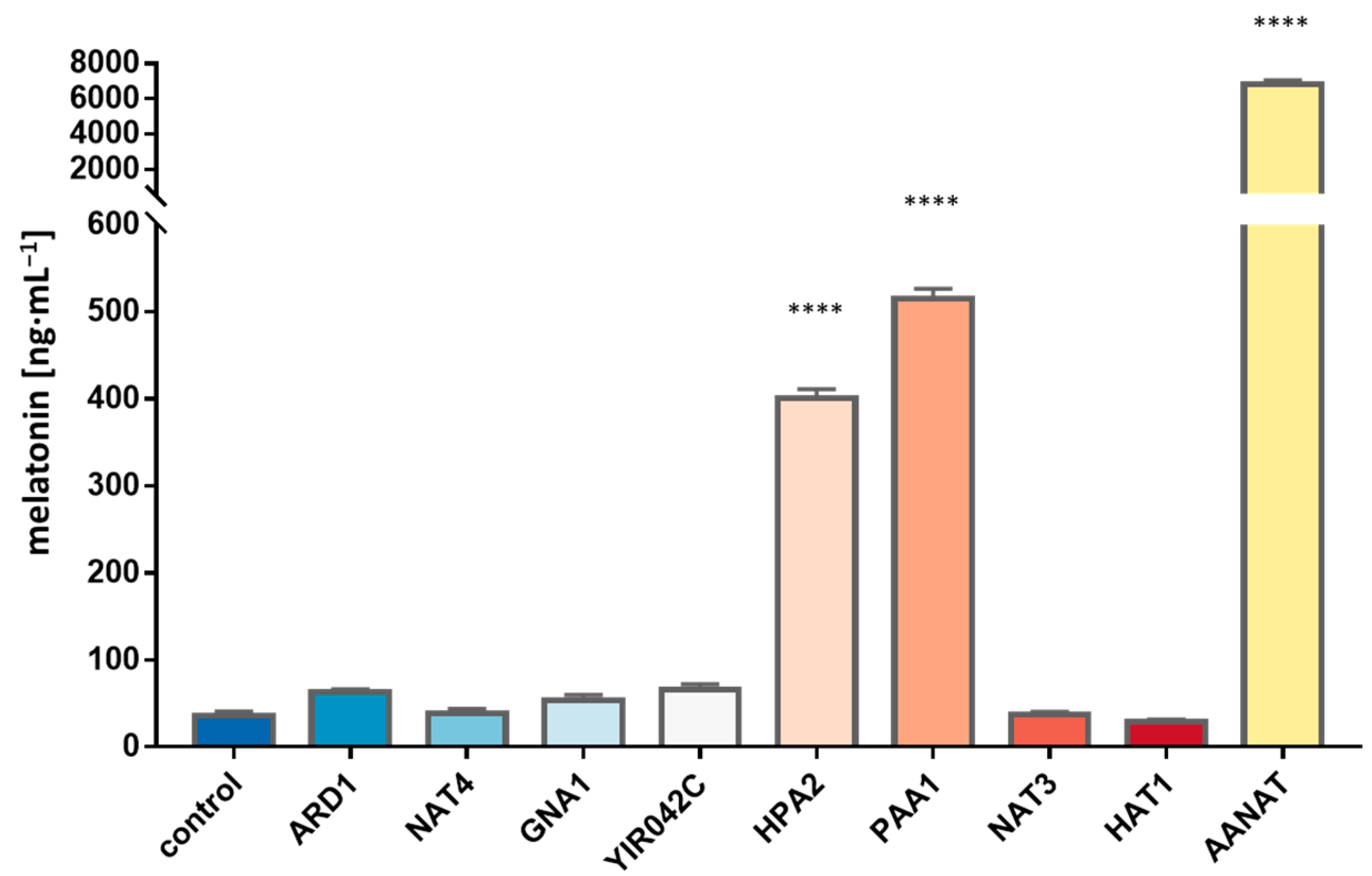
3.3. Search for New N-Acetyltransferases in S. cerevisiae
3.4. Detection of AANAT Activity in the New Gene Candidates by Overexpression in E. coli
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tan, D.X.; Hardeland, R.; Back, K.; Manchester, L.C.; Alatorre-Jimenez, M.A.; Reiter, R.J. On the significance of an alternate pathway of melatonin synthesis via 5-methoxytryptamine: Comparisons across species. J. Pineal Res. 2016, 61, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.X.; Manchester, L.C.; Esteban-Zubero, E.; Zhou, Z.; Reiter, R.J. Melatonin as a potent and inducible endogenous antioxidant: Synthesis and metabolism. Molecules 2015, 20, 18886–18906. [Google Scholar] [CrossRef]
- Sprenger, J.; Hardeland, R.; Fuhrberg, B. Melatonin and Other 5-Methoxylated Indoles in Yeast: Presence in High Concentrations and Dependence on Tryptophan Availability. Cytologia 1999, 64, 209–213. [Google Scholar] [CrossRef]
- Fernández-Pachón, M.S.; Medina, S.; Herrero-Martín, G.; Cerrillo, I.; Berná, G.; Escudero-Lõpez, B.; Ferreres, F.; Martín, F.; García-Parrilla, M.C.; Gil-Izquierdo, A. Alcoholic fermentation induces melatonin synthesis in orange juice. J. Pineal Res. 2014, 56, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Moreno, H.; Calvo, J.R.; Maldonado, M.D. High levels of melatonin generated during the brewing process. J. Pineal Res. 2013, 55, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Naranjo, M.I.; Gil-Izquierdo, A.; Troncoso, A.M.; Cantos-Villar, E.; García-Parrilla, M.C. Melatonin is synthesised by yeast during alcoholic fermentation in wines. Food Chem. 2011, 126, 1608–1613. [Google Scholar] [CrossRef]
- Vigentini, I.; Gardana, C.; Fracassetti, D.; Gabrielli, M.; Foschino, R.; Simonetti, P.; Tirelli, A.; Iriti, M. Yeast contribution to melatonin, melatonin isomers and tryptophan ethyl ester during alcoholic fermentation of grape musts. J. Pineal Res. 2015, 58, 388–396. [Google Scholar] [CrossRef]
- Reiter, R.J.; Tan, D.X.; Rosales-Corral, S.; Galano, A.; Zhou, X.J.; Xu, B. Mitochondria: Central organelles for melatonins antioxidant and anti-Aging actions. Molecules 2018, 23, 509. [Google Scholar] [CrossRef]
- Rosales-Corral, S.A.; Acuña-Castroviejo, D.; Coto-Montes, A.; Boga, J.A.; Manchester, L.C.; Fuentes-Broto, L.; Korkmaz, A.; Ma, S.; Tan, D.X.; Reiter, R.J. Alzheimer’s disease: Pathological mechanisms and the beneficial role of melatonin. J. Pineal Res. 2012, 52, 167–202. [Google Scholar] [CrossRef]
- Tan, D.X.; Hardeland, R.; Manchester, L.C.; Paredes, S.D.; Korkmaz, A.; Sainz, R.M.; Mayo, J.C.; Fuentes-Broto, L.; Reiter, R.J. The changing biological roles of melatonin during evolution: From an antioxidant to signals of darkness, sexual selection and fitness. Biol. Rev. 2010, 85, 607–623. [Google Scholar] [CrossRef]
- Bisquert, R.; Muñiz-Calvo, S.; Guillamón, J.M. Protective role of intracellular Melatonin against oxidative stress and UV radiation in Saccharomyces cerevisiae. Front. Microbiol. 2018, 9, 318. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, J.; González, B.; Sempere, V.; Mas, A.; Torija, M.J.; Beltran, G. Melatonin reduces oxidative stress damage induced by hydrogen peroxide in Saccharomyces cerevisiae. Front. Microbiol. 2017, 8, 1066. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, J.; Grillitsch, K.; Daum, G.; Mas, A.; Torija, M.J.; Beltran, G. Melatonin minimizes the impact of oxidative stress induced by hydrogen peroxide in Saccharomyces and Non-conventional yeast. Front. Microbiol. 2018, 9, 1933. [Google Scholar] [CrossRef]
- Muñiz-Calvo, S.; Bisquert, R.; Fernández-Cruz, E.; García-Parrilla, M.C.; Guillamón, J.M. Deciphering the melatonin metabolism in Saccharomyces cerevisiae by the bioconversion of related metabolites. J. Pineal Res. 2019, 66, e12554. [Google Scholar] [CrossRef]
- Ganguly, S.; Mummaneni, P.; Steinbach, P.J.; Klein, D.C.; Coon, S.L. Characterization of the Saccharomyces cerevisiae Homolog of the Melatonin Rhythm Enzyme Arylalkylamine N-Acetyltransferase (EC 2.3.1.87). J. Biol. Chem. 2001, 276, 47239–47247. [Google Scholar] [CrossRef] [PubMed]
- Germann, S.M.; Baallal Jacobsen, S.A.; Schneider, K.; Harrison, S.J.; Jensen, N.B.; Chen, X.; Stahlhut, S.G.; Borodina, I.; Luo, H.; Zhu, J.; et al. Glucose-based microbial production of the hormone melatonin in yeast Saccharomyces cerevisiae. Biotechnol. J. 2016, 11, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Mumberg, D.; Müller, R.; Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 1995, 156, 119–122. [Google Scholar] [CrossRef]
- Liu, B.; Sutton, A.; Sternglanz, R. A yeast polyamine acetyltransferase. J. Biol. Chem. 2005, 280, 16659–16664. [Google Scholar] [CrossRef]
- Neuwald, A.F.; Landsman, D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 1997, 22, 154–155. [Google Scholar] [CrossRef]
- Reiter, R.J.; Mayo, J.C.; Tan, D.X.; Sainz, R.M.; Alatorre-Jimenez, M.; Qin, L. Melatonin as an antioxidant: Under promises but over delivers. J. Pineal Res. 2016, 253–278. [Google Scholar] [CrossRef]
- Rodriguez, C.; Mayo, J.C.; Sainz, R.M.; Antolín, I.; Herrera, F.; Martín, V.; Reiter, R.J. Regulation of antioxidant enzymes: A significant role for melatonin. J. Pineal Res. 2004, 36, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Galano, A.; Medina, M.E.; Tan, D.X.; Reiter, R.J. Melatonin and its metabolites as copper chelating agents and their role in inhibiting oxidative stress: A physicochemical analysis. J. Pineal Res. 2015, 58, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Lv, Y.; Shi, Y.; Li, T.; Chen, Y.; Zhao, D.; Zhao, Z. The role of phyto-melatonin and related metabolites in response to stress. Molecules 2018, 23, 1887. [Google Scholar] [CrossRef] [PubMed]
- Mulder, N.J.; Kersey, P.; Pruess, M.; Apweiler, R. In silico characterization of proteins: UniProt, InterPro and Integr8. Mol. Biotechnol. 2008, 38, 165–177. [Google Scholar] [CrossRef] [PubMed]



| Name | Relevant Traits | Integration Site/Replicon | Marker | Source |
|---|---|---|---|---|
| pGEX-5X-1 | Taq promoter-GST-MCS | pBR322 (ColE1) | Amp | GE Healthcare |
| pGEX-5X-1 PAA1 | Taq promoter-GST-PAA1 | ColE1 | Amp | This study |
| pGEX-5X-1 AANAT | Taq promoter-GST-BtAANAT | ColE1 | Amp | This study |
| pGEX-5X-1 ARD1 | Taq promoter-GST-ARD1 | ColE1 | Amp | This study |
| pGEX-5X-1 NAT4 | Taq promoter-GST-NAT4 | ColE1 | Amp | This study |
| pGEX-5X-1 GNA1 | Taq promoter-GST-GNA1 | ColE1 | Amp | This study |
| pGEX-5X-1 YIR042C | Taq promoter-GST-YIR042C | ColE1 | Amp | This study |
| pGEX-5X-1 HPA2 | Taq promoter-GST-HPA2 | ColE1 | Amp | This study |
| pGEX-5X-1 NAT3 | Taq promoter-GST-NAT3 | ColE1 | Amp | This study |
| pGEX-5X-1 HAT1 | Taq promoter-GST-HAT1 | ColE1 | Amp | This study |
| p426GPD | TDH3p-MCS-CYC1t | 2μ | URA3 | [17] |
| pCfB2628 | TEFp-HsDDC, PGK1p-BtAANAT | XI-5 | HIS5 | [16] |
| p426GPD PAA1 | TDH3p-PAA1-CYC1t | 2μ | URA3 | This study |
| p426GPD AANAT | TDH3p-BtAANAT-CYC1t | 2μ | URA3 | This study |
| Title 1 | Title 2 |
|---|---|
| PAA1 F BamHI | AGGTCGTGGGATCCCCATGGCCTCCTCAAGTAGCA |
| PAA1 R XhoI | TGCGGCCGCTCGAGCTAGTTGTCGTATTCTTCCTTAAT |
| AANAT F BamHI | AGGTCGTGGGATCCCCATGAGCACCCCGAGCAT |
| AANAT R XhoI | TGCGGCCGCTCGAGTTAACGATCGCTATTACGACGCA |
| GPD Pro F | CGGTAGGTATTGATTGTAATTCTG |
| CYC1-R | GCGTGAATGTAAGCGTGAC |
| pGEX seq F | GGGCTGGCAAGCCACGTTTGGTG |
| pGEX seq R | CCGGGAGCTGCATGTGTCAGAGG |
| PAA1 (qPCR) F | GGTTTCCCACCAAACGAAAG |
| PAA1 (qPCR) R | ACTTCTTTGCCCTCGATCTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bisquert, R.; Planells-Cárcel, A.; Alonso-del-Real, J.; Muñiz-Calvo, S.; Guillamón, J.M. The Role of the PAA1 Gene on Melatonin Biosynthesis in Saccharomyces cerevisiae: A Search of New Arylalkylamine N-Acetyltransferases. Microorganisms 2023, 11, 1115. https://doi.org/10.3390/microorganisms11051115
Bisquert R, Planells-Cárcel A, Alonso-del-Real J, Muñiz-Calvo S, Guillamón JM. The Role of the PAA1 Gene on Melatonin Biosynthesis in Saccharomyces cerevisiae: A Search of New Arylalkylamine N-Acetyltransferases. Microorganisms. 2023; 11(5):1115. https://doi.org/10.3390/microorganisms11051115
Chicago/Turabian StyleBisquert, Ricardo, Andrés Planells-Cárcel, Javier Alonso-del-Real, Sara Muñiz-Calvo, and José Manuel Guillamón. 2023. "The Role of the PAA1 Gene on Melatonin Biosynthesis in Saccharomyces cerevisiae: A Search of New Arylalkylamine N-Acetyltransferases" Microorganisms 11, no. 5: 1115. https://doi.org/10.3390/microorganisms11051115
APA StyleBisquert, R., Planells-Cárcel, A., Alonso-del-Real, J., Muñiz-Calvo, S., & Guillamón, J. M. (2023). The Role of the PAA1 Gene on Melatonin Biosynthesis in Saccharomyces cerevisiae: A Search of New Arylalkylamine N-Acetyltransferases. Microorganisms, 11(5), 1115. https://doi.org/10.3390/microorganisms11051115







