Properties of Multidrug-Resistant Mutants Derived from Heterologous Expression Chassis Strain Streptomyces albidoflavus J1074
Abstract
1. Introduction
2. Materials and Methods
2.1. General Experimental Procedures
2.2. Analysis of Specialized Metabolome of S. albidoflavus Strains under Submerged Conditions
2.3. Sequencing and Analysis of Genomes of Selected Mutants
2.4. Cloning of rpoB Alleles
3. Results
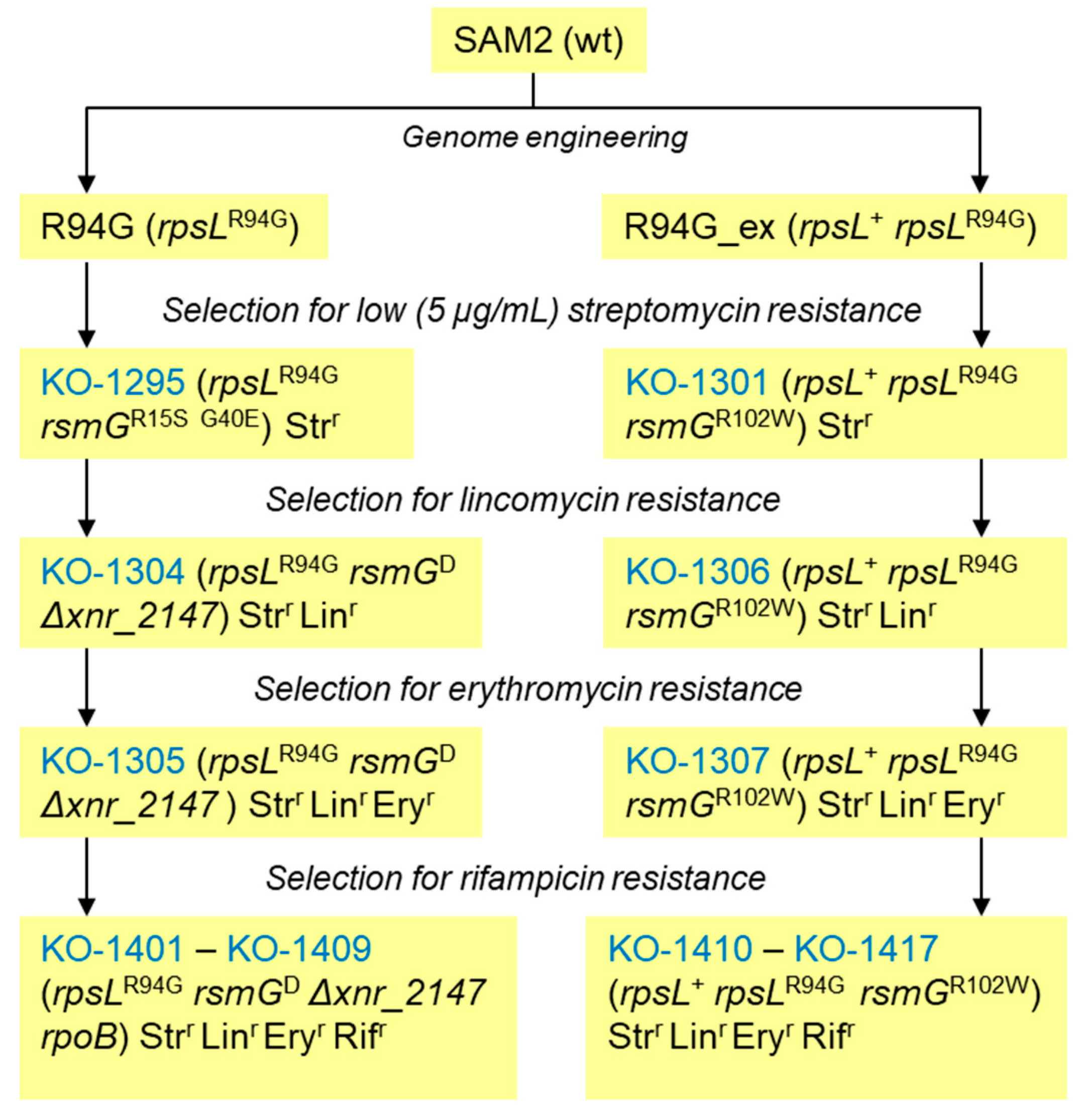
3.1. Generation and Analysis of Erythromycin Resistant Strains KO-1305 and KO-1307
3.2. Antibiotic Resistance and Bioactivities of Rifampicin-Resistant Derivatives of KO-1305 and KO-1307
3.3. Heterologous Expression of Aranciamycin BGC in Selected Mutant Strains
3.4. Genomics of Strains KO-1403, KO-1407, KO-1408, KO-1412
3.5. Construction and Studies of S. albidoflavus rpoB Merodiploids
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Myronovskyi, M.; Luzhetskyy, A. Heterologous production of small molecules in the optimized Streptomyces hosts. Nat. Prod. Rep. 2019, 36, 1281–1294. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, H.; Low-Beinart, L.; Obiajulu, J.; Brady, S.F. Natural product discovery through improved functional metagenomics in Streptomyces. J. Am. Chem. Soc. 2016, 138, 9341–9344. [Google Scholar] [CrossRef] [PubMed]
- Shuai, H.; Myronovskyi, M.; Nadmid, S.; Luzhetskyy, A. Identification of a biosynthetic gene cluster responsible for the production of a new pyrrolopyrimidine natural product—Huimycin. Biomolecules 2020, 10, 1074. [Google Scholar] [CrossRef] [PubMed]
- Lopatniuk, M.; Myronovskyi, M.; Luzhetskyy, A. Streptomyces albus: A new cell factory for non-canonical amino acids incorporation into ribosomally synthesized natural products. ACS Chem. Biol. 2017, 12, 2362–2370. [Google Scholar] [CrossRef]
- Horbal, L.; Marques, F.; Nadmid, S.; Mendes, M.V.; Luzhetskyy, A. Secondary metabolites overproduction through transcriptional gene cluster refactoring. Metab. Eng. 2018, 49, 299–315. [Google Scholar] [CrossRef] [PubMed]
- Horbal, L.; Siegl, T.; Luzhetskyy, A. A set of synthetic versatile genetic control elements for the efficient expression of genes in Actinobacteria. Sci. Rep. 2018, 8, 491. [Google Scholar] [CrossRef] [PubMed]
- Myronovskyi, M.; Rosenkränzer, B.; Nadmid, S.; Pujic, P.; Normand, P.; Luzhetskyy, A. Generation of a cluster-free Streptomyces albus chassis strains for improved heterologous expression of secondary metabolite clusters. Metab. Eng. 2018, 49, 316–324. [Google Scholar] [CrossRef]
- Kallifidas, D.; Jiang, G.; Ding, Y.; Luesch, H. Rational engineering of Streptomyces albus J1074 for the overexpression of secondary metabolite gene clusters. Microb. Cell Fact. 2018, 17, 25. [Google Scholar] [CrossRef]
- Lopatniuk, M.; Myronovskyi, M.; Nottebrock, A.; Busche, T.; Kalinowski, J.; Ostash, B.; Fedorenko, V.; Luzhetskyy, A. Effect of “ribosome engineering” on the transcription level and production of S. albus indigenous secondary metabolites. Appl. Microbiol. Biotechnol. 2019, 103, 7097–7110. [Google Scholar] [CrossRef]
- Koshla, O.; Lopatniuk, M.; Borys, O.; Misaki, Y.; Kravets, V.; Ostash, I.; Shemediuk, A.; Ochi, K.; Luzhetskyy, A.; Fedorenko, V.; et al. Genetically engineered rpsL merodiploidy impacts secondary metabolism and antibiotic resistance in Streptomyces. World J. Microbiol. Biotechnol. 2021, 37, 62. [Google Scholar] [CrossRef]
- Zhu, S.; Duan, Y.; Huang, Y. The application of ribosome engineering to natural product discovery and yield improvement in Streptomyces. Antibiotics 2019, 8, 133. [Google Scholar] [CrossRef] [PubMed]
- Ostash, B.; Misaki, Y.; Dolya, B.; Kharaton, Y.; Busche, T.; Luzhetskyy, A.; Kalinowski, J.; Ochi, K.; Fedorenko, V. Generation and initial characterization of a collection of spontaneous Streptomyces albus J1074 mutants resistant to rifampicin. Factors Exp. Evol. Org. 2020, 27, 139–143. [Google Scholar] [CrossRef]
- Tseduliak, V.M.; Dolia, B.; Ostash, I.; Lopatniuk, M.; Busche, T.; Ochi, K.; Kalinowski, J.; Luzhetskyy, A.; Fedorenko, V.; Ostash, B. Mutations within gene XNR_2147 for TetR-like protein enhance lincomycin resistance and endogenous specialized metabolism of Streptomyces albus J1074. J. Appl. Genet. 2022, 64, 185–195. [Google Scholar] [CrossRef]
- Koshla, O.; Lopatniuk, M.; Rokytskyy, I.; Yushchuk, O.; Dacyuk, Y.; Fedorenko, V.; Luzhetskyy, A.; Ostash, B. Properties of Streptomyces albus J1074 mutant deficient in tRNALeuUAA gene bldA. Arch. Microbiol. 2017, 199, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics: A Laboratory Manual; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Bauer, A.; Kirby, W.; Sherris, J.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 36, 493–496. [Google Scholar] [CrossRef]
- Lee, J.H.; Yoo, J.S.; Kim, Y.; Kim, J.S.; Lee, E.J.; Roe, J.H. The WblC/WhiB7 transcription factor controls intrinsic resistance to translation-targeting antibiotics by altering ribosome composition. mBio 2020, 11, e00625-20. [Google Scholar] [CrossRef]
- Alvarez-Arevalo, M.; Sterndorff, E.B.; Faurdal, D.; Jørgensen, T.S.; Mourched, A.S.; Vuksanovic, O.; Saha, S.; Weber, T. Extraction and Oxford Nanopore sequencing of genomic DNA from filamentous Actinobacteria. STAR Protoc. 2022, 4, 101955. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 21 January 2022).
- Bolger, A.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Hilker, R.; Stadermann, K.; Doppmeier, D.; Kalinowski, J.; Stoye, J.; Straube, J.; Winnebald, R.; Goesmann, A. ReadXplorer—visualization and analysis of mapped sequences. Bioinformatics 2014, 30, 2247–2254. [Google Scholar] [CrossRef]
- Herrmann, S.; Siegl, T.; Luzhetska, M.; Petzke, L.; Jilg, C.; Welle, E.; Erb, A.; Leadlay, P.F.; Bechthold, A.; Luzhetskyy, A. Site-specific recombination strategies for engineering actinomycete genomes. Appl. Environ. Microbiol. 2012, 78, 1804–1812. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Kasahara, K.; Hirose, Y.; Murakami, K.; Kugimiya, R.; Ochi, K. Activation and products of the cryptic secondary metabolite biosynthetic gene clusters by rifampin resistance (rpoB) mutations in actinomycetes. J. Bacteriol. 2013, 195, 2959–2970. [Google Scholar] [CrossRef] [PubMed]
- Ochi, K. Insights into microbial cryptic gene activation and strain improvement: Principle, application and technical aspects. J. Antibiot. 2017, 70, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Zhang, Q.; Ochi, K. Activation of antibiotic biosynthesis by specified mutations in the rpoB gene (encoding the RNA polymerase beta subunit) of Streptomyces lividans. J. Bacteriol. 2002, 184, 3984–3991. [Google Scholar] [CrossRef] [PubMed]
- Marín, L.; Gutiérrez-Del-Río, I.; Villar, C.J.; Lombó, F. De novo biosynthesis of garbanzol and fustin in Streptomyces albus based on a potential flavanone 3-hydroxylase with 2-hydroxylase side activity. Microb. Biotechnol. 2021, 14, 2009–2024. [Google Scholar] [CrossRef] [PubMed]
- Yuzawa, S.; Mirsiaghi, M.; Jocic, R.; Fujii, T.; Masson, F.; Benites, V.T.; Baidoo, E.E.K.; Sundstrom, E.; Tanjore, D.; Pray, T.R.; et al. Short-chain ketone production by engineered polyketide synthases in Streptomyces albus. Nat. Commun. 2018, 9, 4569. [Google Scholar] [CrossRef]
- Gummerlich, N.; Manderscheid, N.; Rebets, Y.; Myronovskyi, M.; Gläser, L.; Kuhl, M.; Wittmann, C.; Luzhetskyy, A. Engineering the precursor pool to modulate the production of pamamycins in the heterologous host S. albus J1074. Metab. Eng. 2021, 67, 11–18. [Google Scholar] [CrossRef]
- Li, X.; Guo, R.; Luan, J.; Fu, J.; Zhang, Y.; Wang, H. Improving spinosad production by tuning expressions of the forosamine methyltransferase and the forosaminyl transferase to reduce undesired less active byproducts in the heterologous host Streptomyces albus J1074. Microb. Cell Fact. 2023, 19, 15. [Google Scholar] [CrossRef]
- Hoshino, K.; Imai, Y.; Mukai, K.; Hamauzu, R.; Ochi, K.; Hosaka, T. A putative mechanism underlying secondary metabolite overproduction by Streptomyces strains with a 23S rRNA mutation conferring erythromycin resistance. Appl. Microbiol. Biotechnol. 2020, 104, 2193–2203. [Google Scholar] [CrossRef]
- Golkar, T.; Zieliński, M.; Berghuis, A.M. Look and outlook on enzyme-mediated macrolide resistance. Front. Microbiol. 2018, 9, 1942. [Google Scholar] [CrossRef]
- Santos-Beneit, F.; Ceniceros, A.; Nikolaou, A.; Salas, J.A.; Gutierrez-Merino, J. Identification of antimicrobial compounds in two Streptomyces sp. strains isolated from beehives. Front. Microbiol. 2022, 13, 742168. [Google Scholar] [CrossRef] [PubMed]
- Santos-Beneit, F. Genome sequencing analysis of Streptomyces coelicolor mutants that overcome the phosphate-depending vancomycin lethal effect. BMC Genom. 2018, 19, 457. [Google Scholar] [CrossRef] [PubMed]



| Gene/Region (Function) | KO-1305 | KO-1307 |
|---|---|---|
| xnr_3039 (FtsW, cell division) | - 1 | 3455194; I→S |
| xnr_3154 (LamG/jellyroll domain protein) | - | 3594936; I→L |
| xnr_4320 (Na+/H+ antiporter) | 4919731; W→R 2 | - |
| xnr_4895 (Transcriptional factor) | 5569092; ∆T 3 | - |
| xnr_4783 (Transglutaminase) | - | 5437784; L→P |
| S. albidoflavus Strain | Growth Inhibition Zones (mm) | ||
|---|---|---|---|
| Thiostrepton (50 µg/disk) | Novobiocin (50 µg/disk) | Ristomycin (50 µg/disk) | |
| SAM2 | 23 ± 2 | 30 ± 3 | 22 ± 2 |
| KO-1305 1 | 28 ± 3 | 30 ± 2 | 28 ± 2 |
| KO-1401 | 27 ± 2 | 27 ± 2 | 25 ± 2 |
| KO-1402 | 34 ± 2 | 26 ± 1 | 31 ± 3 |
| KO-1403 | 35 ± 2 2 | 38 ± 2 | 40 ± 5 |
| KO-1404 | 26 ± 2 | 30 ± 2 | 27 ± 2 |
| KO-1405 | 32 ± 2 | 33 ± 2 | 28 ± 3 |
| KO-1406 | 35 ± 2 | 34 ± 2 | 40 ± 3 |
| KO-1407 | 33 ± 2 | 37 ± 2 | 34 ± 2 |
| KO-1408 | 35 ± 2 | 35 ± 2 | 36 ± 3 |
| KO-1409 | 21 ± 2 | 27 ± 2 | 26 ± 1 |
| KO-1307 1 | 32 ± 3 | 28 ± 2 | 31 ± 3 |
| KO-1410 | 35 ± 2 | 42 ± 2 | 42 ± 2 |
| KO-1411 | 30 ± 2 | 29 ± 2 | 32 ± 2 |
| KO-1412 | 26 ± 2 | 35 ± 2 | 33 ± 2 |
| KO-1413 | 28 ± 2 | 30 ± 2 | 28 ± 1 |
| KO-1414 | 23 ± 1 | 27 ± 1 | 22 ± 1 |
| KO-1415 | 32 ± 2 | 38 ± 1 | 40 ± 2 |
| KO-1416 | 28 ± 2 | 35 ± 3 | 30 ± 3 |
| KO-1417 | 28 ± 2 | 31 ± 3 | 27 ± 3 |
| Medium | SG2 | SFM | TSA | GYM | MM | ||
|---|---|---|---|---|---|---|---|
| Strain | B.c. 1 | D.h. 2 | B.c. | B.c. | B.c. | S.a. 3 | B.c. |
| SAM2 | +++ 4 | ++ | + | + | + | + | + |
| KO-1305 | +++ | + | – | – | + | ++ | – |
| KO-1401 | ++++ | ++ | ND 5 | ND | ND | +++ | ND |
| KO-1402 | +++ | ++ | ND | ND | ND | ++ | ND |
| KO-1403 | +++ | ++ | – | – | ++ | +++ | – |
| KO-1404 | +++ | ++ | ND | ND | ND | +++ | ND |
| KO-1405 | +++ | +++ | ND | ND | ND | ++ | ND |
| KO-1406 | +++ | ++ | – | – | ++ | ++ | + |
| KO-1407 | +++ | +++ | + | ++ | – | +++ | ++ |
| KO-1408 | ++++ | + | + | ++ | – | +++ | ++ |
| KO-1409 | +++ | ++ | ND | ND | ND | ++ | ND |
| KO-1307 | ++ | ++ | – | – | + | + | ND |
| KO-1410 | + | +++ | ND | ND | + | ++ | ND |
| KO-1411 | – | + | ND | ND | + | ++ | ND |
| KO-1412 | ++ | +++ | ND | ND | ++ | ++ | ND |
| KO-1413 | – | + | ND | ND | – | ++ | ND |
| KO-1414 | ++++ | + | ND | ND | + | ++ | ND |
| KO-1415 | +++ | + | ND | ND | + | ++ | ND |
| KO-1416 | ++ | ++ | ND | ND | ++ | +++ | ND |
| KO-1417 | ++ | ++ | ND | ND | + | ++ | ND |
| Gene/Region (Function) | KO-1403 | KO-1407 |
|---|---|---|
| xnr_1600 (secreted protein) | 1,896,611; Y→UAG 1 | - |
| xnr_2792 (unknown) | - | 3,168,482; S→G |
| xnr_4479 (TCS 2 histidine kinase) | 5,095,310; A→V | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dolya, B.; Hryhorieva, O.; Sorochynska, K.; Lopatniuk, M.; Ostash, I.; Tseduliak, V.-M.; Sterndorff, E.B.; Jørgensen, T.S.; Gren, T.; Dacyuk, Y.; et al. Properties of Multidrug-Resistant Mutants Derived from Heterologous Expression Chassis Strain Streptomyces albidoflavus J1074. Microorganisms 2023, 11, 1176. https://doi.org/10.3390/microorganisms11051176
Dolya B, Hryhorieva O, Sorochynska K, Lopatniuk M, Ostash I, Tseduliak V-M, Sterndorff EB, Jørgensen TS, Gren T, Dacyuk Y, et al. Properties of Multidrug-Resistant Mutants Derived from Heterologous Expression Chassis Strain Streptomyces albidoflavus J1074. Microorganisms. 2023; 11(5):1176. https://doi.org/10.3390/microorganisms11051176
Chicago/Turabian StyleDolya, Borys, Olena Hryhorieva, Khrystyna Sorochynska, Maria Lopatniuk, Iryna Ostash, Vasylyna-Marta Tseduliak, Eva Baggesgaard Sterndorff, Tue Sparholt Jørgensen, Tetiana Gren, Yuriy Dacyuk, and et al. 2023. "Properties of Multidrug-Resistant Mutants Derived from Heterologous Expression Chassis Strain Streptomyces albidoflavus J1074" Microorganisms 11, no. 5: 1176. https://doi.org/10.3390/microorganisms11051176
APA StyleDolya, B., Hryhorieva, O., Sorochynska, K., Lopatniuk, M., Ostash, I., Tseduliak, V.-M., Sterndorff, E. B., Jørgensen, T. S., Gren, T., Dacyuk, Y., Weber, T., Luzhetskyy, A., Fedorenko, V., & Ostash, B. (2023). Properties of Multidrug-Resistant Mutants Derived from Heterologous Expression Chassis Strain Streptomyces albidoflavus J1074. Microorganisms, 11(5), 1176. https://doi.org/10.3390/microorganisms11051176







