Mucosal Microbiota from Colorectal Cancer, Adenoma and Normal Epithelium Reveals the Imprint of Fusobacterium nucleatum in Cancerogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. Laboratory Procedures
2.3. Quantitative Real-Time PCR (RT-qPCR)
2.4. Bioinformatics and Statistical Data Analysis
3. Results
3.1. Clinicopathologic Characteristics and Diversity Analysis
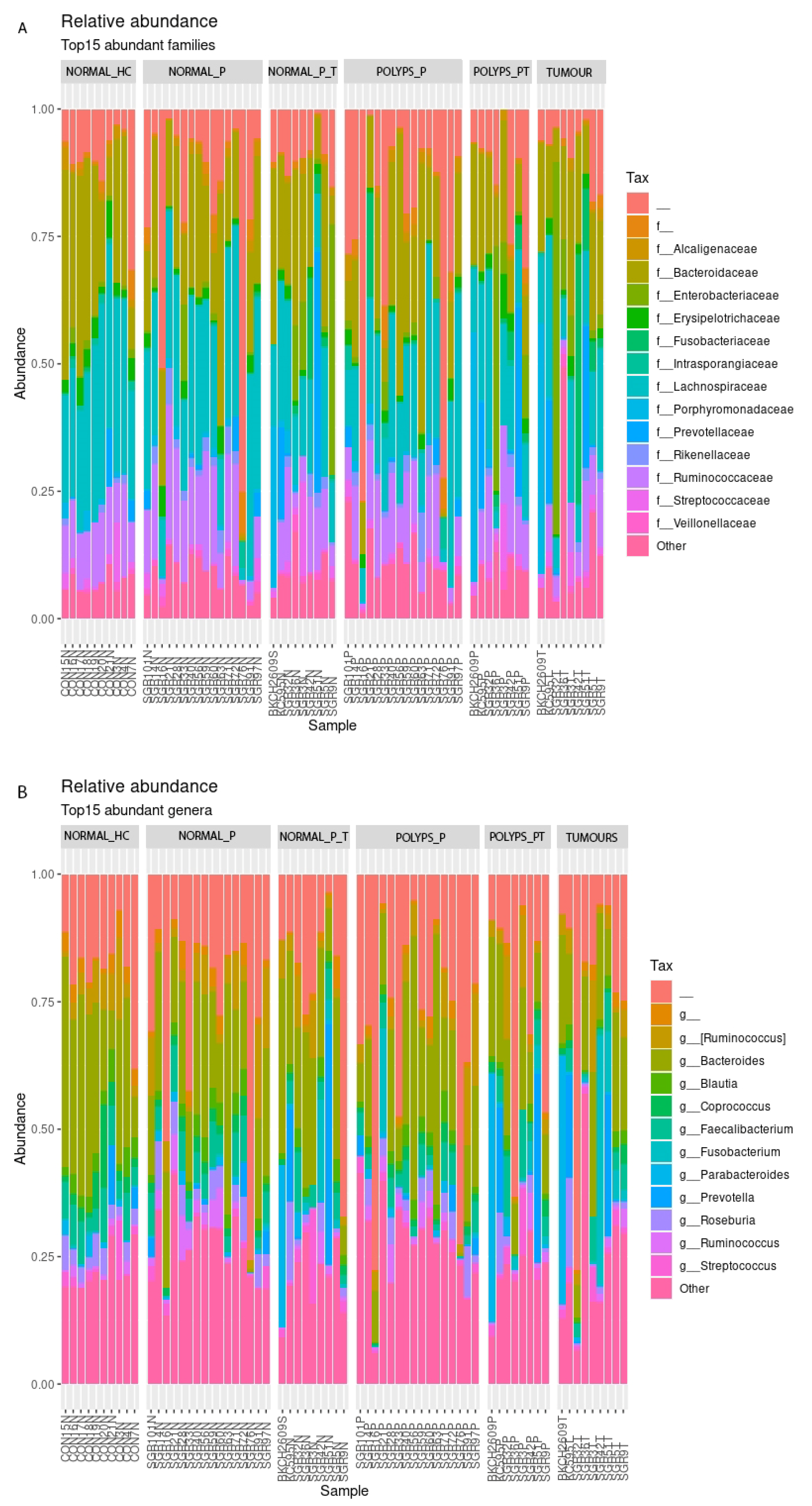
3.2. 16 S rRNA V3-V4 Region Sequencing
3.3. Microbial Diversity and Community Analyses
3.4. Discriminant Taxa from Pairwise Group Comparisons
3.5. Differential Analysis of Inferred Microbial Pathways
3.6. Fusobacterium Nucleatum Detection and Quantification
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Lichtenstein, P.; Holm, N.V.; Verkasalo, P.K.; Iliadou, A.; Kaprio, J.; Koskenvuo, M.; Pukkala, E.; Skytthe, A.; Hemminki, K. Environmental and heritable factors in the causation of cancer–Analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000, 343, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Jasperson, K.W.; Tuohy, T.M.; Neklason, D.W.; Burt, R.W. Hereditary and familial colon cancer. Gastroenterology 2010, 138, 2044–2058. [Google Scholar] [CrossRef] [PubMed]
- Garrett, W.S. Immune recognition of microbial metabolites. Nat. Rev. Immunol. 2020, 20, 91–92. [Google Scholar] [CrossRef]
- Chapelle, N.; Martel, M.; Toes-Zoutendijk, E.; Barkun, A.N.; Bardou, M. Recent advances in clinical practice: Colorectal cancer chemoprevention in the average-risk population. Gut 2020, 69, 2244–2255. [Google Scholar] [CrossRef]
- Wong, S.H.; Yu, J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 690–704. [Google Scholar] [CrossRef]
- Flemer, B.; Lynch, D.B.; Brown, J.M.; Jeffery, I.B.; Ryan, F.J.; Claesson, M.J.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 2017, 66, 633–643. [Google Scholar] [CrossRef]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef]
- Wu, S.; Rhee, K.J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F.; et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef]
- Matson, V.; Chervin, C.S.; Gajewski, T.F. Cancer and the Microbiome-Influence of the Commensal Microbiota on Cancer, Immune Responses, and Immunotherapy. Gastroenterology 2021, 160, 600–613. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.A.; Garrett, W.S. Fusobacterium nucleatum–symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef] [PubMed]
- 16S Metagenomic Sequencing Library Preparation. 2013. Available online: https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 12 March 2023).
- Estaki, M.; Jiang, L.; Bokulich, N.A.; McDonald, D.; González, A.; Kosciolek, T.; Martino, C.; Zhu, Q.; Birmingham, A.; Vázquez-Baeza, Y.; et al. QIIME 2 Enables Comprehensive End-to-End Analysis of Diverse Microbiome Data and Comparative Studies with Publicly Available Data. Curr. Protoc. Bioinform. 2020, 70, e100. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Shannon, C.E.; Weaver, W.; Wiener, N. The Mathematical Theory of Communication. Phys. Today 1950, 3, 31–32. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Pielou, E.C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Jaccard, P. Nouvelles recherches sur la distribution florale. Bull. De La Soc. Vaud. Des Sci. Nat. 1908, 44, 223. [Google Scholar]
- Sørensen, T.; Sørensen, T. A Method of Establishing Groups of Equal Amplitude in Plant Sociology Based on Similarity of Species Content and Ist Application to Analyses of the Vegetation on Danish Commons; I kommission hos E. Munksgaard: Copenhagen, Denmark, 1948. [Google Scholar]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. Gigascience 2013, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Shi, P.; Li, H. Generalized linear models with linear constraints for microbiome compositional data. Biometrics 2019, 75, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. Huttenhower C, Langille MGI. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc database of metabolic pathways and enzymes–A 2019 update. Nucleic Acids Res. 2020, 48, D445–D453. [Google Scholar] [CrossRef]
- Lahti, L.; Shetty, S.; Blake, T. Microbiome R Package; 2019. Available online: http://microbiome.github.io (accessed on 12 March 2023).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, NY, USA, 2016. [Google Scholar]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- O’Brien, M.J.; Winawer, S.J.; Zauber, A.G.; Gottlieb, L.S.; Sternberg, S.S.; Diaz, B.; Dickersin, G.R.; Ewing, S.; Geller, S.; Kasimian, D.; et al. The National Polyp Study. Patient and polyp characteristics associated with high-grade dysplasia in colorectal adenomas. Gastroenterology 1990, 98, 371–379. [Google Scholar]
- Song, M.; Chan, A.T.; Sun, J. Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer. Gastroenterology 2020, 158, 322–340. [Google Scholar] [CrossRef]
- Saus, E.; Iraola-Guzmán, S.; Willis, J.R.; Brunet-Vega, A.; Gabaldón, T. Microbiome and colorectal cancer: Roles in carcinogenesis and clinical potential. Mol. Asp. Med. 2019, 69, 93–106. [Google Scholar] [CrossRef]
- Costa, C.P.D.; Vieira, P.; Mendes-Rocha, M.; Pereira-Marques, J.; Ferreira, R.M.; Figueiredo, C. The Tissue-Associated Microbiota in Colorectal Cancer: A Systematic Review. Cancers 2022, 14, 3385. [Google Scholar] [CrossRef] [PubMed]
- Chénard, T.; Malick, M.; Dubé, J.; Massé, E. The influence of blood on the human gut microbiome. BMC Microbiol. 2020, 20, 44. [Google Scholar] [CrossRef] [PubMed]
- Zwinsová, B.; Petrov, V.A.; Hrivňáková, M.; Smatana, S.; Micenková, L.; Kazdová, N.; Popovici, V.; Hrstka, R.; Šefr, R.; Bencsiková, B. Colorectal Tumour Mucosa Microbiome Is Enriched in Oral Pathogens and Defines Three Subtypes That Correlate with Markers of Tumour Progression. Cancers 2021, 13, 4799. [Google Scholar] [CrossRef]
- Mira-Pascual, L.; Cabrera-Rubio, R.; Ocon, S.; Costales, P.; Parra, A.; Suarez, A.; Moris, F.; Rodrigo, L.; Mira, A.; Collado, M.C. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J. Gastroenterol. 2015, 50, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Loke, M.F.; Chua, E.G.; Gan, H.M.; Thulasi, K.; Wanyiri, J.W.; Thevambiga, I.; Goh, K.L.; Wong, W.F.; Vadivelu, J. Metabolomics and 16S rRNA sequencing of human colorectal cancers and adjacent mucosa. PLoS ONE 2018, 13, e0208584. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, Y.; Shao, L.; Ling, Z. Alterations of the Predominant Fecal Microbiota and Disruption of the Gut Mucosal Barrier in Patients with Early-Stage Colorectal Cancer. Biomed. Res. Int. 2020, 2020, 2948282. [Google Scholar] [CrossRef]
- de Carvalho, A.C.; de Mattos Pereira, L.; Datorre, J.G.; Dos Santos, W.; Berardinelli, G.N.; Matsushita, M.M.; Oliveira, M.A.; Durães, R.O.; Guimarães, D.P.; Reis, R.M. Microbiota Profile and Impact of Fusobacterium nucleatum in Colorectal Cancer Patients of Barretos Cancer Hospital. Front. Oncol. 2019, 9, 813. [Google Scholar] [CrossRef]
- Gao, R.; Kong, C.; Huang, L.; Li, H.; Qu, X.; Liu, Z.; Lan, P.; Wang, J.; Qin, H. Mucosa-associated microbiota signature in colorectal cancer. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 2073–2083. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, J.; Zheng, J.; Hu, G.; Wang, J.; Huang, C.; Lou, L.; Wang, X.; Zeng, Y. Mucosal adherent bacterial dysbiosis in patients with colorectal adenomas. Sci. Rep. 2016, 6, 26337. [Google Scholar] [CrossRef]
- Sun, T.; Liu, S.; Zhou, Y.; Yao, Z.; Zhang, D.; Cao, S.; Wei, Z.; Tan, B.; Li, Y.; Lian, Z.; et al. Evolutionary biologic changes of gut microbiota in an ‘adenoma-carcinoma sequence’ mouse colorectal cancer model induced by 1, 2-Dimethylhydrazine. Oncotarget 2017, 8, 444–457. [Google Scholar] [CrossRef]
- Pignatelli, P.; Iezzi, L.; Pennese, M.; Raimondi, P.; Cichella, A.; Bondi, D.; Grande, R.; Cotellese, R.; Di Bartolomeo, N.; Innocenti, P. The Potential of Colonic Tumor Tissue Fusobacterium nucleatum to Predict Staging and Its Interplay with Oral Abundance in Colon Cancer Patients. Cancers 2021, 13, 1032. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, Y.; Li, J.; Zhao, L.; Yan, W.; Lin, B.; Guo, X.; Wei, Y. Fusobacterium nucleatum Acts as a Pro-carcinogenic Bacterium in Colorectal Cancer: From Association to Causality. Front. Cell. Dev. Biol. 2021, 9, 710165. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Kanno, S.; Nosho, K.; Sukawa, Y.; Mitsuhashi, K.; Kurihara, H.; Igarashi, H.; Takahashi, T.; Tachibana, M.; Takahashi, H.; et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int. J. Cancer 2015, 137, 1258–1268. [Google Scholar] [CrossRef]
- Xia, X.; Wu, W.K.K.; Wong, S.H.; Liu, D.; Kwong, T.N.Y.; Nakatsu, G.; Yan, P.S.; Chuang, Y.M.; Chan, M.W.; Coker, O.O.; et al. Bacteria pathogens drive host colonic epithelial cell promoter hypermethylation of tumor suppressor genes in colorectal cancer. Microbiome 2020, 8, 108. [Google Scholar] [CrossRef]
- Mima, K.; Nishihara, R.; Qian, Z.R.; Cao, Y.; Sukawa, Y.; Nowak, J.A.; Yang, J.; Dou, R.; Masugi, Y.; Song, M.; et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016, 65, 1973–1980. [Google Scholar] [CrossRef]
- Borozan, I.; Zaidi, S.H.; Harrison, T.A.; Phipps, A.I.; Zheng, J.; Lee, S.; Trinh, Q.M.; Steinfelder, R.S.; Adams, J.; Banbury, B.L.; et al. Molecular and Pathology Features of Colorectal Tumors and Patient Outcomes Are Associated with Fusobacterium nucleatum and Its Subspecies animalis. Cancer Epidemiol. Biomark. Prev. 2022, 31, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Serna, G.; Ruiz-Pace, F.; Hernando, J.; Alonso, L.; Fasani, R.; Landolfi, S.; Comas, R.; Jimenez, J.; Elez, E.; Bullman, S.; et al. Fusobacterium nucleatum persistence and risk of recurrence after preoperative treatment in locally advanced rectal cancer. Ann. Oncol. 2020, 31, 1366–1375. [Google Scholar] [CrossRef]
- Peng, B.J.; Cao, C.Y.; Li, W.; Zhou, Y.J.; Zhang, Y.; Nie, Y.Q.; Cao, Y.W.; Li, Y.Y. Diagnostic Performance of Intestinal Fusobacterium nucleatum in Colorectal Cancer: A Meta-Analysis. Chin. Med. J. 2018, 131, 1349–1356. [Google Scholar] [CrossRef]
- Sze, M.A.; Schloss, P.D. Leveraging Existing 16S rRNA Gene Surveys To Identify Reproducible Biomarkers in Individuals with Colorectal Tumors. mBio 2018, 9, e00630-18. [Google Scholar] [CrossRef]
- Zhong, X.; Wang, Y.; Xu, J.; Cao, H.; Zhang, F.; Wang, X. Gut microbiota signatures in tissues of the colorectal polyp and normal colorectal mucosa, and faeces. Front. Cell. Infect. Microbiol. 2023, 12, 1054808. [Google Scholar] [CrossRef]
- Datorre, J.G.; de Carvalho, A.C.; Dos Reis, M.B.; Dos Reis, M.; Matsushita, M.; Santos, F.; Guimarães, D.P.; Reis, R.M. Accuracy and Clinical Relevance of Intra-Tumoral Fusobacterium nucleatum Detection in Formalin-Fixed Paraffin-Embedded (FFPE) Tissue by Droplet Digital PCR (ddPCR) in Colorectal Cancer. Diagnostics 2022, 12, 114. [Google Scholar] [CrossRef]
- Allali, I.; Delgado, S.; Marron, P.I.; Astudillo, A.; Yeh, J.J.; Ghazal, H.; Amzazi, S.; Keku, T.; Azcarate-Peril, M.A. Gut microbiome compositional and functional differences between tumor and non-tumor adjacent tissues from cohorts from the US and Spain. Gut Microbes 2015, 6, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Pastor, J.M.; Salvador, M.; Argandoña, M.; Bernal, V.; Reina-Bueno, M.; Csonka, L.N.; Iborra, J.L.; Vargas, C.; Nieto, J.J.; Cánovas, M. Ectoines in cell stress protection: Uses and biotechnological production. Biotechnol. Adv. 2010, 28, 782–801. [Google Scholar] [CrossRef] [PubMed]
- Castro-Ochoa, K.F.; Vargas-Robles, H.; Chánez-Paredes, S.; Felipe-López, A.; Cabrera-Silva, R.I.; Shibayama, M.; Betanzos, A.; Nava, P.; Galinski, E.A.; Schnoor, M. Homoectoine Protects Against Colitis by Preventing a Claudin Switch in Epithelial Tight Junctions. Dig. Dis. Sci. 2019, 64, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Rani, A.; Saini, K.C.; Bast, F.; Mehariya, S.; Bhatia, S.K.; Lavecchia, R.; Zuorro, A. Microorganisms: A Potential Source of Bioactive Molecules for Antioxidant Applications. Molecules 2021, 26, 1142. [Google Scholar] [CrossRef]

| Characteristics | Healthy Controls | Adenomatous Polyps | CRC and Synchronous Polyps | |
|---|---|---|---|---|
| Individuals | total number | 10 | 16 | 9 |
| ratio of females to males | 5/5 | 3/13 | 2/7 | |
| Age, years | median | 47 | 70 | 66 |
| range | 27–66 | 41–82 | 51–89 | |
| Age of diagnosis | early (<50) | - | 2 | 0 |
| late (>50) | - | 14 | 9 | |
| Polyp localization | proximal to splenic flexure | - | 5 | 3 |
| distal to splenic flexure | - | 11 | 6 | |
| Cancer localization | proximal to splenic flexure | - | - | 3 |
| distal to splenic flexure | - | - | 6 | |
| Concordance localization of cancer and polyp | proximal to splenic flexure | - | - | 6/6 |
| distal to splenic flexure | - | - | 3/3 | |
| Polyp dysplasia | low grade | - | 6 | 4 |
| high grade | - | 10 | 5 | |
| Features of advanced adenoma (AA) | ratio of AA to total | 14/16 | 6/9 | |
| TNM stage | I | - | - | 0 |
| II | - | - | 3 | |
| III | - | - | 4 | |
| IV | - | - | 1 | |
| NA | - | - | 1 | |
| Grading (WHO) | G1 | - | - | 2 |
| G2 | - | - | 6 | |
| G3 | - | - | 0 | |
| NA | - | - | 1 |
| Healthy Colon | |||||
|---|---|---|---|---|---|
| vs. | |||||
| NORMAL_P_T | POLYPS_P_T | TUMOUR | NORMAL_P | POLYPS_P | |
| Pielou’s evenness | 0.478 | 0.478 | 0.236 | 0.833 | 0.486 |
| Faith’s Phylogenetic Distance | 0.723 | 0.615 | 0.932 | 0.616 | 0.723 |
| Number of observed Features | 0.472 | 0.382 | 0.490 | 0.382 | 0.472 |
| Shannon’s Entropy | 1 | 0.481 | 0.481 | 0.237 | 0.583 |
| Healthy Colon | |||||
|---|---|---|---|---|---|
| vs. | |||||
| NORMAL_P_T | POLYPS_P_T | TUMOR | NORMAL_P | POLYPS_P | |
| Bray–Curtis dissimilarity | 0.050 | 0.060 | 0.030 | 0.126 | 0.063 |
| Jaccard similarity | 0.039 | 0.015 | 0.020 | 0.026 | 0.015 |
| Unweighted UniFrac dissimilarity | 0.461 | 0.09 | 0.324 | 0.324 | 0.324 |
| Weighted UniFrac dissimilarity | 0.531 | 0.531 | 0.531 | 0.556 | 0.531 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palmieri, O.; Castellana, S.; Latiano, A.; Latiano, T.; Gentile, A.; Panza, A.; Nardella, M.; Ciardiello, D.; Latiano, T.P.; Corritore, G.; et al. Mucosal Microbiota from Colorectal Cancer, Adenoma and Normal Epithelium Reveals the Imprint of Fusobacterium nucleatum in Cancerogenesis. Microorganisms 2023, 11, 1147. https://doi.org/10.3390/microorganisms11051147
Palmieri O, Castellana S, Latiano A, Latiano T, Gentile A, Panza A, Nardella M, Ciardiello D, Latiano TP, Corritore G, et al. Mucosal Microbiota from Colorectal Cancer, Adenoma and Normal Epithelium Reveals the Imprint of Fusobacterium nucleatum in Cancerogenesis. Microorganisms. 2023; 11(5):1147. https://doi.org/10.3390/microorganisms11051147
Chicago/Turabian StylePalmieri, Orazio, Stefano Castellana, Anna Latiano, Tiziana Latiano, Annamaria Gentile, Anna Panza, Marianna Nardella, Davide Ciardiello, Tiziana Pia Latiano, Giuseppe Corritore, and et al. 2023. "Mucosal Microbiota from Colorectal Cancer, Adenoma and Normal Epithelium Reveals the Imprint of Fusobacterium nucleatum in Cancerogenesis" Microorganisms 11, no. 5: 1147. https://doi.org/10.3390/microorganisms11051147
APA StylePalmieri, O., Castellana, S., Latiano, A., Latiano, T., Gentile, A., Panza, A., Nardella, M., Ciardiello, D., Latiano, T. P., Corritore, G., Mazza, T., Perri, F., & Biscaglia, G. (2023). Mucosal Microbiota from Colorectal Cancer, Adenoma and Normal Epithelium Reveals the Imprint of Fusobacterium nucleatum in Cancerogenesis. Microorganisms, 11(5), 1147. https://doi.org/10.3390/microorganisms11051147









