Bacterial Communities Associated with Houseflies (Musca domestica L.) Inhabiting Hospices in South Africa
Abstract
1. Introduction
2. Materials and Methods
2.1. Housefly Sampling
2.2. DNA Extraction from Houseflies
2.3. Partial 16S rRNA Gene Amplicon Sequencing
2.4. Metagenomic Data Analysis
Bioinformatic and Diversity Analyses
2.5. Screening for Antibiotic Resistance Genes
2.6. PCR for Amplification of Resistance Genes
3. Results
3.1. Housefly Collection
3.2. Summary of Generated ASV
3.3. Taxonomic Classification of Bacteria
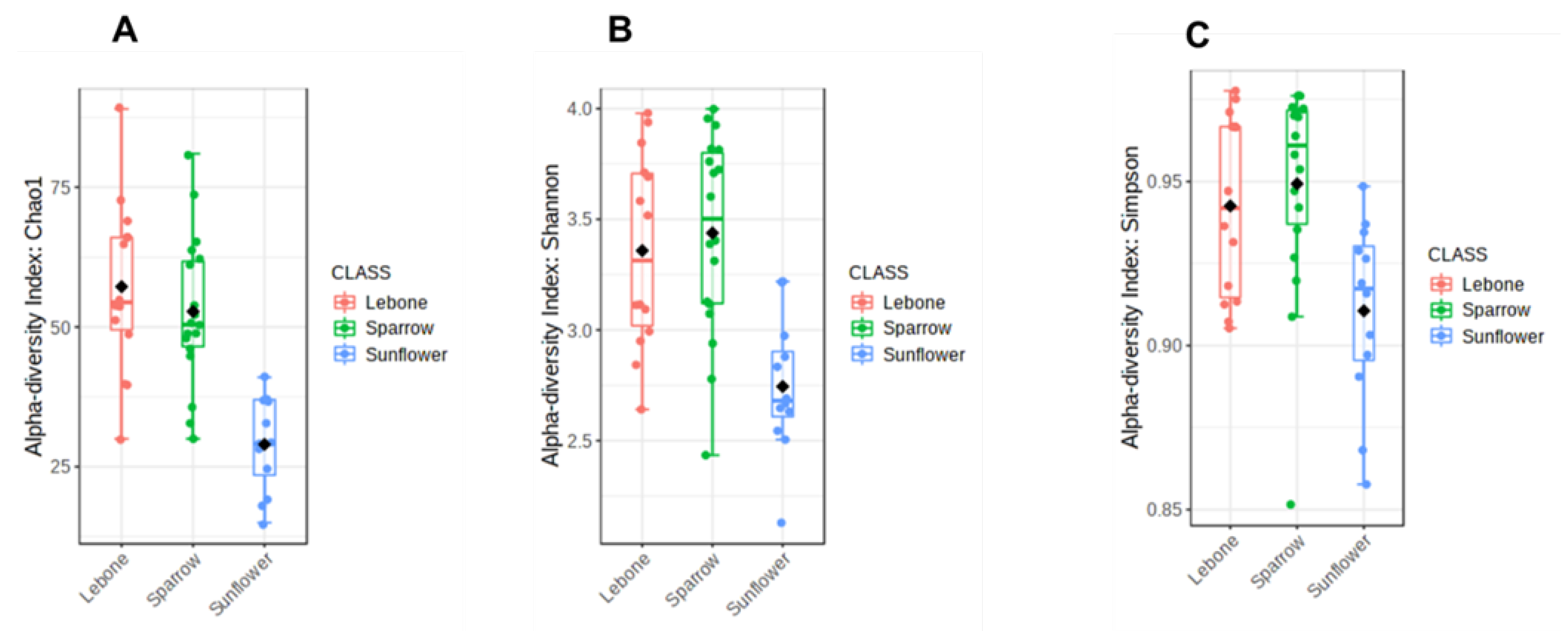
3.4. Alpha Diversity Index
3.5. Beta Diversity Index
3.6. Antibiotic Resistance Genes
3.7. Occurrence of Antibiotic Resistance Genes
3.7.1. β-Lactam Resistance Gene
3.7.2. Macrolide Resistance Genes
3.7.3. Tetracycline Resistance Genes
3.7.4. Integrons
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zurek, L.; Denning, S.S.; Schal, C.; Watson, D.W. Vector Competence of Musca domestica (Diptera: Muscidae) for Yersinia pseudotuberculosis. J. Med. Entomol. 2001, 38, 333–335. [Google Scholar] [CrossRef] [PubMed]
- Khatter, N.A. Transmission of bacterial pathogens by the house fly Musca domestica vicina. Am. J. Res. Commun. 2013, 1, 1–12. [Google Scholar]
- Zurek, K.; Nayduch, D. Bacterial Associations Across House Fly Life History: Evidence for Transstadial Carriage from Managed Manure. J. Insect Sci. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Vaziriannzadeh, B.; Solary, S.S.; Radhar, M.; Hajhassein, R.; Mendinejad, M. Identification of bacteria with possible transmitted by Musca domestica (Diptera: Muscidae) in the region of Ahvaz; SW Iran. J. Microbiol. 2008, 1, 28–31. [Google Scholar]
- Gupta, A.K.; Nayduch, D.; Verma, P.; Shah, B.; Ghate, H.V.; Patole, M.S.; Yogesh, S.S. Phylogenetic characterization of bacteria in the gut of houseflies (Musca domestica L.). FEMS Microbiol. Ecol. 2012, 79, 581–593. [Google Scholar] [CrossRef]
- Nazari, M.; Mahrabi, T.; Hosseini, S.M.; Alikhani, M.Y. Bacterial contamination of adult houseflies (Musca domestica) and sensitivity of these bacteria to various antibiotics; captured from Hamadan City, Iran. J.Clin. Diagn. Res. 2017, 11, 4–7. [Google Scholar] [CrossRef]
- Onwugamba, F.C.; Fitzgerald, J.R.; Rochon, K.; Guadabassi, L.; Alabi, A. The role of ‘filth flies’ in the spread of antimicrobial resistance. Travel Med. Infect. Dis. 2018, 22, 8–17. [Google Scholar] [CrossRef]
- Bouamama, L.; Sorlozano, A.; Laglaoui, A.; Lebbadi, M.; Aarab, A.; Gutierez, J. Antibiotic resistance patterns of bacterial strains isolated from Periplaneta americana and Musca domestica in Tangier; Morocco. J. Infect. Dev. Ctries. 2010, 4, 194–201. [Google Scholar] [CrossRef]
- Zhang, A.; Li, Y.; Guan, Z.; Tuo, H.; Liu, D.; Yang, Y.; Xu, C.; Lei, C.; Wang, H. Characterization of resistance patterns and detection of apramycin resistance genes in Escherichia coli Isolated from chicken feces and houseflies after apramycin administration. Front. Microbiol. 2018, 9, 328. [Google Scholar] [CrossRef]
- Makhele, M.F.; Mulaudzi, F.M. The experiences of Batswana families regarding hospice care of AIDS patients in the Bophirima district; North West province; South Africa. SAHARA J. Soc. Asp. HIV/AIDS 2012, 9, 104–112. [Google Scholar]
- Gwyther, L.; Rawlinson, F. Palliative medicine teaching program at the University of Cape Town: Integrating palliative care principles into practice. J. Pain Symptom Manag. 2007, 33, 558–562. [Google Scholar] [CrossRef]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Kotsilkov, K.; Popova, C.; Boyanova, L.; Setchanova, L.; Mitov, I. Comparison of culture method and real-time PCR for detection of putative period on to pathogenic bacteria in deep periodontal pockets. Biotechnol. Biotechnol. Equip. 2015, 29, 996–1002. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The gut microbiota of insects—Diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [PubMed]
- Muthukumar, A.; Zitterkopf, N.L.; Payne, D. Molecular tools for the detection and characterization of bacterial infections: A review. Labmedicine 2008, 39, 430–436. [Google Scholar] [CrossRef]
- Zhao, W.; Cheng, Y.; Zhang, C.; Zhao, W.; You, Q.; Shen, X.; Guo, W.; Jiaoet, Y. Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci. Rep. 2017, 7, 5636. [Google Scholar] [CrossRef]
- Czekalski, N.; Diez, E.G.; Burgmann, H. Wastewater as a point sourc of antibiotic resistance genes in the sediment of a freshwater lake. International Soc. Microb. Ecol. 2014, 8, 1381–1390. [Google Scholar]
- Anjum, M.F.; Zankari, E.; Hasman, H. Molecular methods for detection of antimicrobial resistance. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Rotty, I.E.; Pinontoan, O.; Tulung, M.; Rumengan, I.; Semuel, M.Y. Molecular identification of house fly; Musca domestica L. (Diptera: Muscidae); using mitochondrial DNA partial genes cytochrome oxidase subunit 1 (COI) in Manado city. Int. J. Entomol. Res. 2018, 3, 168–176. [Google Scholar]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glockner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, 1–11. [Google Scholar] [CrossRef]
- Callahan, B.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible; interactive; scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerkem, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Dhariwal, A.; Chong, J.; Habib, S.; Irah, L.K.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical; visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef] [PubMed]
- Chong, C.; Müller, M.; Pak, H.; Harnett, D.; Huber, F.; Grun, D.; Leleu, M.; Auger, A.; Arnaud, M.; Stevenson, B.J.; et al. Integrated proteogenomic deep sequencing and analytics accurately identify non-canonical peptides in tumor immunopeptidomes. Nat. Commun. 2020, 11, 1293. [Google Scholar] [CrossRef]
- Foster, Z.S.L.; Sharpton, T.J.; Grundwald, N.J. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput. Biol. 2017, 13. [Google Scholar] [CrossRef]
- Rodriguez-Mozaz, S.; Chamorro, S.; Marti, E.; Huerta, B.; Gros, M.; Sanchez-Melsio, A.; Borrego, C.M.; Barcelo, D.; Balcazar, J.L. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015, 69, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Hu, X.L.; Xu, T.; Zhang, H.C.; Sheng, D.; Yin, D.Q. Prevalence of antibiotic resistance genes and their relationship with antibiotic in the Huangpu River and the drinking water sources; Shanghai; China. Sci. Total Environ. 2013, 458–460, 267–272. [Google Scholar] [CrossRef]
- Bahrndorff, S.; De Jonge, N.; Skovgård, H.; Nielsen, J.L. Bacterial communities associated with houseflies (Musca domestica L.) sampled within and between farms. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- de Jonge, N.; Michaelsen, T.Y.; Ejbye-Ernst, R.; Jensen, A.; Nielsen, M.E.; Bahrndorff, S.; Nielsen, J.L. Housefly (Musca domestica L.) associated microbiota across different life stages. Sci. Rep. 2020, 10, 7842. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, N.; Martens, R.; Tebbe, C.C. Origin and diversity of metabolically active gut bacteria from laboratory-bred larvae of Manduca sexta (Sphingidae, Lepidoptera, Insecta). Appl. Environ. Microbiol. 2008, 74, 7189–7196. [Google Scholar] [CrossRef]
- Priya, N.G.; Ojha, A.; Kajla, M.K.; Raj, A.; Rajagopal, R. Host plant induced variation in gut bacteria of Helicoverpa armigera. PLoS ONE 2012, 7, e30768. [Google Scholar]
- Chandel, K.; Mendki, M.J.; Parikh, R.Y.; Kulkarni, G.; Tikar, S.N.; Sukumaran, D.; Prakash, S.; Parashar, B.D.; Shouche, Y.S.; Veer, V. Midgut microbial community of Culex quinquefasciatus mosquito populations from india. PLoS ONE 2013, 8, e80453. [Google Scholar] [CrossRef] [PubMed]
- Hendriksma, H.P.; Kuting, M.; Hartel, S.; Nather, A.; Dohrmann, A.B.; Steffan-Dewenter, I.; Tebbe, C.C. Effect of stacked insecticidal cry proteins from maize pollen on nurse bees (Apis mellifera Carnica) and their gut bacteria. PLoS ONE 2013, 8, e59589. [Google Scholar] [CrossRef] [PubMed]
- Moro, C.V.; Tran, F.-H.; Raharimalala, F.N.; Ravelonandro, P.; Mavingui, P. Diversity of culturable bacteria including Pantoea in wild mosquito Aedes albopictus. BMC Microbiol. 2013, 13, 70. [Google Scholar]
- Gupta, A.K.; Rastogi, G.; Nayduch, D.; Sawant, S.S.; Bhonde, R.R.; Shouche, Y.S. Molecular phylogenetic profiling of gut-associated bacteria in larvae and adults of flesh flies. Med. Vet. Entomol. 2014, 28, 345–354. [Google Scholar] [CrossRef]
- Junqueira, A.C.M.; Ratan, A.; Acerbi, E.; Drautz-Moses, D.I.; Premkrishnan, B.N.V.; Costea, P.I.; Linz, B.; Purbojati, R.W.; Paulo, D.F.; Gaultier, N.E.; et al. The microbiomes of blowflies and houseflies as bacterial transmission reservoirs. Sci. Rep. 2017, 7, 16324. [Google Scholar] [CrossRef]
- Khamesipour, F.; Lankarani, K.B.; Honarvar, B.; Kwenti, T.E. A systematic review of human pathogens carried by the housefly (Musca domestica L.). BMC Public Health 2018, 18, 1049. [Google Scholar] [CrossRef]
- Park, R.; Dzialo, M.C.; Spaepen, S.; Nsabimana, D.; Gielens, K.; Devriese, H.; Crauwels, S.; Tito, R.Y.; Raes, J.; Lievens, B.; et al. Microbial communities of the house fly Musca domestica vary with geographical location and habitat. Microbiome 2019, 7, 147. [Google Scholar] [CrossRef]
- Singh, B.; Crippen, T.L.; Zheng, L.; Fields, A.T.; Yu, Z.; Ma, Q.; Wood, T.K.; Dowd, S.E.; Flores, M.; Tomberlin, J.K.; et al. A metagenomic assessment of the bacteria associated with Lucilia sericata and Lucilia cuprina (Diptera: Calliphoridae). Appl. Microbiol. Biotechnol. 2014, 99, 869–883. [Google Scholar] [CrossRef]
- Grubel, P.; Hoffman, J.S.; Chong, F.K.; Burstein, N.E.; Mepani, C.; Cave, D.R. Vector potential of houseflies (Musca domestica) for Helicobacter pylori. J. Clin Microbiol. 1997, 35, 1300–1303. [Google Scholar] [CrossRef]
- Esmaeili, S.; Mohabati, A.M.; Khalil, M.; Mostafavi, E.; Moradnejad, P. Genetic evidence of Coxiella burnetii infection in acute febrile illnesses in Iran. PLoS Negl. Trop. Dis. 2019, 13, e0007181. [Google Scholar] [CrossRef] [PubMed]
- Hucko, M. The role of the housefly (Musca domestica L.) in the transmission of Coxiella burnetii. Folia Parasitol. 1984, 31, 177–181. [Google Scholar]
- He, Y.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J.J. Antibiotic resistance genes from livestock waste: Occurrence; dissemination; and treatment. Clean Water 2020, 3, 4. [Google Scholar] [CrossRef]
- Colomer-Lluch, M.; Jofre, J.; Muniesa, M. Antibiotic Resistance Genes in the Bacteriophage DNA Fraction of Environmental Samples. PLoS ONE 2011, 6, e17549. [Google Scholar] [CrossRef] [PubMed]
- Eftekhar, F.; Rastegar, M.; Golalipoor, M.; Mansour, S.N. Detection of extended spectrum beta-lactamases in urinary isolates of Klebsiella pneumonia in relation to BlaSHV.; BlaTEM.; BlaCTX-M gene carriage. Iran. J. Public Health 2012, 41, 127–132. [Google Scholar] [PubMed]
- Yazdi, M.; Nazemi, A.; Mirinargasi, M.; Jafarpour, M.; Sharifi, S.H. Genotypic versus phenotypic methods to detect extended-spectrum beta lactamases (ESBL’s) in uropathogenic Escherichia coli. Ann. Biol. Res. 2012, 3, 2454–2458. [Google Scholar]
- Pitout, J.D.D.; Nordman, P.; Laupland, K.B.; Poirel, L. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 2005, 56, 52–59. [Google Scholar] [CrossRef]
- Ramos, M.M.B.; Gaetti-Jardim, E.C.; Gaetti-Jardim, J.E. Resistance to tetracycline and β-lactams and distribution of resistance markers in enteric microorganisms and pseudomonads isolated from the oral cavity. J. Appl. Oral Sci. 2009, 17, 13–18. [Google Scholar] [CrossRef]
- Hemmatinezhad, B.; Ommi, D.; Taktaz-Hafshejani, T.; Khamesipour, F. Molecular detection and antimicrobial resistance of Pseudomonas aeruginosa from houseflies (Musca domestica) in Iran. J. Venom. Anim. Toxins 2015, 21, 18. [Google Scholar] [CrossRef]
- Choi, J.; Rieke, E.L.; Moorman, T.B.; Soupir, M.L.; Heather, K.; Allen, H.L.; Smith, S.; Howe, A. Practical implications of erythromycin resistance gene diversity on surveillance and monitoring of resistance. Fed. Eur. Microbiol. Soc. Microbiol. Ecol. 2018, 94, 1–11. [Google Scholar] [CrossRef]
- Jahantigh, M.; Samadi, K.; Dizaji, R.E.; Salari, S. Antimicrobial resistance and prevalence of tetracycline resistance genes in Escherichia coli isolated from lesions of colibacillosis in broiler chickens in Sistan, Iran. BMC Vet. Res. 2020, 16, 267. [Google Scholar] [CrossRef] [PubMed]
- Akter, S.; Sabuj, A.A.M.; Haque, Z.F.; Rahman, M.T.; Kafi, M.A.; Saha, S. Detection of antibiotic-resistant bacteria and their resistance genes from houseflies. Vet. World 2020, 13, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Hubeny, J.; Buta, M.; Zieliński, W.; Harnisz, M.; Korzeniewska, E.; Nowrotek, M.; Płaza, G. The Prevalence of tet(A) and tet(M) Tetracycline Resistance Genes in Municipal Wastewater. J. Ecol. Eng. 2019, 20, 1–6. [Google Scholar] [CrossRef]
- Tansirichaiya, S.; Rahman, M.A.; Antepowicz, A.; Mullany, P.; Roberts, A.P. Detection of Novel Integrons in the Metagenome of Human Saliva. PLoS ONE 2016, 11, e0157605. [Google Scholar] [CrossRef] [PubMed]
- Barlow, R.S.; Desmarchelier, P.M.; Gobius, K.S. Isolation and characterizationof integron-containing bacteria without antibiotic selection. Antimicrob. Agents Chemother. 2004, 48, 838–842. [Google Scholar] [CrossRef]
- Labbate, M.; Case, R.J.; Stokes, H.W. The integron/gene cassette system: An active player in bacterial adaptation. Methods Mol. Biol. 2009, 532, 103–125. [Google Scholar]
- Deng, Y.; Bao, X.; Ji, L.; Chen, L.; Liu, J.; Miao, J.; Chen, D.; Bian, H.; Li, Y.; Yu, G. Resistance integrons: Class 1; 2 and 3 integrons. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 45. [Google Scholar] [CrossRef]
- Huang, J.; Lan, F.; Lu, Y.; Li, B. Characterization of integrons and antimicrobial resistance in Escherichia coli sequence TYPE 131 isolates. Can. J. Infect. Dis. Med. Microbiol. 2020, 2020, 3826186. [Google Scholar] [CrossRef]
- Ng, L.K.; Mulvey, M.R.; Martin, I.; Peters, G.A.; Johnson, W. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob. Agents Chemother. 1999, 43, 3018–3021. [Google Scholar] [CrossRef]
- Peerayeh, S.N.; Karmostaji, A. Molecular identification of resistance determinants, integrons and genetic relatedness of extensively drug resistant Acinetobacter baumannii isolated from hospitals in Tehran, Iran. J. Microbiol. 2015, 8, e27021. [Google Scholar] [CrossRef]
- Mignard, S.; Flandrois, J.P. 16S rRNA sequencing in routine bacterial identification: A 30-month experiment. J. Microbiol Methods 2006, 67, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Bisi-Johnson, M.; Obi, C.; Vasaikar, S.D.; Baba, K.A.; Hattori, T. Molecular basis of virulence in clinical isolates of Escherichia coli and Salmonella species from a tertiary hospital in the Eastern Cape, South Africa. Gut Path. 2011, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Aher, J.; Roy, A.; Kumar, P. Molecular detection of virulence genes associated with pathogenicity of Klebsiella spp. isolated from the respiratory tract of apparently healthy as well as sick goats. Israel J. Vet. Med. 2012, 67, 249–252. [Google Scholar]



| Resistance Gene Detected | Lebone Hospice n = 15 (%) | Sparrow Hospice n = 15 (%) | Sunflower Hospice n = 15 (%) | p-Value |
|---|---|---|---|---|
| Penicillin antibiotic resistance genes | 0.231 | |||
| ermB | 2 (13.3%) | 1 (6.6%) | 0 (0%) | |
| mecA | 0 (0%) | 0 (0%) | 0 (0%) | |
| vanA | 0 (0%) | 0 (0%) | 0 (0%) | |
| β-lactam antibiotic resistance genes | 0.125 | |||
| blaCARB | 0 (0%) | 0 (0%) | 0 (0%) | |
| blaTEM | 4 (26.7%) | 0 (0%) | 2 (13.3%) | |
| blaSHV | 6 (40.0%) | 0 (0%) | 5 (33.3%) | |
| ampC | 0 (0%) | 0 (0%) | 0 (0%) | |
| Tetracycline antibiotic resistance genes | 0.122 | |||
| tetA | 9 (60%) | 3 (20.0%) | 5 (33.3%) | |
| tetW | 0 (0%) | 0 (0%) | 0(0%) | |
| tetX | 0 (0%) | 0 (0%) | 0 (0%) | |
| Integrons | 0.783 | |||
| intI | 8 (53.3%) | 6 (40.0%) | 7 (46.7%) | |
| intII | 9 (60.0%) | 2 (13.3%) | 8 (53.3%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monyama, M.C.; Taioe, O.M.; Nkhebenyane, J.S.; van Wyk, D.; Ramatla, T.; Thekisoe, O.M.M. Bacterial Communities Associated with Houseflies (Musca domestica L.) Inhabiting Hospices in South Africa. Microorganisms 2023, 11, 1440. https://doi.org/10.3390/microorganisms11061440
Monyama MC, Taioe OM, Nkhebenyane JS, van Wyk D, Ramatla T, Thekisoe OMM. Bacterial Communities Associated with Houseflies (Musca domestica L.) Inhabiting Hospices in South Africa. Microorganisms. 2023; 11(6):1440. https://doi.org/10.3390/microorganisms11061440
Chicago/Turabian StyleMonyama, Maropeng C., Oriel M. Taioe, Jane S. Nkhebenyane, Deidre van Wyk, Tsepo Ramatla, and Oriel M. M. Thekisoe. 2023. "Bacterial Communities Associated with Houseflies (Musca domestica L.) Inhabiting Hospices in South Africa" Microorganisms 11, no. 6: 1440. https://doi.org/10.3390/microorganisms11061440
APA StyleMonyama, M. C., Taioe, O. M., Nkhebenyane, J. S., van Wyk, D., Ramatla, T., & Thekisoe, O. M. M. (2023). Bacterial Communities Associated with Houseflies (Musca domestica L.) Inhabiting Hospices in South Africa. Microorganisms, 11(6), 1440. https://doi.org/10.3390/microorganisms11061440







