Genome Analysis of a Variant of Streptomyces coelicolor M145 with High Lipid Content and Poor Ability to Synthetize Antibiotics
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Constructions of KO Mutants of sco0982 in S. coelicolor
2.3. Construction of the Plasmid to Over-Express sco0984
2.4. Assay of Extracellular ACT Production
2.5. Determination of Total Lipid Content Using Attenuated Total Reflectance–Fourier Transform Infrared Spectroscopy (ATR-FTIR) Measurements
2.6. Atomic Force Microscopic Measurements Coupled to IR Spectroscopy (AFM-IR)
2.7. Esterified Fatty Acids Quantification Using GC-MS
2.8. Comparative Analysis of the Genome of S. coelicolor M145 and of Its TD Variant
3. Results
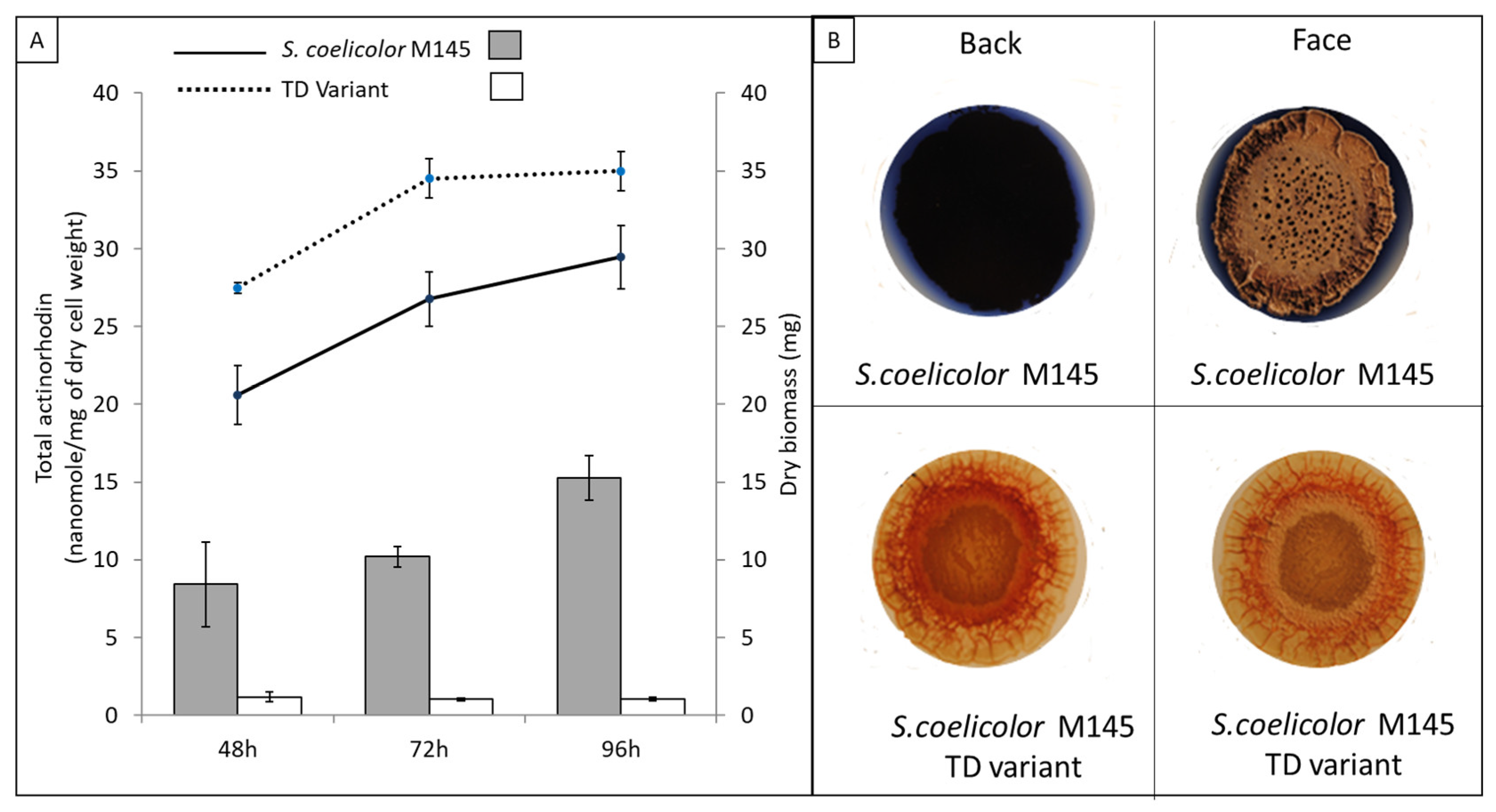
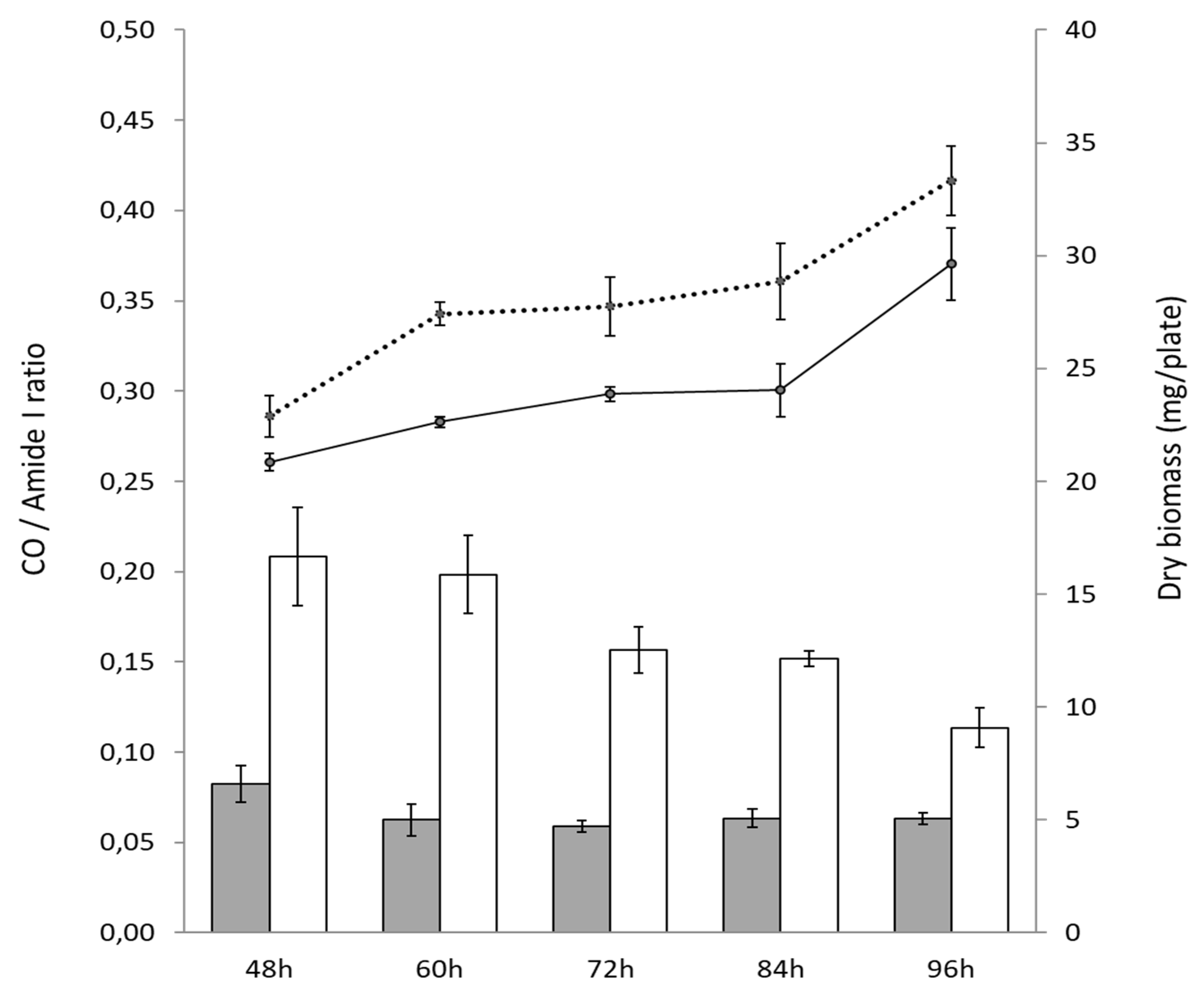
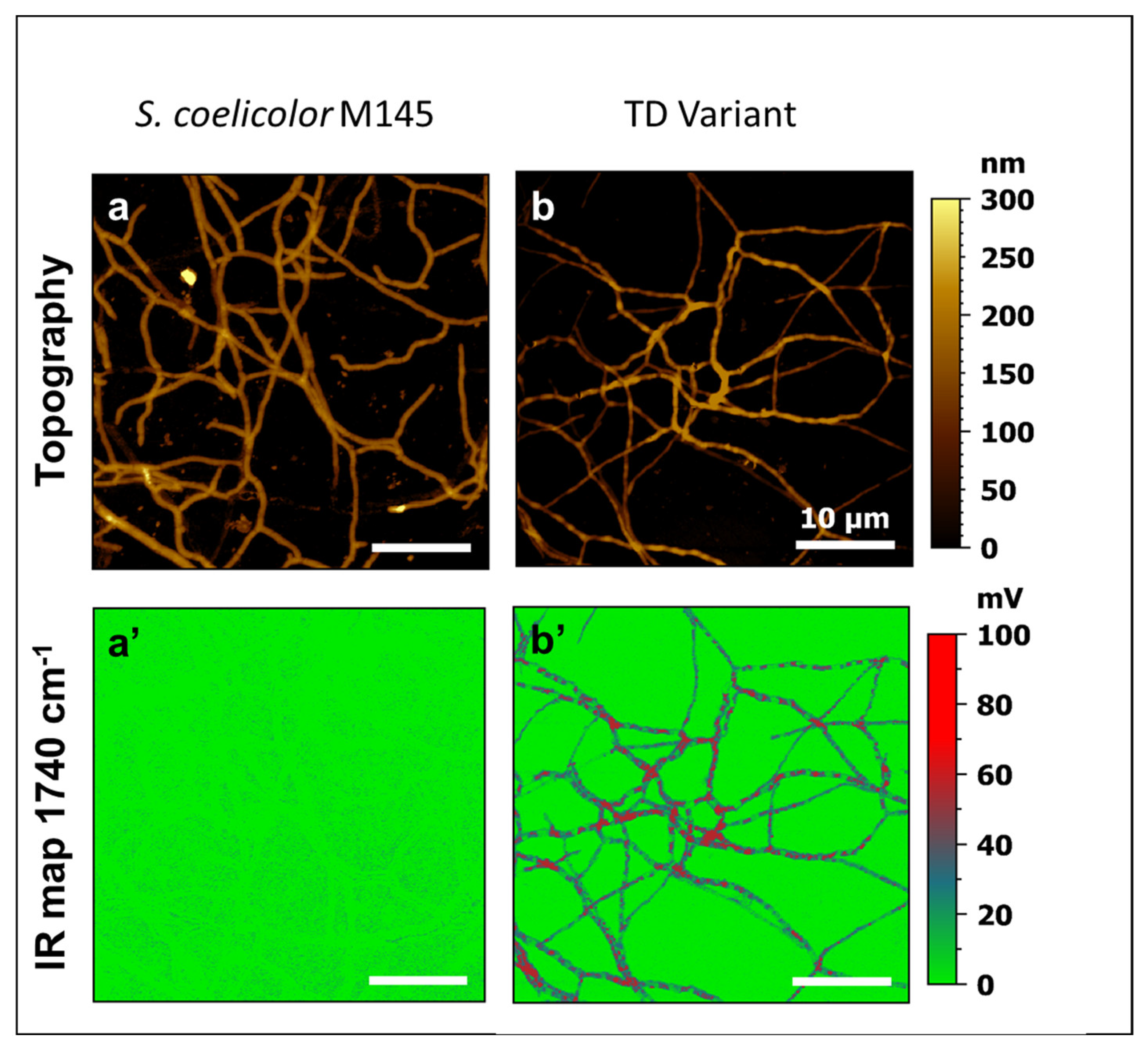
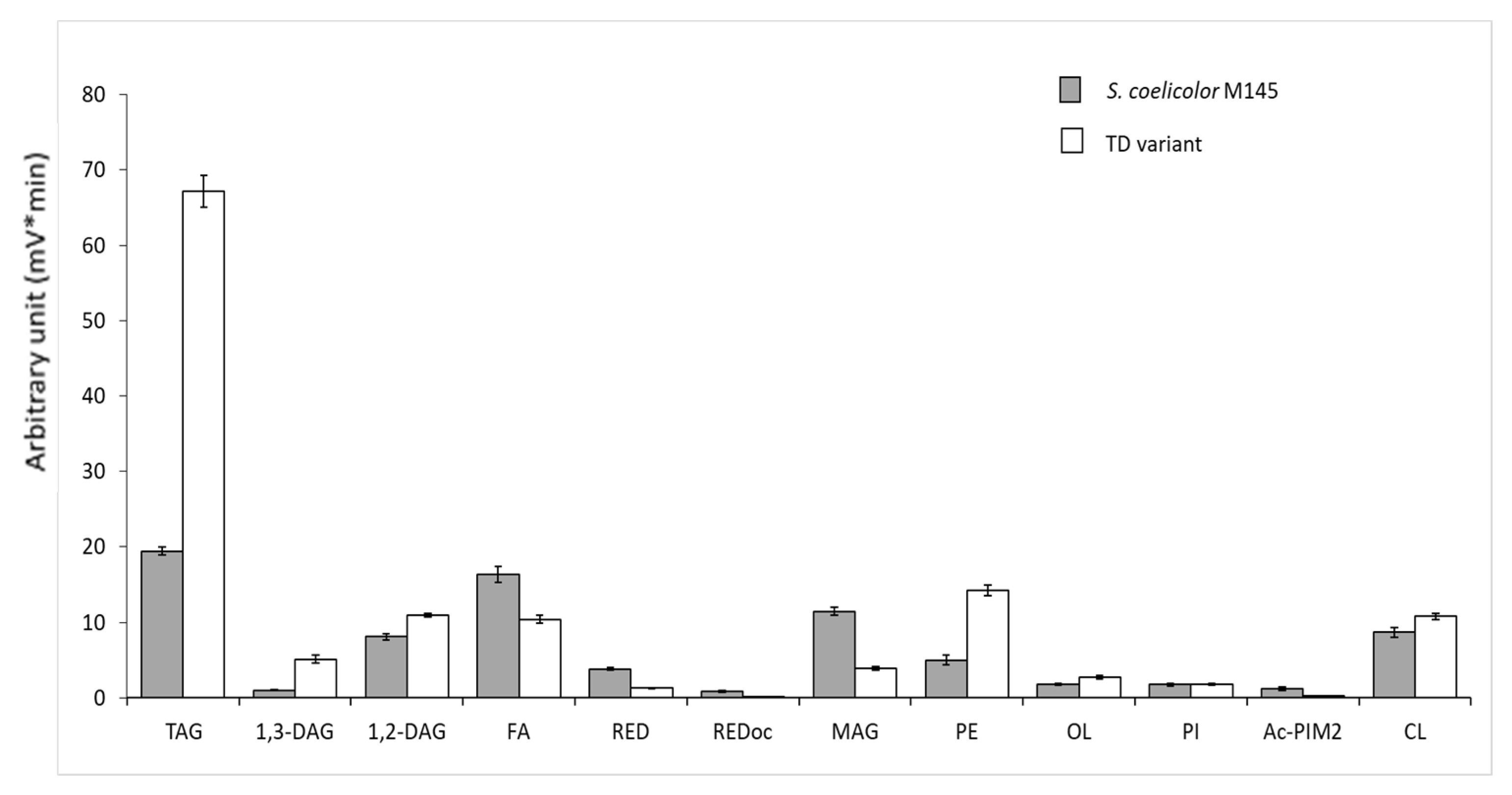
3.1. A Pale-Blue Variant of S. coelicolor Weakly Produces ACT and RED and Has a High Triacylglycerol and Phosphatidylethanolamine Content
3.2. Genomic Structure of the Pale-Blue TD Variant of S. coelicolor
- -
- A total of 32 were deleted individually (Table S2).
- -
- A total of 263 deleted genes were part of large deletions including the terminal arms of the chromosome (spanning over 101 genes: from SCO0001 to SCO0081 and SCO7827 to SCO7846). This was probably associated with circularization of the chromosome, a rearrangement event commonly observed in Streptomyces grown in laboratory conditions [26,27].
- -
- A total of 386 genes were deleted through 16 large deletions resulting from the loss of mobile genetic elements (MGEs) such as phages, plasmids or integrated conjugative elements (AICEs), etc.
- -
- A total of 60 of the 90 insertion sequences (ISs) (2/3) present in the strain were deleted and among them 21 ISs (including 2 ISs composed of 2 genes, therefore 23 genes) had probably been excised autonomously and 37 were carried by the large deletions mentioned above.
3.2.1. Deletion of the Region Encompassing Genes of the Glyoxylate Cycle
3.2.2. Deletion of Two Pairs of Genes Encoding Proteins of the TCA Cycle
3.2.3. Deletion of Genes Encoding Proteins Involved in Nitrogen Assimilation
3.2.4. Deletion of Genes Involved in the Biosynthesis of Specialized Metabolites
3.2.5. Deletion of sco7335 Encoding a Putative Amylase
3.2.6. Deletion of Genes Encoding Proteins Involved in Lipid Biosynthesis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bentley, S.D.; Chater, K.F.; Cerdeno-Tarraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar]
- Van Keulen, G.; Dyson, P.J. Production of specialized metabolites by Streptomyces coelicolor A3(2). Adv. Appl. Microbiol. 2014, 89, 217–266. [Google Scholar] [CrossRef]
- Okamoto, S.; Taguchi, T.; Ochi, K.; Ichinose, K. Biosynthesis of actinorhodin and related antibiotics: Discovery of alternative routes for quinone formation encoded in the act gene cluster. Chem. Biol. 2009, 16, 226–236. [Google Scholar] [CrossRef]
- Esnault, C.; Dulermo, T.; Smirnov, A.; Askora, A.; David, M.; Deniset-Besseau, A.; Holland, I.B.; Virolle, M.J. Strong antibiotic production is correlated with highly active oxidative metabolism in Streptomyces coelicolor M145. Sci. Rep. 2017, 7, 200. [Google Scholar] [CrossRef] [PubMed]
- Lejeune, C.; Sago, L.; Cornu, D.; Redeker, V.; Virolle, M.J. A Proteomic Analysis Indicates That Oxidative Stress Is the Common Feature Triggering Antibiotic Production in Streptomyces coelicolor and in the pptA Mutant of Streptomyces lividans. Front. Microbiol. 2021, 12, 813993. [Google Scholar] [CrossRef] [PubMed]
- Millan-Oropeza, A.; Henry, C.; Blein-Nicolas, M.; Aubert-Frambourg, A.; Moussa, F.; Bleton, J.; Virolle, M.J. Quantitative Proteomics Analysis Confirmed Oxidative Metabolism Predominates in Streptomyces coelicolor versus Glycolytic Metabolism in Streptomyces lividans. J. Proteome Res. 2017, 16, 2597–2613. [Google Scholar] [CrossRef]
- De la Peña Mattozzi, M.; Kang, Y.; Keasling, J.D. Feast: Choking on Acetyl-CoA, the Glyoxylate Shunt, and Acetyl-CoA-Driven Metabolism. In Handbook of Hydrocarbon and Lipid Microbiology; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar] [CrossRef]
- Dolan, S.K.; Welch, M. The Glyoxylate Shunt, 60 Years On. Annu. Rev. Microbiol. 2018, 72, 309–330. [Google Scholar] [CrossRef] [PubMed]
- Gui, L.; Sunnarborg, A.; Pan, B.; LaPorte, D.C. Autoregulation of iclR, the gene encoding the repressor of the glyoxylate bypass operon. J. Bacteriol. 1996, 178, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.A.; Laing, E.; Allenby, N.; Bucca, G.; Brenner, V.; Harrison, M.; Kierzek, A.M.; Smith, C.P. Metabolic and evolutionary insights into the closely-related species Streptomyces coelicolor and Streptomyces lividans deduced from high-resolution comparative genomic hybridization. BMC Genom. 2010, 11, 682. [Google Scholar] [CrossRef]
- Lejeune, C.; Abreu, S.; Chaminade, P.; Dulermo, T.; David, M.; Werten, S.; Virolle, M.J. Impact of Phosphate Availability on Membrane Lipid Content of the Model Strains, Streptomyces lividans and Streptomyces coelicolor. Front. Microbiol. 2021, 12, 623919. [Google Scholar] [CrossRef]
- Skeggs, P.A.; Holmes, D.J.; Cundliffe, E. Cloning of aminoglycoside-resistance determinants from Streptomyces tenebrarius and comparison with related genes from other actinomycetes. J. Gen. Microbiol. 1987, 133, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Boubakri, H.; Seghezzi, N.; Duchateau, M.; Gominet, M.; Kofronova, O.; Benada, O.; Mazodier, P.; Pernodet, J.L. The Absence of Pupylation (Prokaryotic Ubiquitin-like Protein Modification) Affects Morphological and Physiological Differentiation in Streptomyces coelicolor. J. Bacteriol. 2015, 197, 3388–3399. [Google Scholar] [CrossRef] [PubMed]
- Mazodier, P.; Petter, R.; Thompson, C. Intergeneric conjugation between Escherichia coli and Streptomyces species. J. Bacteriol. 1989, 171, 3583–3585. [Google Scholar] [CrossRef]
- Nguyen, H.C.; Karray, F.; Lautru, S.; Gagnat, J.; Lebrihi, A.; Huynh, T.D.; Pernodet, J.L. Glycosylation steps during spiramycin biosynthesis in Streptomyces ambofaciens: Involvement of three glycosyltransferases and their interplay with two auxiliary proteins. Antimicrob. Agents Chemother. 2010, 54, 2830–2839. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Pülher, A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat. Biotech. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. (Eds.) Practical Streptomyces Genetics; John Innes Foundation: Norfolk, UK, 2000. [Google Scholar]
- Millan-Oropeza, A.; Rebois, R.; David, M.; Moussa, F.; Dazzi, A.; Bleton, J.; Virolle, M.J.; Deniset-Besseau, A. Attenuated Total Reflection Fourier Transform Infrared (ATR FT-IR) for Rapid Determination of Microbial Cell Lipid Content: Correlation with Gas Chromatography-Mass Spectrometry (GC-MS). Appl. Spectrosc. 2017, 71, 2344–2352. [Google Scholar] [CrossRef] [PubMed]
- Deniset-Besseau, A.; Prater, C.B.; Virolle, M.J.; Dazzi, A. Monitoring TriAcylGlycerols Accumulation by Atomic Force Microscopy Based Infrared Spectroscopy in Streptomyces Species for Biodiesel Applications. J. Phys. Chem. Lett. 2014, 5, 654–658. [Google Scholar] [CrossRef]
- Dazzi, A.; Prater, G.P.; Hu, Q.; Chase, D.B.; Rabolt, J.F.; Marcott, C. AFM-IR: Combining atomic force microscopy and infrared spectroscopy for nanoscale chemical characterization. Appl. Spectrosc. 2012, 66, 1365–1384. [Google Scholar]
- Dazzi, A.; Prater, C.B. AFM-IR: Technology and Applications in Nanoscale Infrared Spectroscopy and Chemical Imaging. Chem. Rev. 2017, 117, 5146–5173. [Google Scholar] [CrossRef]
- Mathurin, J.; Deniset-Besseau, A.; Bazin, D.; Dartois, E.; Wagner, M.; Dazzi, A. Photothermal AFM-IR spectroscopy and imaging: Status, challenges, and trends. J. Appl. Phys. 2022, 131, 010901. [Google Scholar] [CrossRef]
- Lu, F.; Jin, M.; Belkin, M.A. Tip-enhanced infrared nanospectroscopy via molecular expansion force detection. Nat. Photonics 2014, 8, 307–312. [Google Scholar] [CrossRef]
- Lepage, G.; Roy, C.C. Direct transesterification of all classes of lipids in a one-step reaction. J. Lipid Res. 1986, 27, 114–120. [Google Scholar] [CrossRef] [PubMed]
- David, M.; Lejeune, C.; Abreu, S.; Thibessard, A.; Leblond, P.; Chaminade, P.; Virolle, M.J. Negative Correlation between Lipid Content and Antibiotic Activity in Streptomyces: General Rule and Exceptions. Antibiotics 2020, 9, 280. [Google Scholar] [CrossRef] [PubMed]
- Fischer, G.; Decaris, B.; Leblond, P. Occurrence of deletions, associated with genetic instability in Streptomyces ambofaciens, is independent of the linearity of the chromosomal DNA. J. Bacteriol. 1997, 179, 4553–4558. [Google Scholar] [CrossRef][Green Version]
- Nindita, Y.; Nishikawa, T.; Arakawa, K.; Wang, G.; Ochi, K.; Qin, Z.; Kinashi, H. Chromosomal circularization of the model Streptomyces species, Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 2013, 347, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Chapleau, M.; Guertin, J.F.; Farrokhi, A.; Lerat, S.; Burrus, V.; Beaulieu, C. Identification of genetic and environmental factors stimulating excision from Streptomyces scabiei chromosome of the toxicogenic region responsible for pathogenicity. Mol. Plant. Pathol. 2016, 17, 501–509. [Google Scholar] [CrossRef]
- Stratton, K.J.; Bush, M.J.; Chandra, G.; Stevenson, C.E.M.; Findlay, K.C.; Schlimpert, S. Genome-Wide Identification of the LexA-Mediated DNA Damage Response in Streptomyces venezuelae. J. Bacteriol. 2022, 204, e0010822. [Google Scholar] [CrossRef]
- Bacon, J.; Dover, L.G.; Hatch, K.A.; Zhang, Y.; Gomes, J.M.; Kendall, S.; Wernisch, L.; Stoker, N.G.; Butcher, P.D.; Besra, G.S.; et al. Lipid composition and transcriptional response of Mycobacterium tuberculosis grown under iron-limitation in continuous culture: Identification of a novel wax ester. Microbiology 2007, 153, 1435–1444. [Google Scholar] [CrossRef][Green Version]
- Devadasu, E.; Subramanyam, R. Enhanced Lipid Production in Chlamydomonas reinhardtii Caused by Severe Iron Deficiency. Front. Plant. Sci. 2021, 12, 615577. [Google Scholar] [CrossRef]
- Fischer, M.; Alderson, J.; van Keulen, G.; White, J.; Sawers, R.G. The obligate aerobe Streptomyces coelicolor A3(2) synthesizes three active respiratory nitrate reductases. Microbiology 2010, 156, 3166–3179. [Google Scholar] [CrossRef]
- Lejeune, C.; Cornu, D.; Sago, L.; Redeker, V.; Virolle, M.-J. Comparative Proteomic Analysis of Transcriptional and Regulatory Proteins Abundances in S. lividans and S. coelicolor Suggests a Link between Various Stresses and Antibiotic Production. Int. J. Mol. Sci. 2022, 23, 14792. [Google Scholar] [CrossRef]
- Cescut, J.; Fillaudeau, L.; Molina-Jouve, C.; Uribelarrea, J.L. Carbon accumulation in Rhodotorula glutinis induced by nitrogen limitation. Biotechnol. Biofuels 2014, 7, 164. [Google Scholar] [CrossRef] [PubMed]
- Wierzchowska, K.; Zieniuk, B.; Nowak, D.; Fabiszewska, A. Phosphorus and Nitrogen Limitation as a Part of the Strategy to Stimulate Microbial Lipid Biosynthesis. Appl. Sci. 2021, 11, 11819. [Google Scholar]
- Jiang, P.L.; Pasaribu, B.; Chen, C.S. Nitrogen-deprivation elevates lipid levels in Symbiodinium spp. by lipid droplet accumulation: Morphological and compositional analyses. PLoS ONE 2014, 9, e87416. [Google Scholar] [CrossRef]
- Cruz-Morales, P.; Kopp, J.F.; Martinez-Guerrero, C.; Yanez-Guerra, L.A.; Selem-Mojica, N.; Ramos-Aboites, H.; Feldmann, J.; Barona-Gomez, F. Phylogenomic Analysis of Natural Products Biosynthetic Gene Clusters Allows Discovery of Arseno-Organic Metabolites in Model Streptomycetes. Genome Biol. Evol. 2016, 8, 1906–1916. [Google Scholar] [CrossRef]
- Schneider, D.; Bruton, C.J.; Chater, K.F. Duplicated gene clusters suggest an interplay of glycogen and trehalose metabolism during sequential stages of aerial mycelium development in Streptomyces coelicolor A3(2). Mol. Gen. Genet. 2000, 263, 543–553. [Google Scholar] [CrossRef]
- Bhutada, G.; Kavscek, M.; Ledesma-Amaro, R.; Thomas, S.; Rechberger, G.N.; Nicaud, J.M.; Natter, K. Sugar versus fat: Elimination of glycogen storage improves lipid accumulation in Yarrowia lipolytica. FEMS Yeast Res. 2017, 17, fox020. [Google Scholar] [CrossRef]
- Goncalves, E.C.; Koh, J.; Zhu, N.; Yoo, M.J.; Chen, S.; Matsuo, T.; Johnson, J.V.; Rathinasabapathi, B. Nitrogen starvation-induced accumulation of triacylglycerol in the green algae: Evidence for a role for ROC40, a transcription factor involved in circadian rhythm. Plant. J. 2016, 85, 743–757. [Google Scholar] [CrossRef]
- Imlay, J.A. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef]
- Imlay, J.A. Where in the world do bacteria experience oxidative stress? Environ. Microbiol. 2019, 21, 521–530. [Google Scholar] [CrossRef]
- Millan-Oropeza, A.; Henry, C.; Lejeune, C.; David, M.; Virolle, M.J. Expression of genes of the Pho regulon is altered in Streptomyces coelicolor. Sci. Rep. 2020, 10, 8492. [Google Scholar] [CrossRef]
- Virolle, M.J. A Challenging View: Antibiotics Play a Role in the Regulation of the Energetic Metabolism of the Producing Bacteria. Antibiotics 2020, 9, 83. [Google Scholar] [CrossRef] [PubMed]
- Seghezzi, N.; Darbon, E.; Martel, C.; David, M.; Lejeune, C.; Esnault, C.; Virolle, M.-J. The Generation of an Artificial ATP Deficit Triggers Antibiotic Production in Streptomyces lividans. Antibiotics 2022, 11, 1157. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.; Jung, J.; Jang, I.A.; Madsen, E.L.; Park, W. Role of Glyoxylate Shunt in Oxidative Stress Response. J. Biol. Chem. 2016, 291, 11928–11938. [Google Scholar] [CrossRef]
- Sarkhel, R.; Apoorva, S.; Priyadarsini, S.; Sridhar, H.B.; Bhure, S.K.; Mahawar, M. Malate synthase contributes to the survival of Salmonella Typhimurium against nutrient and oxidative stress conditions. Sci. Rep. 2022, 12, 15979. [Google Scholar] [CrossRef]
- Rizwana, R.; Hahn, P.J. CpG methylation reduces genomic instability. J. Cell. Sci. 1999, 112 Pt 24, 4513–4519. [Google Scholar] [CrossRef] [PubMed]
- Nichol, K.; Pearson, C.E. CpG methylation modifies the genetic stability of cloned repeat sequences. Genome Res. 2002, 12, 1246–1256. [Google Scholar] [CrossRef] [PubMed]
- Pisciotta, A.; Sampino, A.M.; Presentato, A.; Galardini, M.; Manteca, A.; Alduina, R. The DNA cytosine methylome revealed two methylation motifs in the upstream regions of genes related to morphological and physiological differentiation in Streptomyces coelicolor A(3)2 M145. Sci. Rep. 2023, 13, 7038. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dulermo, T.; Lejeune, C.; Aybeke, E.; Abreu, S.; Bleton, J.; David, M.; Deniset-Besseau, A.; Chaminade, P.; Thibessard, A.; Leblond, P.; et al. Genome Analysis of a Variant of Streptomyces coelicolor M145 with High Lipid Content and Poor Ability to Synthetize Antibiotics. Microorganisms 2023, 11, 1470. https://doi.org/10.3390/microorganisms11061470
Dulermo T, Lejeune C, Aybeke E, Abreu S, Bleton J, David M, Deniset-Besseau A, Chaminade P, Thibessard A, Leblond P, et al. Genome Analysis of a Variant of Streptomyces coelicolor M145 with High Lipid Content and Poor Ability to Synthetize Antibiotics. Microorganisms. 2023; 11(6):1470. https://doi.org/10.3390/microorganisms11061470
Chicago/Turabian StyleDulermo, Thierry, Clara Lejeune, Ece Aybeke, Sonia Abreu, Jean Bleton, Michelle David, Ariane Deniset-Besseau, Pierre Chaminade, Annabelle Thibessard, Pierre Leblond, and et al. 2023. "Genome Analysis of a Variant of Streptomyces coelicolor M145 with High Lipid Content and Poor Ability to Synthetize Antibiotics" Microorganisms 11, no. 6: 1470. https://doi.org/10.3390/microorganisms11061470
APA StyleDulermo, T., Lejeune, C., Aybeke, E., Abreu, S., Bleton, J., David, M., Deniset-Besseau, A., Chaminade, P., Thibessard, A., Leblond, P., & Virolle, M.-J. (2023). Genome Analysis of a Variant of Streptomyces coelicolor M145 with High Lipid Content and Poor Ability to Synthetize Antibiotics. Microorganisms, 11(6), 1470. https://doi.org/10.3390/microorganisms11061470







