Exopolysaccharides Producing Bacteria: A Review
Abstract
1. Introduction
2. Bacterial Cellulose-Producing Bacteria
3. Levan-Producing Bacteria
4. Xanthan-Producing Bacteria
5. Conclusions and Future Perspective
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ahuja, V.; Bhatt, A.K.; Banu, J.R.; Kumar, V.; Kumar, G.; Yang, Y.H.; Bhatia, S.K. Microbial Exopolysaccharide Composites in Biomedicine and Healthcare: Trends and Advances. Polymers 2023, 15, 1801. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Dey, P. Bacterial Exopolysaccharides as Emerging Bioactive Macromolecules: From Fundamentals to Applications. Res. Microbiol. 2022, 29, 104024. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, Y.A.-G.; El-Naggar, M.E.; Abdel-Megeed, A.; El-Newehy, M. Recent Advancements in Microbial Polysaccharides: Synthesis and Applications. Polymers 2021, 13, 4136. [Google Scholar] [CrossRef] [PubMed]
- Mohd Nadzir, M.; Nurhayati, R.W.; Idris, F.N.; Nguyen, M.H. Biomedical Applications of Bacterial Exopolysaccharides: A Review. Polymers 2021, 13, 530. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Upadhyay, L.S.B. Microbial exopolysaccharides: Synthesis pathways, types and their commercial applications. Int. J. Biol. Macromol. 2020, 157, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Barcelos, M.C.S.; Vespermann, K.A.C.; Pelissari, F.M.; Molina, G. Current status of biotechnological production and applications of microbial exopolysaccharides. Crit. Rev. Food Sci. Nutr. 2019, 60, 1475–1495. [Google Scholar] [CrossRef]
- Osemwegie, O.O.; Adetunji, C.O.; Ayeni, E.A.; Adejobi, O.I.; Arise, R.O.; Nwonuma, C.O.; Oghenekaro, A.O. Exopolysaccharides from bacteria and fungi: Current status and perspectives in Africa. Heliyon 2020, 6, e04205. [Google Scholar] [CrossRef]
- Sutherland, I.W. Bacterial exopolysaccharides. Adv. Microb. Physiol. 1972, 8, 143–213. [Google Scholar] [CrossRef]
- Xiao, M.; Ren, X.; Yu, Y.; Gao, W.; Zhu, C.; Sun, H.; Kong, Q.; Fu, X.; Mou, H. Fucose-containing bacterial exopolysaccharides: Sources, biological activities, and food applications. Food Chem. X 2022, 13, 100233. [Google Scholar] [CrossRef]
- Schmid, J.; Sieber, V.; Rehm, B. Bacterial exopolysaccharides: Biosynthesis pathways and engineering strategies. Front. Microbiol. 2015, 6, 496. [Google Scholar] [CrossRef]
- Pereira, S.B.; Sousa, A.; Santos, M.; Araújo, M.; Serôdio, F.; Granja, P.; Tamagnini, P. Strategies to obtain designer polymers based on cyanobacterial extracellular polymeric substances (EPS). Int. J. Mol. Sci. 2019, 20, 5693. [Google Scholar] [CrossRef] [PubMed]
- Manan, S.; Wajid Ullah, M.; Ul-Islam, M.; Shi, Z.; Gauthier, M.; Yang, G. Bacterial cellulose: Molecular regulation of biosynthesis, supramolecular assembly, and tailored structural and functional properties. Prog. Mater. Sci. 2022, 129, 100972. [Google Scholar] [CrossRef]
- Aditya, T.; Allain, J.P.; Jaramillo, C.; Restrepo, A.M. Surface Modification of Bacterial Cellulose for Biomedical Applications. Int. J. Mol. Sci. 2022, 23, 610. [Google Scholar] [CrossRef] [PubMed]
- Aziz, T.; Farid, A.; Haq, F.; Kiran, M.; Ullah, A.; Zhang, K.; Li, C.; Ghazanfar, S.; Sun, H.; Ullah, R.; et al. A Review on the Modification of Cellulose and Its Applications. Polymers 2022, 14, 3206. [Google Scholar] [CrossRef] [PubMed]
- Angelin, J.; Kavitha, M. Exopolysaccharides from probiotic bacteria and their health potential. Int. J. Biol. Macromol. 2020, 162, 853–865. [Google Scholar] [CrossRef] [PubMed]
- Wünsche, J.; Schmid, J. Acetobacteraceae as exopolysaccharide producers: Current state of knowledge and further perspectives. Front. Bioeng. Biotechnol. 2023, 11, 1166618. [Google Scholar] [CrossRef]
- Díaz-Cornejo, S.; Otero, M.C.; Banerjee, A.; Gordillo-Fuenzalida, F. Biological properties of exopolysaccharides produced by Bacillus spp. Microbiol. Res. 2023, 268, 127276. [Google Scholar] [CrossRef]
- Qi, M.; Zheng, C.; Wu, W.; Yu, G.; Wang, P. Exopolysaccharides from Marine Microbes: Source, Structure and Application. Mar. Drugs 2022, 20, 512. [Google Scholar] [CrossRef]
- Ibrahim, H.A.H.; Abou Elhassayeb, H.E.; El-Sayed, W.M.M. Potential functions and applications of diverse microbial exopolysaccharides in marine environments. J. Genet. Eng. Biotechnol. 2022, 20, 151. [Google Scholar] [CrossRef]
- Jurášková, D.; Ribeiro, S.C.; Silva, C.C.G. Exopolysaccharides Produced by Lactic Acid Bacteria: From Biosynthesis to Health-Promoting Properties. Food 2022, 11, 156. [Google Scholar] [CrossRef]
- Sørensen, H.M.; Rochfort, K.D.; Maye, S.; MacLeod, G.; Brabazon, D.; Loscher, C.; Freeland, B. Exopolysaccharides of Lactic Acid Bacteria: Production, Purification and Health Benefits towards Functional Food. Nutrients 2022, 14, 2938. [Google Scholar] [CrossRef] [PubMed]
- Khalil, M.A.; Sonbol, F.I.; Al-Madboly, L.A.; Aboshady, T.A.; Alqurashi, A.S.; Ali, S.S. Exploring the Therapeutic Potentials of Exopolysaccharides Derived from Lactic Acid Bacteria and Bifidobacteria: Antioxidant, Antitumor, and Periodontal Regeneration. Front. Microbiol. 2022, 13, 803688. [Google Scholar] [CrossRef] [PubMed]
- Werning, M.L.; Hernández-Alcántara, A.M.; Ruiz, M.J.; Soto, L.P.; Dueñas, M.T.; López, P.; Frizzo, L.S. Biological Functions of Exopolysaccharides from Lactic Acid Bacteria and Their Potential Benefits for Humans and Farmed Animals. Foods 2022, 11, 1284. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, J.; Shimizu, H.; Hisa, K.; Matsuzaki, C.; Inuki, S.; Ando, Y.; Nishida, A.; Izumi, A.; Yamano, M.; Ushiroda, C.; et al. Host metabolic benefits of prebiotic exopolysaccharides produced by Leuconostoc mesenteroides. Gut Microbes 2023, 15, 2161271. [Google Scholar] [CrossRef] [PubMed]
- Casillo, A.; Lanzetta, R.; Parrilli, M.; Corsaro, M.M. Exopolysaccharides from marine and marine extremophilic bacteria: Structures, properties, ecological roles and applications. Mar. Drugs 2018, 16, 69. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Salem, D.R.; Sani, R.K. Extremophilic exopolysaccharides: A review and new perspectives on engineering strategies and applications. Carbohydr. Polym. 2019, 5, 8–26. [Google Scholar] [CrossRef]
- Isfahani, F.M.; Tahmourespour, A.; Hoodaji, M.; Ataabadi, M.; Mohammadi, A. Characterizing the new bacterial isolates of high yielding exopolysaccharides under hypersaline conditions. J. Clean. Prod. 2018, 185, 922–928. [Google Scholar] [CrossRef]
- Knowles, E.J.; Castenholz, R.W. Effect of exogenous extracellular polysaccharides on the desiccation and freezing tolerance of rock-inhabiting phototrophic microorganisms. FEMS Microbiol. Ecol. 2008, 66, 261–270. [Google Scholar] [CrossRef]
- Pallach, M.; Marchetti, R.; Di Lorenzo, F.; Fabozzi, A.; Giraud, E.; Gully, D.; Paduano, L.; Molinaro, A.; D’Errico, G.; Silipo, A. Zymomonas mobilis exopolysaccharide structure and role in high ethanol tolerance. Carbohydr. Polym. 2018, 201, 293–299. [Google Scholar] [CrossRef]
- Kuschmierz, L.; Meyer, M.; Bräsen, C.; Wingender, J.; Schmitz, O.J.; Siebers, B. Exopolysaccharide composition and size in Sulfolobus acidocaldarius biofilms. Front. Microbiol. 2022, 13, 982745. [Google Scholar] [CrossRef]
- Secor, P.R.; Michaels, L.A.; Bublitz, D.C.; Jennings, L.K.; Singh, P.K. The Depletion Mechanism Actuates Bacterial Aggregation by Exopolysaccharides and Determines Species Distribution & Composition in Bacterial Aggregates. Front. Cell Infect. Microbiol. 2022, 12, 869736. [Google Scholar] [CrossRef] [PubMed]
- Balducci, E.; Papi, F.; Capialbi, D.E.; Del Bino, L. Polysaccharides’ Structures and Functions in Biofilm Architecture of Antimicrobial-Resistant (AMR) Pathogens. Int. J. Mol. Sci. 2023, 24, 4030. [Google Scholar] [CrossRef] [PubMed]
- Timilsina, S.; Potnis, N.; Newberry, E.A.; Liyanapathiranage, P.; Iruegas-Bocardo, F.; White, F.F.; Goss, E.M.; Jones, J.B. Xanthomonas diversity, virulence, and plant-pathogen interactions. Nat. Rev. Microbiol. 2020, 18, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.L.; Tang, D.J.; Dubrow, Z.E.; Bogdanove, A.; An, S.Q. Xanthomonas campestris Pathovars. Trends Microbiol. 2021, 29, 182–183. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.A.; Onyeziri, M.C.; Fuqua, C. Function and regulation of Agrobacterium tumefaciens cell surface structures that promote attachment. Agrobacterium Biol. 2018, 418, 143–184. [Google Scholar] [CrossRef]
- Ahmed, B.; Jailani, A.; Lee, J.H.; Lee, J. Inhibition of growth, biofilm formation, virulence, and surface attachment of Agrobacterium tumefaciens by cinnamaldehyde derivatives. Front. Microbiol. 2022, 13, 1001865. [Google Scholar] [CrossRef]
- Khalid, M.Y.; Arif, Z.U. Novel biopolymer-based sustainable composites for food packaging applications: A narrative review. Food Packag. Shelf Life 2022, 33, 100892. [Google Scholar] [CrossRef]
- Liu, L.; Li, M.; Yu, M.; Shen, M.; Wang, Q.; Yu, Y.; Xie, J. Natural polysaccharides exhibit anti-tumor activity by targeting gut microbiota. Int. J. Biol. Macromol. 2019, 121, 743–751. [Google Scholar] [CrossRef]
- Chakraborty, I.; Sen, I.K.; Mondal, S.; Rout, D.; Bhanja, S.K.; Maity, G.N.; Maity, P. Bioactive polysaccharides from natural sources: A review on the antitumor and immunomodulating activities. Biocatal. Agric. Biotechnol. 2019, 22, 101425. [Google Scholar] [CrossRef]
- Noufal, Z.M.; Sivaperumal, P.; Elumalai, P.J. Extraction, characterization, and anticancer potential of extracellular polymeric substances from marine actinobacteria of Streptomyces species. Adv. Pharm. Technol. Res. 2022, 13 (Suppl. 1), S125–S129. [Google Scholar] [CrossRef]
- Vinothkanna, A.; Sathiyanarayanan, G.; Rai, A.K.; Mathivanan, K.; Saravanan, K.; Sudharsan, K.; Kalimuthu, P.; Ma, Y.; Sekar, S. Exopolysaccharide Produced by Probiotic Bacillus albus DM-15 Isolated from Ayurvedic Fermented Dasamoolarishta: Characterization, Antioxidant, and Anticancer Activities. Front. Microbiol. 2022, 13, 832109. [Google Scholar] [CrossRef] [PubMed]
- Andrew, M.; Jayaraman, G. Structural features of microbial exopolysaccharides in relation to their antioxidant activity. Carbohydr. Res. 2020, 487, 107881. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Shah, M.D.; Shah, L.; Lee, P.C.; Khan, I. Bacterial polysaccharides-A big source for prebiotics and therapeutics. Front. Nutr. 2022, 9, 1031935. [Google Scholar] [CrossRef] [PubMed]
- Chaisuwan, W.; Jantanasakulwong, K.; Wangtueai, S.; Phimolsiripol, Y.; Chaiyaso, T.; Techapun, C.; Phongthai, S.; You, S.; Regenstein, J.M.; Seesuriyachan, P. Microbial exopolysaccharides for immune enhancement: Fermentation, modifications and bioactivities. Food Biosci. 2020, 35, 100564. [Google Scholar] [CrossRef]
- Moscovici, M.; Balas, C. Bacterial polysaccharides versatile medical uses. In Polysaccharides of Microbial Origin: Biomedical Applications; Oliveira, J.M., Randhouani, H., Reis, R.L., Eds.; Springer: Cham, Switzerland, 2022; pp. 859–891. [Google Scholar]
- Zhai, P.; Peng, X.; Li, B.; Liu, Y.; Sun, H.; Li, X. The application of hyaluronic acid in bone regeneration. Int. J. Biol. Macromol. 2020, 151, 1224–1239. [Google Scholar] [CrossRef]
- Su, C.; Chen, Y.; Tian, S.; Lu, C.; Lv, Q. Research Progress on Emerging Polysaccharide Materials Applied in Tissue Engineering. Polymers 2022, 14, 3268. [Google Scholar] [CrossRef]
- Hu, H.; Xu, F.J. Rational design and latest advances of polysaccharide-based hydrogels for wound healing. Biomater. Sci. 2020, 8, 2084–2101. [Google Scholar] [CrossRef]
- Wu, L.; He, Y.; Mao, H.; Gu, Z. Bioactive hydrogels based on polysaccharides and peptides for soft tissue wound management. J. Mater. Chem. B 2022, 10, 7148–7160. [Google Scholar] [CrossRef]
- Asgher, M.; Qamar, S.A.; Iqbal, H.M.N. Microbial exopolysaccharide-based nanocarriers with unique multi-functionalities for biomedical sectors. Biologia 2021, 76, 673–685. [Google Scholar] [CrossRef]
- Qiu, A.; Wang, Y.; Zhang, G.; Wang, H. Natural Polysaccharide-Based Nanodrug Delivery Systems for Treatment of Diabetes. Polymers 2022, 14, 3217. [Google Scholar] [CrossRef]
- Islam, S.U.; Ul-Islam, M.; Ahsan, H.; Ahmed, M.B.; Shehzad, A.; Fatima, A.; Sonn, J.K.; Lee, Y.S. Potential applications of bacterial cellulose and its composites for cancer treatment. Int. J. Biol. Macromol. 2020, 168, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Cacicedo, M.L.; Islan, G.A.; León, I.E.; Álvarez, V.A.; Chourpa, I.; Allard-Vannier, E.; Castro, G.R. Bacterial cellulose hydrogel loaded with lipid nanoparticles for localized cancer treatment. Colloids Surf. B Biointerfaces 2018, 170, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.K.; Du, S.; Wang, X.; Jiao, Y.; Yin, L.; Zhang, Y.; Guan, Y.Q. Bacterial cellulose based composites enhanced transdermal drug targeting for breast cancer treatment. Chem. Eng. J. 2019, 370, 749–759. [Google Scholar] [CrossRef]
- Inoue, B.S.; Streit, S.; Schneider, A.L.D.S.; Meier, M.M. Bioactive bacterial cellulose membrane with prolonged release of chlorhexidine for dental medical application. Int. J. Biol. Macromol. 2020, 148, 1098–1108. [Google Scholar] [CrossRef]
- Gangalla, R.; Gattu, S.; Palaniappan, S.; Ahamed, M.; Macha, B.; Thampu, R.K.; Fais, A.; Cincotti, A.; Gatto, G.; Dama, M.; et al. Structural Characterisation and Assessment of the Novel Bacillus amyloliquefaciens RK3 Exopolysaccharide on the Improvement of Cognitive Function in Alzheimer’s Disease Mice. Polymers 2021, 13, 2842. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.-J.; Wang, T.; Lang, J.-Q.; Zhou, N.; Ma, M.-G. Multifunctional Cellulose and Cellulose-Based (Nano) Composite Adsorbents. Front. Bioeng. Biotechnol. 2022, 10, 891034. [Google Scholar] [CrossRef] [PubMed]
- Varsha, M.; Senthil Kumar, P.; Senthil Rathi, B. A Review on Recent Trends in the Removal of Emerging Contaminants from Aquatic Environment Using Low-Cost Adsorbents. Chemosphere 2022, 287, 132270. [Google Scholar] [CrossRef]
- Galdino, C.J.S.; Maia, A.D.; Meira, H.M.; Souza, T.C.; Amorim, J.D.; Almeida, F.C.; Costa, A.F.; Sarubbo, L.A. Use of a bacterial cellulose filter for the removal of oil from wastewater. Process. Biochem. 2020, 91, 288–296. [Google Scholar] [CrossRef]
- Kalpana, R.; Angelaalincy, M.J.; Kamatchirajan, B.V.; Vasantha, V.S.; Ashokkumar, B.; Ganesh, V.; Varalakshmi, P. Exopolysaccharide from Bacillus cereus VK1: Enhancement, characterization and its potential application in heavy metal removal. Colloids Surf. B Biointerfaces 2018, 171, 327–334. [Google Scholar] [CrossRef]
- Banerjee, A.; Sarkar, S.; Govil, T.; González-Faune, P.; Cabrera-Barjas, G.; Bandopadhyay, R.; Salem, D.R.; Sani, R.K. Extremophilic Exopolysaccharides: Biotechnologies and Wastewater Remediation. Front. Microbiol. 2021, 12, 721365. [Google Scholar] [CrossRef]
- Salama, A.; Abouzeid, R.; Leong, W.S.; Jeevanandam, J.; Samyn, P.; Dufresne, A.; Bechelany, M.; Barhoum, A. Nanocellulose-Based Materials for Water Treatment: Adsorption, Photocatalytic Degradation, Disinfection, Antifouling, and Nanofiltration. Nanomaterials 2021, 11, 3008. [Google Scholar] [CrossRef] [PubMed]
- Ul-Islam, M.; Khan, S.; Ullah, M.W.; Park, J.K. Bacterial cellulose composites: Synthetic strategies and multiple applications in bio-medical and electro-conductive fields. Biotechnol. J. 2015, 10, 1847–1861. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Ul-Islam, M.; Khattak, W.A.; Park, J.K. Overview of bacterial cellulose composites: A multipurpose advanced material. Carbohydr. Polym. 2013, 98, 1585–1598. [Google Scholar] [CrossRef] [PubMed]
- Mbituyimana, B.; Liu, L.; Ye, W.; Ode Boni, B.O.; Zhang, K.; Chen, J.; Thomas, S.; Revin, V.V.; Shi, Z.; Yang, G. Bacterial cellulose-based composites for biomedical and cosmetic applications: Research progress and existing products. Carbohydr. Polym. 2021, 273, 118565. [Google Scholar] [CrossRef] [PubMed]
- Liyaskina, E.V.; Rakova, N.A.; Kitykina, A.A.; Rusyaeva, V.V.; Toukach, P.V.; Fomenkov, A.; Vainauskas, S.; Roberts, R.J.; Revin, V.V. Production and characterization of the exopolysaccharide from strain Paenibacillus polymyxa 2020. PLoS ONE 2021, 16, e0253482. [Google Scholar] [CrossRef] [PubMed]
- Efremenko, E.; Senko, O.; Maslova, O.; Stepanov, N.; Aslanli, A.; Lyagin, I. Biocatalysts in Synthesis of Microbial Polysaccharides: Properties and Development Trends. Catalysts 2022, 12, 1377. [Google Scholar] [CrossRef]
- Revin, V.V.; Liyaskina, E.V.; Parchaykina, M.V.; Kuzmenko, T.P.; Urgaeva, I.V.; Revin, V.D.; Ullah, M.W. Bacterial Cellulose-Based Polymer Nanocomposites: A Review. Polymers 2022, 14, 4670. [Google Scholar] [CrossRef]
- Xie, Z.; Meng, K.; Yang, X.; Liu, J.; Yu, J.; Zheng, C.; Cao, W.; Liu, H. Identification of a quorum sensing system regulating capsule polysaccharide production and biofilm formation in Streptococcus zooepidemicus. Front. Cell. Infect. Microbiol. 2019, 9, 121. [Google Scholar] [CrossRef]
- Wang, X.L.; Chen, N.; Li, J.; Han, C.F.; Wang, S.; Hao, L.M.; Jia, S.R.; Han, P.P. The effects of quorum sensing molecule farnesol on the yield and activity of extracellular polysaccharide from Grifola frondosa in liquid fermentation. Int. J. Biol. Macromol. 2021, 191, 377–384. [Google Scholar] [CrossRef]
- Popa, L.; Ghica, M.V.; Tudoroiu, E.-E.; Ionescu, D.G.; Dinu-Pîrvu, C.E. Bacterial Cellulose—A Remarkable Polymer as a Source for Biomaterials Tailoring. Materials 2022, 15, 1054. [Google Scholar] [CrossRef]
- Navya, P.V.; Gayathri, V.; Samanta, D.; Sampath, S. Macromolecules Bacterial Cellulose: A Promising Biopolymer with Interesting Properties and Applications. Int. J. Biol. Macromol. 2022, 220, 435–461. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.M.; Shin, E.J. The Nanofication and Functionalization of Bacterial Cellulose and Its Applications. Nanomaterials 2020, 10, 406. [Google Scholar] [CrossRef] [PubMed]
- Ullah, M.W.; Rojas, O.J.; McCarthy, R.R.; Yang, G. Editorial: Nanocellulose: A Multipurpose Advanced Functional Material. Front. Bioeng. Biotechnol. 2021, 9, 738779. [Google Scholar] [CrossRef] [PubMed]
- Ul-Islam, M.; Ahmad, F.; Fatima, A.; Shah, N.; Yasir, S.; Ahmad, M.W.; Manan, S.; Ullah, M.W. Ex situ Synthesis and Characterization of High Strength Multipurpose Bacterial Cellulose- Aloe vera Hydrogels. Front. Bioeng. Biotechnol. 2021, 9, 601988. [Google Scholar] [CrossRef]
- Kaczmarek, M.; Jedrzejczak-Krzepkowska, M.; Ludwicka, K. Comparative Analysis of Bacterial Cellulose Membranes Synthesized by Chosen Komagataeibacter Strains and Their Application Potential. Int. J. Mol. Sci. 2022, 23, 3391. [Google Scholar] [CrossRef] [PubMed]
- Revin, V.V.; Liyas’kina, E.V.; Sapunova, N.B.; Bogatyreva, A.O. Isolation and characterization of the strains producing bacterial cellulose. Microbiology 2020, 14, 86–95. [Google Scholar] [CrossRef]
- Nie, W.; Zheng, X.; Feng, W.; Liu, Y.; Li, Y.; Liang, X. Characterization of Bacterial Cellulose Produced by Acetobacter Pasteurianus MGC-N8819 Utilizing Lotus Rhizome. LWT 2022, 165, 113763. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kim, J.Y.; Cha, M.Y.; Kang, H.C. Characteristics of biocellulose by Gluconobacter uchimurae GYS15. J. Soc. Cosmet. Sci. Korea 2016, 42, 247–255. [Google Scholar] [CrossRef]
- Barnhart, D.M.; Su, S.; Farrand, S.K. A signaling pathway involving the diguanylate cyclase CelR and the response regulator DivK controls cellulose synthesis in Agrobacterium tumefaciens. J. Bacteriol. 2014, 196, 1257–1274. [Google Scholar] [CrossRef]
- Hasanin, M.S.; Abdelraof, M.; Hashem, A.H.; El Saied, H. Sustainable bacterial cellulose production by Achromobacter using mango peel waste. Microb. Cell Fact. 2023, 22, 24. [Google Scholar] [CrossRef]
- Ji, K.; Wang, W.; Zeng, B.; Chen, S.; Zhao, Q.; Chen, Y.; Li, G.; Ma, T. Bacterial cellulose synthesis mechanism of facultative anaerobe Enterobacter sp. FY-07. Sci. Rep. 2016, 6, 21863. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Ji, K.; Zhang, Y.; Liu, X.; Dai, X.; Zhi, B.; Cao, Y.; Liu, D.; Wu, M.; Li, G.; et al. Microbial enhanced oil recovery through deep profile control using a conditional bacterial cellulose-producing strain derived from Enterobacter sp. FY-07. Microb. Cell. Fact. 2020, 19, 59. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Mendoza, D.; Romero-Jiménez, L.; Rodríguez-Carvajal, M.Á.; Lorite, M.J.; Muñoz, S.; Olmedilla, A.; Sanjuán, J. The Role of Two Linear β-Glucans Activated by c-di-GMP in Rhizobium etli CFN42. Biology 2022, 11, 1364. [Google Scholar] [CrossRef] [PubMed]
- Ardré, M.; Dufour, D.; Rainey, P.B. Causes and Biophysical Consequences of Cellulose Production by Pseudomonas fluorescens SBW25 at the Air-Liquid Interface. J. Bacteriol. 2019, 201, e00110-19. [Google Scholar] [CrossRef]
- Fratty, I.S.; Shachar, D.; Katsman, M.; Yaron, S. The activity of BcsZ of Salmonella Typhimurium and its role in Salmonella-plants interactions. Front. Cell. Infect. Microbiol. 2022, 12, 967796. [Google Scholar] [CrossRef]
- Bagewadi, Z.K.; Bhavikatti, J.S.; Muddapur, U.M.; Yaraguppi, D.A.; Mulla, S.I. Statistical Optimization and Characterization of Bacterial Cellulose Produced by Isolated Thermophilic Bacillus licheniformis Strain ZBT2. Carbohydr. Res. 2020, 491, 107979. [Google Scholar] [CrossRef]
- Li, G.; Wang, L.; Deng, Y.; Wei, Q. Research progress of the biosynthetic strains and pathways of bacterial cellulose. J. Ind. Microbiol. Biotechnol. 2022, 49, kuab071. [Google Scholar] [CrossRef]
- Komagata, K.; Lino, T.; Yamada, Y. The Family Acetobacteraceae. In The Prokaryotes Alphaproteobacteria and Betaproteobacteria; De Long, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2014; pp. 3–78. [Google Scholar]
- Yamada, Y.; Yukphan, P.; Lan Vu, H.T.; Muramatsu, Y.; Ochaikul, D.; Tanasupawat, S.; Nakagawa, Y. Description of Komagataeibacter gen. nov., with proposals of new combinations (Acetobacteraceae). J. Gen. Appl. Microbiol. 2012, 58, 397–404. [Google Scholar] [CrossRef]
- Yamada, Y. Systematics of Acetic Acid Bacteria. In Acetic Acid Bacteria: Ecology and Physiology; Matsushita, K., Toyama, H., Tonouchi, N., Okamoto-Kainuma, A., Eds.; Springer: Tokyo, Japan, 2016; pp. 1–50. [Google Scholar]
- Liu, L.X.; Liu, S.X.; Wang, Y.M.; Bi, J.C.; Chen, H.M.; Deng, J.; Zhang, C.; Hu, Q.S.; Li, C.F. Komagataeibacter cocois sp. nov., a novel cellulose-producing strain isolated from coconut milk. Int. J. Syst. Evol. Microbiol. 2018, 68, 3125–3131. [Google Scholar] [CrossRef]
- Naloka, K.; Yukphan, P.; Matsutani, M.; Matsushita, K.; Theeragool, G. Komagataeibacter diospyri sp. nov., a novel species of thermotolerant bacterial nanocellulose-producing bacterium. Int. J. Syst. Evol. Microbiol. 2020, 70, 251–258. [Google Scholar] [CrossRef]
- Skraban, J.; Cleenwerck, I.; Vandamme, P.; Fanedl, L.; Trcek, J. Genome sequences and description of novel exopolysaccharides producing species Komagataeibacter pomaceti sp. nov. and reclassification of Komagataeibacter kombuchae (Dutta and Gachhui 2007) Yamada et al., 2013 as a later heterotypic synonym of Komagataeibacter hansenii (Gossele et al. 1983) Yamada et al., 2013. Syst. Appl. Microbiol. 2018, 41, 581–592. [Google Scholar] [PubMed]
- Yamada, Y.; Yukpan, P.; Vu, H.T.L.; Muramatsu, Y.; Ochaikul, D.; Nakagawa, Y. Subdivision of the genus Gluconacetobacter Yamada, Hoshino and Ishikawa 1998: The proposal of Komagatabacter gen. nov., for strains accomodated to the Gluconacetobacter xylinus group in the α-Proteobacteria. Ann. Microbiol. 2012, 62, 849–859. [Google Scholar] [CrossRef]
- Yamada, Y. Transfer of Gluconacetobacter kakiaceti, Gluconacetobacter medellinensis and Gluconacetobacter maltaceti to the genus Komagataeibacter as Komagataeibacter kakiaceti comb. nov., Komagataeibacter medellinensis comb. nov. and Komagataeibacter maltaceti comb. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 1670–1672. [Google Scholar] [PubMed]
- Zhang, Q.; Poehlein, A.; Hollensteiner, J.; Daniela, R. Draft Genome Sequence of Komagataeibacter maltaceti LMG 1529 T, a Vinegar-Producing Acetic Acid Bacterium Isolated from Malt Vinegar Brewery Acetifiers. Genome Announc. 2018, 6, e00330-18. [Google Scholar] [CrossRef] [PubMed]
- Hordt, A.; Lopez, M.G.; Meier-Kolthoff, J.P.; Schleuning, M.; Weinhold, L.M.; Tindall, B.J.; Gronow, S.; Kyrpides, N.C.; Woyke, T.; Goker, M. Analysis of 1,000+ Type-Strain Genomes Substantially Improves Taxonomic Classification of Alphaproteobacteria. Front. Microbiol. 2020, 11, 468. [Google Scholar] [CrossRef] [PubMed]
- Marič, L.; Cleenwerck, I.; Accetto, T.; Vandamme, P.; Trček, J. Description of Komagataeibacter melaceti sp. nov. and Komagataeibacter melomenusus sp. nov. Isolated from Apple Cider Vinegar. Microorganisms 2020, 8, 1178. [Google Scholar] [CrossRef] [PubMed]
- Ogino, H.; Azuma, Y.; Hosoyama, A.; Nakazawa, H.; Matsutani, M.; Hasegawa, A.; Otsuyama, K.; Matsushita, K.; Fujita, N.; Shirai, M. Complete genome sequence of NBRC 3288, a unique cellulose-nonproducing strain of Gluconacetobacter xylinus isolated from vinegar. J. Bacteriol. 2011, 193, 6997–6998. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xu, X.; Chen, X.; Yuan, F.; Sun, B.; Xu, Y.; Yang, J.; Sun, D. Complete genome sequence of the cellulose-producingstrain Komagataeibacter nataicola RZS01. Sci. Rep. 2017, 7, 4431. [Google Scholar] [CrossRef]
- Florea, M.; Reeve, B.; Abbott, J.; Freemont, P.S.; Ellis, T. Genome sequence and plasmid transformation of the model high-yield bacterial cellulose producer Gluconacetobacter hansenii ATCC 53582. Sci. Rep. 2016, 6, 23635. [Google Scholar] [CrossRef]
- Kubiak, K.; Kurzawa, M.; Jędrzejczak-Krzepkowska, M.; Ludwicka, K.; Krawczyk, M.; Migdalski, A.; Kacprzak, M.M.; Loska, D.; Krystynowicz, A.; Bielecki, S. Complete genome sequence of Gluconacetobacter xylinus E25 strain-Valuable and effective producer of bacterial nanocellulose. J. Biotechnol. 2014, 176, 18–19. [Google Scholar] [CrossRef]
- Liu, M.; Liu, L.; Jia, S. Complete genome analysis of Gluconacetobacter xylinus CGMCC 2955 for elucidating bacterial cellulose biosynthesis and metabolic regulation. Sci. Rep. 2018, 8, 6266. [Google Scholar] [CrossRef] [PubMed]
- Ryngajllo, M.; Kubiak, K.; Jędrzejczak-Krzepkowska, M.; Jacek, P.; Bielecki, S. Comparative genomics of the Komagataeibacter strains-Efficient bionanocellulose producers. Microbiologyopen 2018, 8, e00731. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Gao, H.; Liao, B.; Wu, J.; Zhang, W.; Huang, J.; Liu, M.; Huang, J.; Chang, Z.; Jin, M.; et al. Characterization and optimization of production of bacterial cellulose from strain CGMCC 17276 based on whole-genome analysis. Carbohydr. Polym. 2020, 232, 115788. [Google Scholar] [CrossRef] [PubMed]
- Cannazza, P.; Rissanen, A.J.; Guizelini, D.; Losoi, P.; Sarlin, E.; Romano, D.; Santala, V.; Mangayil, R. Characterization of Komagataeibacter Isolate Reveals New Prospects in Waste Stream Valorization for Bacterial Cellulose Production. Microorganisms. 2021, 9, 2230. [Google Scholar] [CrossRef]
- Mangayil, R.; Rissanen, A.J.; Pammo, A.; Guizelini, D.; Losoi, P.; Sarlin, E.; Tuukkanen, S.; Santala, V. Characterization of a novel bacterial cellulose producer for the production of eco-friendly piezoelectric-responsive films from a minimal medium containing waste carbon. Cellulose 2021, 28, 671–689. [Google Scholar] [CrossRef]
- Hollensteiner, J.; Poehlein, A.; Kloskowski, P.; Ali, T.T.; Daniel, R. Genome Sequence of Komagataeibacter saccharivorans Strain JH1, Isolated from Fruit Flies. Microbiol. Resour. Announc. 2020, 9, e00098-20. [Google Scholar] [CrossRef] [PubMed]
- Nagmetova, G.; Berdimuratova, K.; Amirgazin, A.; Shevtsov, V.; Ismailova, A.; Ramankulov, Y.; Shevtsov, A.; Kurmanbayev, A. Draft genome sequence of a potential commercial biocellulose producer, strain Komagataeibacter europaeus GH1. Microbiol. Resour. Announc. 2020, 9, e00963-20. [Google Scholar] [CrossRef]
- Cannazza, P.; Rissanen, A.J.; Sarlin, E.; Guizelini, D.; Minardi, C.; Losoi, P.; Molinari, F.; Romano, D.; Mangayil, R. Characterization, genome analysis and genetic tractability studies of a new nanocellulose producing Komagataeibacter intermedius isolate. Sci. Rep. 2022, 12, 20520. [Google Scholar] [CrossRef]
- Ryngajłło, M.; Jędrzejczak-Krzepkowska, M.; Kubiak, K.; Ludwicka, K.; Bielecki, S. Towards control of cellulose biosynthesis by Komagataeibacter using systems-level and strain engineering strategies: Current progress and perspectives. Appl. Microbiol. Biotechnol. 2020, 104, 6565–6585. [Google Scholar] [CrossRef]
- Prudnikova, S.V.; Shidlovsky, I.P. The New Strain of Acetic Acid Bacteria Komagataeibacter xylinus B-12068–Producer of Bacterial Cellulose for Biomedical Applications. J. Sib. Fed. Univ. Biology 2017, 10, 246–254. [Google Scholar] [CrossRef]
- Krystynowicz, A.; Koziołkiewicz, M.; Wiktorowska-Jezierska, A.; Bielecki, S.; Klemenska, E.; Masny, A.; Płucienniczak, A. Molecular basis of cellulose biosynthesis disappearance in submerged culture of Acetobacter xylinum. Acta Biochim. Pol. 2005, 52, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Strap, J.L.; Latos, A.; Shim, I.; Bonetta, D.T. Characterization of pellicle inhibition in Gluconacetobacter xylinus 53582 by a small molecule, pellicin, identified by a chemical genetics screen. PLoS ONE 2011, 6, e28015. [Google Scholar] [CrossRef] [PubMed]
- Gromovykh, T.I.; Sadykova, V.S.; Lutcenko, S.V.; Dmitrenok, A.S.; Feldman, N.B.; Danilchuk, T.N.; Kashirin, V.V. Bacterial cellulose synthesized by Gluconacetobacter hansenii for medical applications. Appl. Biochem. Microbiol. 2017, 53, 60–67. [Google Scholar] [CrossRef]
- Volova, T.; Prudnikova, S.V.; Sukovatyi, A.G.; Shishatskaya, E.I. Production and properties of bacterial cellulose by the strain Komagataeibacter xylinus B-12068. Appl. Microbiol. Biotechnol. 2018, 102, 7417–7428. [Google Scholar] [CrossRef]
- Semjonovs, P.; Ruklisha, M.; Paegle, L.; Saka, M.; Treimane, R.; Skute, M.; Rozenberga, L.; Vikele, L.; Sabovics, M.; Cleenwerck, I. Cellulose synthesis by Komagataeibacter rhaeticus strain P 1463 isolated from Kombucha. Appl. Microbiol. Biotechnol. 2017, 101, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Jacek, P.; da Silva, F.A.S.; Dourado, F.; Bielecki, S.; Gama, M. Optimization and characterization of bacterial nanocellulose produced by Komagataeibacter rhaeticus K3. Carbohydr. Polym. Technol. Appl. 2021, 2, 100022. [Google Scholar] [CrossRef]
- Dos Santos, R.A.; Berretta, A.A.; Barud, H.S. Draft Genome Sequence of Komagataeibacter intermedius Strain AF2, a Producer of Cellulose, Isolated from Kombucha Tea. Genome Announc. 2015, 3, e01404-15. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Rutherfurd-Markwick, K.; Zhang, X.-X.; Mutukumira, A.N. Kombucha: Production and Microbiological Research. Foods 2022, 11, 3456. [Google Scholar] [CrossRef]
- Landis, E.A.; Fogarty, E.; Edwards, J.C.; Popa, O.; Eren, A.M.; Wolfe, B.E. Microbial Diversity and Interaction Specificity in Kombucha Tea Fermentations. mSystems 2022, 7, e0015722. [Google Scholar] [CrossRef]
- Kahraman-Ilıkkan, Ö. Microbiome composition of kombucha tea from Türkiye using high-throughput sequencing. J. Food Sci. Technol. 2023, 60, 1826–1833. [Google Scholar] [CrossRef]
- De Filippis, F.; Troise, A.D.; Vitaglione, P.; Ercolini, D. Different temperatures select distinctive acetic acid bacteria species and promotes organic acids production during Kombucha tea fermentation. Food Microbiol. 2018, 73, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Arıkan, M.; Mitchell, A.L.; Finn, R.D.; Gürel, F. Microbial composition of Kombucha determined using amplicon sequencing and shotgun metagenomics. J. Food Sci. 2020, 85, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Harrison, K.; Curtin, C. Microbial composition of SCOBY starter cultures used by commercial kombucha brewers in North America. Microorganisms 2021, 9, 1060. [Google Scholar] [CrossRef] [PubMed]
- Andrés-Barrao, C.; Saad, M.M.; Cabello Ferrete, E.; Bravo, D.; Chappuis, M.L.; Ortega Pérez, R.; Junier, P.; Perret, X.; Barja, F. Metaproteomics and ultrastructure characterization of Komagataeibacter spp. involved in high-acid spirit vinegar production. Food Microbiol. 2016, 55, 112–122. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, J.N.; Wee, Y.J.; Park, D.H.; Ryu, H.W. Production of bacterial cellulose by Gluconacetobacter sp. RKY5 isolated from persimmon vinegar. Appl. Biochem. Biotechnol. 2006, 131, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Castro, C.; Zuluaga, R.; Álvarez, C.; Putaux, J.L.; Caro, G.; Rojas, O.J.; Mondragon, I.; Gañán, P. Bacterial cellulose produced by a new acid-resistant strain of Gluconacetobacter genus. Carbohydr. Polym. 2012, 89, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Cepec, E.; Trček, J. Antimicrobial Resistance of Acetobacter and Komagataeibacter Species Originating from Vinegars. Int. J. Environ. Res. Public Health 2022, 19, 463. [Google Scholar] [CrossRef] [PubMed]
- Greser, A.B.; Avcioglu, N.H. Optimization and Physicochemical Characterization of Bacterial Cellulose by Komagataeibacter nataicola and Komagataeibacter maltaceti Strains Isolated from Grape, Thorn Apple and Apple Vinegars. Arch. Microbiol. 2022, 204, 465. [Google Scholar] [CrossRef]
- Lin, S.P.; Huang, Y.-H.; Hsu, K.-D.; Lai, Y.J.; Chen, Y.K.; Cheng, K.C. Isolation and identification of cellulose-producing strain Komagataeibacter intermedius from fermented fruit juice. Carbohydr. Polym. 2016, 151, 827–833. [Google Scholar] [CrossRef]
- Dellaglio, F.; Cleenwerck, I.; Felis, G.E. Description of Gluconacetobacter swingsii sp. nov. and Gluconacetobacter rhaeticus sp. nov., isolated from Italian apple fruit. Int. J. Syst. Evol. Microbiol. 2005, 55, 2365–2370. [Google Scholar] [CrossRef]
- Kim, S.S.; Lee, S.Y.; Park, K.J.; An, H.J.; Hyun, J.M.; Choi, Y.H. Gluconacetobacter sp. gel_SEA623-2, bacterial cellulose producing bacterium isolated from citrus fruit juice. Saudi J. Biol. Sci. 2017, 24, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Jahan, F.; Kumar, V.; Rawat, G.; Saxena, R.K. Production of microbial cellulose by a bacterium isolated from fruit. Biochem. Biotechnol. 2012, 167, 1157–1171. [Google Scholar] [CrossRef] [PubMed]
- Augimeri, R.V.; Varley, A.J.; Strap, J.L. Establishing a Role for Bacterial Cellulose in Environmental Interactions: Lessons Learned from Diverse Biofilm-Producing Proteobacteria. Front. Microbiol. 2015, 6, 1282. [Google Scholar] [CrossRef] [PubMed]
- Römling, U.; Galperin, M.Y. Bacterial cellulose biosynthesis: Diversity of operons, subunits, products, and functions. Trends Microbiol. 2015, 23, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Castiblanco, L.F.; Sundin, G.W. Cellulose production, activated by cyclic di-GMP through BcsA and BcsZ, is a virulence factor and an essential determinant of the three-dimensional architectures of biofilms formed by Erwinia amylovora Ea1189. Mol. Plant Pathol. 2018, 19, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Varghese, A.; Ray, S.; Verma, T.; Nandi, D. Multicellular String-Like Structure Formation by Salmonella Typhimurium Depends on Cellulose Production: Roles of Diguanylate Cyclases, YedQ and YfiN. Front. Microbiol. 2020, 11, 613704. [Google Scholar] [CrossRef] [PubMed]
- Lamprokostopoulou, A.; Römling, U. Yin and Yang of Biofilm Formation and Cyclic di-GMP Signaling of the Gastrointestinal Pathogen Salmonella enterica Serovar Typhimurium. J. Innate Immun. 2022, 14, 275–292. [Google Scholar] [CrossRef]
- Lee, K.Y.; Buldum, G.; Mantalaris, A.; Bismarck, A. More than meets the eye in bacterial cellulose: Biosynthesis, bioprocessing, and applications in advanced fiber. Macromol. Biosci. 2014, 14, 10–32. [Google Scholar] [CrossRef]
- Valera, M.J.; Mas, A.; Streit, W.R.; Mateo, E. GqqA, a novel protein in Komagataeibacter europaeus involved in bacterial quorum quenching and cellulose formation. Microb. Cell Fact. 2016, 24, 88. [Google Scholar] [CrossRef]
- Isenberg, R.Y.; Christensen, D.G.; Visick, K.L.; Mandel, M.J. High Levels of Cyclic Diguanylate Interfere with Beneficial Bacterial Colonization. mBio 2022, 13, e0167122. [Google Scholar] [CrossRef]
- Thongsomboon, W.; Serra, D.O.; Possling, A.; Hadjineophytou, C.; Hengge, R.; Cegelski, L. Phosphoethanolamine Cellulose: A Naturally Produced Chemically Modified Cellulose. Science 2018, 359, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Bajeli, S.; Kaushal, D.; Radotra, B.D.; Kumar, A. Biofilm formation in the lung contributes to virulence and drug tolerance of Mycobacterium tuberculosis. Nat. Commun. 2021, 12, 1606. [Google Scholar] [CrossRef] [PubMed]
- McNamara, J.T.; Morgan, J.L.; Zimmer, J. A molecular description of cellulose biosynthesis. Annu. Rev. Biochem. 2015, 84, 895–921. [Google Scholar] [CrossRef] [PubMed]
- Abidi, W.; Torres-Sánchez, L.; Siroy, A.; Krasteva, P.V. Weaving of bacterial cellulose by the Bcs secretion systems. FEMS Microbiol. Rev. 2022, 46, fuab051. [Google Scholar] [CrossRef] [PubMed]
- Singhania, R.R.; Patel, A.K.; Tsai, M.L.; Chen, C.W.; Di Dong, C. Genetic modification for enhancing bacterial cellulose production and its applications. Bioengineered 2021, 12, 6793–6807. [Google Scholar] [CrossRef] [PubMed]
- Moradi, M.; Jacek, P.; Farhangfar, A.; Guimarães, J.T.; Forough, M. The role of genetic manipulation and in situ modifications on production of bacterial nanocellulose: A review. Int. J. Biol. Macromol. 2021, 183, 635–650. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.H.; Teng, H.Y.; Lee, C.K. Knockout of glucose dehydrogenase gene in Gluconacetobacter xylinus for bacterial cellulose production enhancement. Biotechnol Bioprocess Eng. 2015, 20, 18–25. [Google Scholar] [CrossRef]
- Imai, T.; Sun, S.J.; Horikawa, Y.; Wada, M.; Sugiyama, J. Functional reconstitution of cellulose synthase in Escherichia coli. Biomacromolecules 2014, 15, 4206–4213. [Google Scholar] [CrossRef] [PubMed]
- Al-Janabi, S.S.; Shawky, H.; El-Waseif, A.A.; Farrag, A.A.; Abdelghany, T.M.; El-Ghwas, D.E. Stable, efficient, and cost-effective system for the biosynthesis of recombinant bacterial cellulose in Escherichia coli DH5_ platform. J. Gen. Eng. Biotechnol. 2022, 20, 90. [Google Scholar] [CrossRef]
- Jozala, A.F.; de Lencastre-Novaes, L.C.; Lopes, A.M.; de Carvalho Santos-Ebinuma, V.; Mazzola, P.G.; Pessoa, A., Jr.; Grotto, D.; Gerenutti, M.; Chaud, M.V. Bacterial nanocellulose production and application: A 10-year overview. Appl. Microbiol. Biotechnol. 2016, 100, 2063–2072. [Google Scholar] [CrossRef]
- Revin, V.; Liyaskina, E.; Nazarkina, M.; Bogatyreva, A.; Shchankin, M. Cost-Effective Production of Bacterial Cellulose Using Acidic Food Industry by-Products. Braz. J. Microbiol. 2018, 49, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Ul-Islam, M.; Ullah, M.W.; Khan, S.; Park, J.K. Production of bacterial cellulose from alternative cheap and waste resources: A step for cost reduction with positive environmental aspects. Korean J. Chem. Eng. 2020, 37, 925–937. [Google Scholar] [CrossRef]
- Czaja, W.; Romanovicz, D.; Brown, R.M. Structural investigations of microbial cellulose produced in stationary and agitated culture. Cellulose 2004, 11, 403–411. [Google Scholar] [CrossRef]
- Bi, J.-C.; Liu, S.-X.; Li, C.-F.; Li, J.; Liu, L.-X.; Deng, J.; Yang, Y.-C. Morphology and structure characterization of bacterial celluloses produced by different strains in agitated culture. J. Appl. Microbiol. 2014, 117, 1305–1311. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.U.; Ullah, M.W.; Khan, S.; Shah, N.; Park, J.K. Strategies for cost-effective and enhanced production of bacterial cellulose. Int. J. Biol. Macromol. 2017, 102, 1166–1173. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Sun, Q.; Han, Z.; Li, J.; Liao, B.; Hu, L.; Huang, J.; Zou, C.; Jia, C.; Huang, J.; et al. Comparison of bacterial nanocellulose produced by different strains under static and agitated culture conditions. Carbohydr. Polym. 2020, 227, 115323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, X.; Qi, X.; Ren, L.; Qiang, T. Isolation and identification of a bacterial cellulose synthesizing strain from kombucha in different conditions: Gluconacetobacter xylinus ZHCJ618. Food Sci. Biotechnol. 2018, 27, 705–713. [Google Scholar] [CrossRef]
- Ullah, M.W.; Ul-Islam, M.; Khan, S.; Kim, Y.; Park, J.K. Innovative production of bio-cellulose using a cell-free system derived from a single cell line. Carbohydr. Polym. 2015, 132, 286–294. [Google Scholar] [CrossRef]
- Ullah, M.W.; Khattak, W.A.; Ul-Islam, M.; Khan, S.; Park, J.K. Metabolic engineering of synthetic cell-free systems: Strategies and applications. Biochem. Eng. J. 2016, 105, 391–405. [Google Scholar] [CrossRef]
- Kim, Y.; Ullah, M.W.; Ul-Islam, M.; Khan, S.; Jang, J.H.; Park, J.K. Self-assembly of bio-cellulose nanofibrils through intermediate phase in a cell-free enzyme system. Biochem. Eng. J. 2019, 142, 135–144. [Google Scholar] [CrossRef]
- Liu, K.; Catchmark, J.M. Bacterial cellulose/hyaluronic acid nanocomposites production through co-culturing Gluconacetobacter hansenii and Lactococcus lactis in a two-vessel circulating system. Bioresour. Technol. 2019, 290, 121715. [Google Scholar] [CrossRef] [PubMed]
- Cubas, A.L.V.; Provin, A.P.; Dutra, A.R.A.; Mouro, C.; Gouveia, I.C. Advances in the Production of Biomaterials through Kombucha Using Food Waste: Concepts, Challenges, and Potential. Polymers 2023, 15, 1701. [Google Scholar] [CrossRef] [PubMed]
- Sionkowska, A.; Mezykowska, O.; Piatek, J. Bacterial nanocelullose in biomedical applications: A review. Polym. Int. 2019, 68, 1841–1847. [Google Scholar] [CrossRef]
- Choi, S.M.; Rao, K.M.; Zo, S.M.; Shin, E.J.; Han, S.S. Bacterial Cellulose and Its Applications. Polymers 2022, 14, 1080. [Google Scholar] [CrossRef] [PubMed]
- Gorgieva, S. Bacterial Cellulose as a Versatile Platform for Research and Development of Biomedical Materials. Processes 2020, 8, 624. [Google Scholar] [CrossRef]
- Nicu, R.; Ciolacu, F.; Ciolacu, D.E. Advanced Functional Materials Based on Nanocellulose for Pharmaceutical/Medical Applications. Pharmaceutics 2021, 13, 1125. [Google Scholar] [CrossRef] [PubMed]
- Pandit, A.; Kumar, R. A Review on Production, Characterization and Application of Bacterial Cellulose and Its Biocomposites. J. Polym. Environ. 2021, 29, 2738–2755. [Google Scholar] [CrossRef]
- Swingler, S.; Gupta, A.; Gibson, H.; Kowalczuk, M.; Heaselgrave, W.; Radecka, I. Recent advances and applications of bacterial cellulose in biomedicine. Polymers 2021, 13, 412. [Google Scholar] [CrossRef]
- Randhawa, A.; Dutta, S.D.; Ganguly, K.; Patil, T.V.; Patel, D.K.; Lim, K.-T. A Review of Properties of Nanocellulose, Its Synthesis, and Potential in Biomedical Applications. Appl. Sci. 2022, 12, 7090. [Google Scholar] [CrossRef]
- Avcioglu, N.H. Bacterial cellulose: Recent progress in production and industrial applications. World J. Microbiol. Biotechnol. 2022, 38, 86. [Google Scholar] [CrossRef]
- Ullah, M.W.; Ul-Islam, M.; Wahid, F.; Yang, G. Editorial: Nanocellulose: A Multipurpose Advanced Functional Material, Volume II. Front. Bioeng. Biotechnol. 2022, 10, 931256. [Google Scholar] [CrossRef] [PubMed]
- El-Gendi, H.; Tarek, T.H.; Ray, J.B.; Saleh, A.K. Recent Advances in Bacterial Cellulose: A Low-Cost Effective Production Media, Optimization Strategies and Applications. Cellulose 2022, 29, 7495–7533. [Google Scholar] [CrossRef]
- Norizan, M.N.; Shazleen, S.S.; Alias, A.H.; Sabaruddin, F.A.; Asyraf, M.R.M.; Zainudin, E.S.; Abdullah, N.; Samsudin, M.S.; Kamarudin, S.H.; Norrrahim, M.N.F. Nanocellulose-Based Nanocomposites for Sustainable Applications: A Review. Nanomaterials 2022, 12, 3483. [Google Scholar] [CrossRef] [PubMed]
- Poulose, A.; Parameswaranpillai, J.; George, J.J.; Gopi, J.A.; Krishnasamy, S.; Dominic, C.D.M.; Hameed, N.; Salim, N.V.; Radoor, S.; Sienkiewicz, N. Nanocellulose: A Fundamental Material for Science and Technology Applications. Molecules 2022, 27, 8032. [Google Scholar] [CrossRef] [PubMed]
- Singhania, R.R.; Patel, A.K.; Tseng, Y.S.; Kumar, V.; Chen, C.W.; Haldar, D.; Saini, J.K.; Dong, C.-D. Developments in Bioprocess for Bacterial Cellulose Production. Bioresour. Technol. 2022, 344, 126343. [Google Scholar] [CrossRef]
- Wahid, F.; Huang, L.-H.; Zhao, X.-Q.; Li, W.-C.; Wang, Y.-Y.; Jia, S.-R.; Zhong, C. Bacterial cellulose and its potential for biomedical applications. Biotechnol. Adv. 2021, 53, 107856. [Google Scholar] [CrossRef]
- Kushwaha, A.; Goswami, L.; Kim, B.S. Nanomaterial-based therapy for wound healing. Nanomaterials 2022, 12, 618. [Google Scholar] [CrossRef]
- Niculescu, A.G.; Grumezescu, A.M. An up-to-date review of biomaterials application in wound management. Polymers 2022, 14, 421. [Google Scholar] [CrossRef]
- Moradpoor, H.; Mohammadi, H.; Safaei, M.; Mozaffari, H.R.; Sharifi, R.; Gorji, P.; Sulong, A.B.; Muhamad, N.; Ebadi, M. Recent Advances on Bacterial Cellulose-Based Wound Management: Promises and Challenges. Int. J. Polym. Sci. 2022, 1, 1214734. [Google Scholar] [CrossRef]
- Jankau, J.; Błażyńska-Spychalska, A.; Kubiak, K.; Jędrzejczak-Krzepkowska, M.; Pankiewicz, T.; Ludwicka, K.; Dettlaff, A.; Pęksa, R. Bacterial Cellulose Properties Fulfilling Requirements for a Biomaterial of Choice in Reconstructive Surgery and Wound Healing. Front. Bioeng. Biotechnol. 2022, 9, 805053. [Google Scholar] [CrossRef]
- Zahel, P.; Beekmann, U.; Eberlein, T.; Schmitz, M.; Werz, O.; Kralisch, D. Bacterial Cellulose-Adaptation of a Nature-Identical Material to the Needs of Advanced Chronic Wound Care. Pharmaceuticals 2022, 15, 683. [Google Scholar] [CrossRef] [PubMed]
- Horue, M.; Silva, J.M.; Berti, I.R.; Brandão, L.R.; Barud, H.D.S.; Castro, G.R. Bacterial Cellulose-Based Materials as Dressings for Wound Healing. Pharmaceutics 2023, 15, 424. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.M.; Ngoc Le, T.T.; Nguyen, A.T.; Thien Le, H.N.; Pham, T.T. Biomedical materials for wound dressing: Recent advances and applications. RSC Adv. 2023, 13, 5509–5528. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Xi, Y.; Weng, Y. Recent Advances in Cellulose-Based Hydrogels for Tissue Engineering Applications. Polymers 2022, 14, 3335. [Google Scholar] [CrossRef] [PubMed]
- Tsiklin, I.L.; Shabunin, A.V.; Kolsanov, A.V.; Volova, L.T. In Vivo Bone Tissue Engineering Strategies: Advances and Prospects. Polymers 2022, 14, 3222. [Google Scholar] [CrossRef]
- Iravani, S.; Varma, R.S. Cellulose-Based Composites as Scaffolds for Tissue Engineering: Recent Advances. Molecules 2022, 27, 8830. [Google Scholar] [CrossRef]
- Janmohammadi, M.; Nazemi, Z.; Salehi, A.O.M.; Seyfoori, A.; John, J.V.; Nourbakhsh, M.S.; Akbari, M. Cellulose-based composite scaffolds for bone tissue engineering and localized drug delivery. Bioact. Mater. 2022, 20, 137–163. [Google Scholar] [CrossRef]
- Lin, L.; Jiang, S.; Yang, J.; Qiu, J.; Jiao, X.; Yue, X.; Ke, X.; Yang, G.; Zhang, L. Application of 3D-bioprinted nanocellulose and cellulose derivative-based bio-inks in bone and cartilage tissue engineering. Int. J. Bioprint. 2022, 9, 637. [Google Scholar] [CrossRef]
- Raut, M.P.; Asare, E.; Syed Mohamed, S.M.D.; Amadi, E.N.; Roy, I. Bacterial Cellulose-Based Blends and Composites: Versatile Biomaterials for Tissue Engineering Applications. Int. J. Mol. Sci. 2023, 24, 986. [Google Scholar] [CrossRef]
- Lunardi, V.B.; Soetaredjo, F.E.; Putro, J.N.; Santoso, S.P.; Yuliana, M.; Sunarso, J.; Ju, Y.-H.; Ismadji, S. Nanocelluloses: Sources, Pretreatment, Isolations, Modification, and Its Application as the Drug Carriers. Polymers 2021, 13, 2052. [Google Scholar] [CrossRef]
- Huo, Y.; Liu, Y.; Xia, M.; Du, H.; Lin, Z.; Li, B.; Liu, H. Nanocellulose-Based Composite Materials Used in Drug Delivery Systems. Polymers 2022, 14, 2648. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ghosh, B.; Sarkar, K. Nanocellulose as sustainable biomaterials for drug delivery. Sens. Int. 2022, 3, 100135. [Google Scholar] [CrossRef]
- Mensah, A.; Chen, Y.; Christopher, N.; Wei, Q. Membrane Technological Pathways and Inherent Structure of Bacterial Cellulose Composites for Drug Delivery Bioengineering. Bioengineering 2022, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Castaño, M.; Martínez, E.; Osorio, M.; Castro, C. Development of Genistein Drug Delivery Systems Based on Bacterial Nanocellulose for Potential Colorectal Cancer Chemoprevention: Effect of Nanocellulose Surface Modification on Genistein Adsorption. Molecules 2022, 27, 7201. [Google Scholar] [CrossRef] [PubMed]
- Kamal, T.; Ul-Islam, M.; Fatima, A.; Ullah, M.W.; Manan, S. Cost-Effective Synthesis of Bacterial Cellulose and Its Applications in the Food and Environmental Sectors. Gels 2022, 8, 552. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ahmed, S.; Sameen, D.E.; Wang, Y.; Lu, R.; Dai, J.; Li, S.; Qin, W. A review of cellulose and its derivatives in biopolymer-based for food packaging application. Trends Food Sci. Technol. 2021, 112, 532–546. [Google Scholar] [CrossRef]
- Jiang, Z.; Ngai, T. Recent Advances in Chemically Modified Cellulose and Its Derivatives for Food Packaging Applications: A Review. Polymers 2022, 14, 1533. [Google Scholar] [CrossRef]
- Oliveira, T.J.; Segato, T.C.M.; Machado, G.P.; Grotto, D.; Jozala, A.F. Evolution of Bacterial Cellulose in Cosmetic Applications: An Updated Systematic Review. Molecules 2022, 27, 8341. [Google Scholar] [CrossRef]
- Sharma, P.; Mittal, M.; Yadav, A.; Aggarwal, N.K. Bacterial cellulose: Nano-biomaterial for biodegradable face masks—A greener approach towards environment. Environ. Nanotechnol. Monit. Manag. 2023, 19, 100759. [Google Scholar] [CrossRef]
- Suárez-Avendaño, D.; Martínez-Correa, E.; Cañas-Gutierrez, A.; Castro-Riascos, M.; Zuluaga-Gallego, R.; Gañán-Rojo, P.; Peresin, M.; Pereira, M.; Castro-Herazo, C. Comparative Study on the Efficiency of Mercury Removal from Wastewater Using Bacterial Cellulose Membranes and Their Oxidized Analogue. Front. Bioeng. Biotechnol. 2022, 10, 815892. [Google Scholar] [CrossRef]
- Revin, V.V.; Dolganov, A.V.; Liyaskina, E.V.; Nazarova, N.B.; Balandina, A.V.; Devyataeva, A.A.; Revin, V.D. Characterizing Bacterial Cellulose Produced by Komagataeibacter sucrofermentans H-110 on Molasses Medium and Obtaining a Biocomposite Based on It for the Adsorption of Fluoride. Polymers 2021, 13, 1422. [Google Scholar] [CrossRef] [PubMed]
- Köse, K.; Mavlan, M.; Youngblood, J.P. Applications and impact of nanocellulose based adsorbents. Cellulose 2020, 27, 2967–2990. [Google Scholar] [CrossRef]
- Alves, A.A.; Silva, W.E.; Belian, M.F.; Lins, L.S.G.; Galembeck, A. Bacterial Cellulose Membranes for Environmental Water Remediation and Industrial Wastewater Treatment. Int. J. Environ. Sci. Technol. 2020, 17, 3997–4008. [Google Scholar] [CrossRef]
- Gomes, N.O.; Carrilho, E.; Machado, S.A.S.; Sgobbi, L.F. Bacterial cellulose-based electrochemical sensing platform: A smart material for miniaturized biosensors. Electrochim. Acta 2020, 349, 136341. [Google Scholar] [CrossRef]
- Yu, H.; Tian, Y.; Dirican, M.; Fang, D.; Yan, C.; Xie, J.; Jia, D.; Liu, Y.; Li, C.; Cui, M.; et al. Flexible, transparent and tough silver nanowire/nanocellulose electrodes for flexible touch screen panels. Carbohydr. Polym. 2021, 273, 118539. [Google Scholar] [CrossRef] [PubMed]
- Kamel, S.; Khattab, T.A. Recent Advances in Cellulose-Based Biosensors for Medical Diagnosis. Biosensors 2020, 10, 67. [Google Scholar] [CrossRef]
- Kotsiri, Z.; Vidic, J.; Vantarakis, A. Applications of Biosensors for Bacteria and Virus Detection in Food and Water–A Systematic Review. J. Environ. Sci. 2022, 111, 367–379. [Google Scholar] [CrossRef]
- Torres, F.G.; Troncoso, O.P.; Gonzales, K.N.; Sari, R.M.; Gea, S. Bacterial cellulose based biosensors. Med. Devices Sens. 2020, 3, e10102. [Google Scholar] [CrossRef]
- Faraco, T.A.; Fontes, M.L.; Paschoalin, R.T.; Claro, A.M.; Gonçalves, I.S.; Cavicchioli, M.; Farias, R.L.; Cremona, M.; Ribeiro, S.J.L.; Barud, H.D.S.; et al. Review of Bacterial Nanocellulose as Suitable Substrate for Conformable and Flexible Organic Light-Emitting Diodes. Polymers 2023, 15, 479. [Google Scholar] [CrossRef]
- de Assis, S.C.; Morgado, D.L.; Scheidt, D.T.; de Souza, S.S.; Cavallari, M.R.; Ando Junior, O.H.; Carrilho, E. Review of Bacterial Nanocellulose-Based Electrochemical Biosensors: Functionalization, Challenges, and Future Perspectives. Biosensors 2023, 13, 142. [Google Scholar] [CrossRef]
- Provin, A.P.; Cubas, A.L.V.; Dutra, A.R.d.A.; Schulte, N.K. Textile Industry and Environment: Can the Use of Bacterial Cellulose in the Manufacture of Biotextiles Contribute to the Sector? Clean Technol. Environ. Policy 2021, 23, 2813–2825. [Google Scholar] [CrossRef]
- Ağçeli, G.K.; Cihangir, N. Nano-sized biopolymer levan: Its antimicrobial, anti-biofilm and anti-cancer effects. Carbohydr. Res. 2020, 494, 108068. [Google Scholar] [CrossRef] [PubMed]
- Vieira, A.M.; Zahed, F.; Crispim, A.C.; de Souza Bento, E.; França, R.F.O.; Pinheiro, I.O.; Pardo, L.A.; Carvalho, B.M. Production of levan from Bacillus subtilis var. natto and apoptotic effect on SH-SY5Y neuroblastoma cells. Carbohydr. Polym. 2021, 273, 118613. [Google Scholar] [CrossRef] [PubMed]
- Charoenwongpaiboon, T.; Wangpaiboon, K.; Septham, P.; Jiamvoraphong, N.; Issaragrisil, S.; Pichyangkura, R.; Lorthongpanich, C. Production and bioactivities of nanoparticulated and ultrasonic-degraded levan generated by Erwinia tasmaniensis levansucrase in human osteosarcoma cells. Int. J. Biol. Macromol. 2022, 221, 1121–1129. [Google Scholar] [CrossRef] [PubMed]
- Hertadi, R.; Umriani Permatasari, N.; Ratnaningsih, E. Box-Wilson Design for Optimization of in vitro Levan Production and Levan Application as Antioxidant and Antibacterial Agents. Iran. Biomed. J. 2021, 25, 202–212. [Google Scholar] [CrossRef]
- Young, I.D.; Latousakis, D.; Juge, N. The Immunomodulatory Properties of β-2,6 Fructans: A Comprehensive Review. Nutrients 2021, 13, 1309. [Google Scholar] [CrossRef] [PubMed]
- Bahroudi, S.; Shabanpour, B.; Combie, J.; Shabani, A.; Salimi, M. Levan exerts health benefit effect through alteration in bifidobacteria population. Iran. Biomed. J. 2020, 24, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Öner, E.T.; Hernández, L.; Combie, J. Review of Levan polysaccharide: From a century of past experiences to future prospects. Biotechnol Adv. 2016, 34, 827–844. [Google Scholar] [CrossRef] [PubMed]
- González-Garcinuño, A.; Tabernero, A.; Domínguez, A.; Galán, M.A.; Martin del Valle, E.M. Levan and levansucrases: Polymer, enzyme, microorganisms and biomedical applications. Biocatal. Biotransform. 2017, 36, 233–244. [Google Scholar] [CrossRef]
- Srikanth, R.; Reddy, C.H.S.S.S.; Siddartha, G.; Ramaiah, M.J.; Uppuluri, K.B. Review on production, characterization and applications of microbial levan. Carbohydr. Polym. 2015, 120, 102–114. [Google Scholar] [CrossRef]
- Hövels, M.; Kosciow, K.; Kniewel, J.; Jakob, F.; Deppenmeier, U. High yield production of levan-type fructans by Gluconobacter japonicus LMG 1417. Int. J. Biol. Macromol. 2020, 164, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Jakob, F.; Gebrande, C.; Bichler, R.M.; Vogel, R.F. Insights into the pH-dependent, extracellular sucrose utilization and concomitant levan formation by Gluconobacter albidus TMW 2.1191. Antonie Van Leeuwenhoek 2020, 113, 863–873. [Google Scholar] [CrossRef] [PubMed]
- Anguluri, K.; La China, S.; Brugnoli, M.; De Vero, L.; Pulvirenti, A.; Cassanelli, S.; Gullo, M. Candidate Acetic Acid Bacteria Strains for Levan Production. Polymers 2022, 14, 2000. [Google Scholar] [CrossRef] [PubMed]
- Kornmann, H.; Duboc, P.; Marison, I.; von Stockar, U. Influence of nutritional factors on the nature, yield, and composition of exopolysaccharides produced by Gluconacetobacter xylinus I- 2281. Appl. Environ. Microbiol. 2003, 69, 6091–6098. [Google Scholar] [CrossRef] [PubMed]
- Melo, I.R.; Pimentel, M.F.; Lopes, C.E. Ca-lazans GMT. Application of fractional factorial design to levan production by Zymomonas mobilis. Braz. J. Microbiol. 2007, 38, 45–51. [Google Scholar] [CrossRef]
- de Oliveira, M.R.; da Silva, R.S.S.F.; Buzato, J.B.; Celligoi, M.A.P.C. Study of levan production by Zymomonas mobilis using regional low-cost carbohydrate sources. Biochem. Eng. J. 2007, 37, 177–183. [Google Scholar] [CrossRef]
- Lorenzetti, M.F.S.; Moro, M.R.; Garcia-Cruz, C.H.J. Alginate/PVA beads for levan production by Zymomonas mobilis. Food Process Eng. 2015, 38, 31–36. [Google Scholar] [CrossRef]
- Korany, S.M.; El-Hendawy, H.H.; Sonbol, H.; Hamada, M.A. Partial characterization of levan polymer from Pseudomonas fluorescens with significant cytotoxic and antioxidant activity. Saudi J. Biol. Sci. 2021, 28, 6679–6689. [Google Scholar] [CrossRef]
- Erkorkmaz, B.A.; Kırtel, O.; Ateş Duru, Ö.; Toksoy Öner, E. Development of a cost-effective production process for Halomonas levan. Bioprocess Biosyst Eng. 2018, 41, 1247–1259. [Google Scholar] [CrossRef]
- Poli, A.; Nicolaus, B.; Denizci, A.A.; Yavuzturk, B.; Kazan, D. Halomonas smyrnensis sp. nov., a moderately halophilic, exopolysaccharide-producing bacterium. Int. J. Syst. Evol. Microbiol. 2013, 63, 10–18. [Google Scholar] [CrossRef]
- Yuan, X.; Eldred, L.I.; Sundin, G.W. Exopolysaccharides amylovoran and levan contribute to sliding motility in the fire blight pathogen Erwinia amylovora. Environ. Microbiol. 2022, 24, 4738–4754. [Google Scholar] [CrossRef] [PubMed]
- Abid, Y.; Azabou, S.; Casillo, A.; Gharsallah, H.; Jemil, N.; Lanzetta, R.; Attia, H.; Corsaro, M.M. Isolation and structural characterization of levan produced by probiotic Bacillus tequilensis-GM from Tunisian fermented goat milk. Int. J. Biol. Macromol. 2019, 133, 786–794. [Google Scholar] [CrossRef] [PubMed]
- Thakham, N.; Thaweesak, S.; Teerakulkittipong, N.; Traiosot, N.; Kaikaew, A.; Lirio, G.A.; Jangiam, W. Structural characterization of functional ingredient Levan synthesized by Bacillus siamensis isolated from traditional fermented food in Thailand. Int. J. Food Sci. 2020, 2020, 7352484. [Google Scholar] [CrossRef]
- Phengnoi, P.; Thakham, N.; Rachphirom, T.; Teerakulkittipong, N.; Lirio, G.A.; Jangiam, W. Characterization of levansucrase produced by novel Bacillus siamensis and optimization of culture condition for levan biosynthesis. Heliyon 2022, 8, e12137. [Google Scholar] [CrossRef] [PubMed]
- Nasir, A.; Ahmad, W.; Sattar, F.; Ashfaq, I.; Lindemann, S.R.; Chen, M.H.; Van den Ende, W.; Öner, E.T.; Kirtel, O.; Khaliq, S.; et al. Production of a high molecular weight levan by Bacillus paralicheniformis, an industrially and agriculturally important isolate from the buffalo grass rhizosphere. Antonie Van Leeuwenhoek 2022, 115, 1101–1112. [Google Scholar] [CrossRef] [PubMed]
- Wagh, V.S.; Said, M.S.; Bennale, J.S.; Dastager, S.G. Isolation and structural characterization of exopolysaccharide from marine Bacillus sp. and its optimization by Microbioreactor. Carbohydr. Polym. 2022, 285, 119241. [Google Scholar] [CrossRef] [PubMed]
- El Halmouch, Y.; Ibrahim, H.A.H.; Dofdaa, N.M.; Mabrouk, M.E.M.; El-Metwally, M.M.; Nehira, T.; Ferji, K.; Ishihara, Y.; Matsuo, K.; Ibrahim, M.I.A. Complementary spectroscopy studies and potential activities of levan-type fructan produced by Bacillus paralicheniformis ND2. Carbohydr. Polym. 2023, 311, 120743. [Google Scholar] [CrossRef] [PubMed]
- Gojgic-Cvijovic, G.; Jakovljevic, D.; Loncarevic, B.; Todorovic, N.; Pergal, M.V.; Ciric, J.; Loos, K.; Beškoski, V.P.; Vrvic, M. Production of levan by Bacillus licheniformis NS032 in sugar beet molasses-based medium. Int. J. Biol. Macromol. 2018, 121, 142–151. [Google Scholar] [CrossRef]
- Nasir, A.; Sattar, F.; Ashfaq, I.; Lindemann, S.R.; Chen, M.H.; Van den Ende, W.; Öner, E.T.; Kirtel, O.; Khaliq, S.; Ghauri, M.A.; et al. Production and characterisation of a high molecular weight levan and fructooligosaccharides from a rhizospheric isolate of Bacillus aryabhattai. LWT Food Sci. Technol. 2020, 123, 109093. [Google Scholar] [CrossRef]
- Wu, F.-C.; Chou, S.-Z.; Shih, I.-L. Factors affecting the production and molecular weight of levan of Bacillus subtilis natto in batch and fed-batch culture in fermenter. J. Taiwan Inst. Chem. Eng. 2013, 44, 846–853. [Google Scholar] [CrossRef]
- Shih, I.L.; Yu, Y.T.; Shieh, C.J.; Hsieh, C.Y. Selective production and characterization of levan by Bacillus subtilis (Natto) Takahashi. J. Agric. Food Chem. 2005, 53, 8211–8215. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, L.F.; Bazani Cabral De Melo, F.C.; Martins Paiva, W.J.; Borsato, D.; Corradi Custodio Da Silva, M.L.; Pedrine Colabone Celligoi, M.A. Characterization and optimization of levan production by Bacillus subtilis Natto. Rom Biotech. Lett. 2013, 18, 8413–8422. [Google Scholar]
- Grinev, V.S.; Tregubova, K.V.; Anis’kov, A.A.; Sigida, E.N.; Shirokov, A.A.; Fedonenko, Y.P.; Yegorenkova, I.V. Isolation, structure, and potential biotechnological applications of the exopolysaccharide from Paenibacillus polymyxa 92. Carbohydr. Polym. 2020, 232, 115780. [Google Scholar] [CrossRef] [PubMed]
- Mead, D.A.; Lucas, S.; Copeland, A.; Lapidus, A.; Cheng, J.-F.; Bruce, D.C.; Goodwin, L.A.; Pitluck, S.; Chertkov, O.; Zhang, X.; et al. Complete Genome Sequence of Paenibacillus strain Y4.12MC10, a Novel Paenibacillus lautus strain Isolated from Obsidian Hot Spring in Yellowstone National Park. Stand. Genom. Sci. 2012, 6, 366–385. [Google Scholar] [CrossRef]
- Yang, S.; Jin, D.; Li, H.; Jiang, L.; Cui, J.; Huang, W.; Rang, J.; Li, Y.; Xia, L. Screening of new Paenibacillus polymyxa S3 and its disease resistance of grass carp (Ctenopharyngodon idellus). J. Fish Dis. 2023, 46, 17–29. [Google Scholar] [CrossRef] [PubMed]
- de Lemos, E.A.; Procópio, L.; da Mota, F.F.; Jurelevicius, D.; Rosado, A.S.; Seldin, L. Molecular characterization of Paenibacillus antarcticus IPAC21, a bioemulsifier producer isolated from Antarctic soil. Front. Microbiol. 2023, 14, 1142582. [Google Scholar] [CrossRef]
- Mamphogoro, T.P.; Kamutando, C.N.; Maboko, M.M.; Babalola, O.O.; Aiyegoro, O.A. Whole-Genome Sequence of Paenibacillus polymyxa Strain SRT9.1, a Promising Plant Growth-Promoting Bacterium. Microbiol. Resour. Announc. 2022, 11, e0109721. [Google Scholar] [CrossRef] [PubMed]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.-C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Factories. 2016, 15, 203. [Google Scholar] [CrossRef]
- Liang, T.-W.; Wang, S.-L. Recent advances in exopolysaccharides from Paenibacillus spp.: Production, isolation, structure, and bioactivities. Marine Drugs. 2015, 13, 1847–1863. [Google Scholar] [CrossRef]
- Dsouza, M.; Taylor, M.W.; Turner, S.J.; Aislabie, J. Genome-based comparative analyses of Antarctic and temperate species of Paenibacillus. PLoS ONE 2014, 9, e108009. [Google Scholar] [CrossRef]
- Xiao, B.; Sun, Y.F.; Lian, B.; Chen, T.M. Complete genome sequence and comparative genome analysis of the Paenibacillus mucilaginosus K02. Microb. Pathog. 2016, 93, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Wang, R.; Wang, D.; Su, J.; Zheng, S.X.; Wang, G.J. Paenibacillus selenitireducens sp. nov., a selenite-reducing bacterium isolated from a selenium mineral soil. Int. J. Syst. Evol. Microbiol. 2014, 64, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Lal, S.; Tabacchioni, S. Ecology and biotechnological potential of Paenibacillus polymyxa: A minireview. Ind. J. Microbiol. 2009, 49, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.F.; Jeong, H.; Park, S.-Y.; Kim, S.-B.; Park, Y.K.; Choi, S.K.; Ryu, C.M.; Hur, C.G.; Ghim, S.Y.; Oh, T.K.; et al. Genome Sequence of the Polymyxin-Producing Plant-Probiotic Rhizobacterium Paenibacillus polymyxa E681. J. Bacteriol. 2010, 192, 6103–6104. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Wang, C.; Ding, Y.; Li, L.; Shen, D.; Jiang, X.; Guan, D.; Cao, F.; Chen, H.; Feng, R.; et al. Complete genome sequence of Paenibacillus polymyxa SC2, a strain of plant growth-promoting rhizobacterium with broad-spectrum antimicrobial activity. J. Bacteriol. 2011, 193, 311–312. [Google Scholar] [CrossRef]
- Jeong, H.; Park, S.Y.; Chung, W.H.; Kim, S.H.; Kim, N.; Park, S.H. Draft genome sequence of the Paenibacillus polymyxa type strain (ATCC 842T), a plant growth-promoting bacterium. J. Bacteriol. 2011, 193, 5026–5027. [Google Scholar] [CrossRef]
- Li, Y.; Li, Q.; Li, Y.; Gao, J.; Fan, X. Draft Genome Sequence of Paenibacillus polymyxa KF-1, an Excellent Producer of Microbicides. Genome Announc. 2016, 4, e00727-16. [Google Scholar] [CrossRef]
- Ahmad, W.; Nasir, A.; Sattar, F.; Ashfaq, I.; Chen, M.H.; Hayat, A.; Rehman, M.U.; Zhao, S.; Khaliq, S.; Ghauri, M.A.; et al. Production of bimodal molecular weight levan by a Lactobacillus reuteri isolate from fish gut. Folia Microbiol. 2022, 67, 21–31. [Google Scholar] [CrossRef]
- Han, J.; Feng, H.; Wang, X.; Liu, Z.; Wu, Z. Levan from Leuconostoc citreum BD1707: Production optimization and changes in molecular weight distribution during cultivation. BMC Biotechnol. 2021, 21, 14. [Google Scholar] [CrossRef] [PubMed]
- Al-Qaysi, S.A.S.; Al-Haideri, H.; Al-Shimmary, S.M.; Abdulhameed, J.M.; Alajrawy, O.I.; Al-Halbosiy, M.M.; Moussa, T.A.A.; Farahat, M.G. Bioactive Levan-Type Exopolysaccharide Produced by Pantoea agglomerans ZMR7: Characterization and Optimization for Enhanced Production. J. Microbiol. Biotechnol. 2021, 31, 696–704. [Google Scholar] [CrossRef]
- Kang, H.K.; Seo, M.Y.; Seo, E.S.; Kim, D.; Chung, S.Y.; Kimura, A.; Day, D.F.; Robyt, J.F. Cloning and expression of levansucrase from Leuconostoc mesenteroides B-512 FMC in Escherichia coli. Biochim. Biophys. Acta 2005, 21, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Rairakhwada, D.; Seo, J.-W.; Seo, M.; Kwon, O.; Rhee, S.-K.; Kim, C.H. Gene cloning, characterization, and heterologous expression of levansucrase from Bacillus amyloliquefaciens. J. Ind. Microbiol. Biotechnol. 2010, 37, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Ávila-Fernández, Á.; Montiel, S.; Rodríguez-Alegría, M.E.; Caspeta, L.; López Munguía, A. Simultaneous enzyme production, Levan-type FOS synthesis and sugar by-products elimination using a recombinant Pichia pastoris strain expressing a levansucrase-endolevanase fusion enzyme. Microb. Cell Fact. 2023, 22, 18. [Google Scholar] [CrossRef]
- Ko, H.; Bae, J.H.; Sung, B.H.; Kim, M.J.; Kim, C.H.; Oh, B.R.; Sohn, J.H. Efficient production of levan using a recombinant yeast Saccharomyces cerevisiae hypersecreting a bacterial levansucrase. J. Ind. Microbiol. Biotechnol. 2019, 46, 1611–1620. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.; Sung, B.H.; Kim, M.J.; Sohn, J.H.; Bae, J.H. Fructan Biosynthesis by Yeast Cell Factories. J. Microbiol. Biotechnol. 2022, 32, 1373–1381. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Tong, Q.; Guo, X.; Cong, H.; Zhao, Z.; Liang, W.; Li, J.; Zhu, P.; Yang, H. Complete secretion of recombinant Bacillus subtilis levansucrase in Pichia pastoris for production of high molecular weight levan. Int. J. Biol. Macromol. 2022, 214, 203–211. [Google Scholar] [CrossRef]
- Donot, F.; Fontana, A.; Baccou, J.C.; Schorr-Galindo, S. Microbial exopolysaccharides: Main examples of synthesis, excretion, genetics and extraction. Carbohydr. Polym. 2012, 87, 951–962. [Google Scholar] [CrossRef]
- Dogsa, I.; Brloznik, M.; Stopar, D.; Mandic-Mulec, I. Exopolymer diversity and the role of levan in Bacillus subtilis biofilms. PLoS ONE 2013, 8, e62044. [Google Scholar] [CrossRef]
- Velázquez-Hernández, M.L.; Baizabal-Aguirre, V.M.; Cruz-Vázquez, F.; Trejo-Contreras, M.J.; Fuentes-Ramírez, L.E.; Bravo-Patiño, A.; Cajero-Juárez, M.; Chávez-Moctezuma, M.P.; Valdez-Alarcón, J.J. Gluconacetobacter diazotrophicus levansucrase is involved in tolerance to NaCl, sucrose and desiccation, and in biofilm formation. Arch. Microbiol. 2011, 193, 137–149. [Google Scholar] [CrossRef]
- Mehmood, A.; Abdallah, K.; Khandekar, S.; Zhurina, D.; Srivastava, A.; Al-Karablieh, N.; Alfaro-Espinoza, G.; Pletzer, D.; Ullrich, M.S. Expression of extra-cellular levansucrase in Pseudomonas syringae is controlled by the in planta fitness-promoting metabolic repressor HexR. BMC Microbiol. 2015, 15, 48. [Google Scholar] [CrossRef]
- Sims, I.M.; Frese, S.A.; Walter, J.; Loach, D.; Wilson, M.; Appleyard, K.; Eason, J.; Livingston, M.; Baird, M.; Cook, G.; et al. Structure and functions of exopolysaccharide produced by gut commensal Lactobacillus reuteri 100-23. ISME J. 2011, 5, 1115–1124. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Wu, D.; Li, X.; Lu, J. Levan from Bacillus amyloliquefaciens JN4 acts as a prebiotic for enhancing the intestinal adhesion capacity of Lactobacillus reuteri JN101. Int. J. Biol. Macromol. 2020, 146, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Kolida, S.; Charalampopoulos, D.; Rastall, R.A. An evaluation of the prebiotic potential of microbial levans from Erwinia sp. 10119. J. Funct. Foods 2020, 64, 103668. [Google Scholar] [CrossRef]
- Wang, M.; Cheong, K.L. Preparation, Structural Characterisation, and Bioactivities of Fructans: A Review. Molecules 2023, 28, 1613. [Google Scholar] [CrossRef] [PubMed]
- Lekakarn, H.; Bunterngsook, B.; Jaikaew, P.; Kuantum, T.; Wansuksri, R.; Champreda, V. Functional Characterization of Recombinant Endo-Levanase (LevBk) from Bacillus koreensis HL12 on Short-Chain Levan-Type Fructooligosaccharides Production. Protein J. 2022, 41, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Koşarsoy Ağçeli, G.; Hammamchi, H.; Cihangir, N. Novel levan/bentonite/essential oil films: Characterization and antimicrobial activity. J. Food Sci. Technol. 2022, 59, 249–256. [Google Scholar] [CrossRef]
- Sezer, A.D.; Kazak, H.; Öner, E.T.; Akbuga, J. Levan-based nanocarrier system for peptide and protein drug delivery: Optimization and influence of experimental parameters on the nanoparticle characteristics. Carbohydr. Polym. 2011, 84, 358–363. [Google Scholar] [CrossRef]
- Akturk, O. The anticancer activity of doxorubicin-loaded levan-functionalized gold nanoparticles synthesized by laser ablation. Int. J. Biol. Macromol. 2022, 196, 72–85. [Google Scholar] [CrossRef]
- Tabernero, A.; Cardea, S. Microbial Exopolysaccharides as Drug Carriers. Polymers 2020, 12, 2142. [Google Scholar] [CrossRef]
- Kim, S.-J.; Bee, P.K.; Chung, B.H. Self-assembled levan nanoparticles for targeted breast cancer imaging. Chem. Commun. 2015, 51, 107–110. [Google Scholar] [CrossRef]
- Ahmed, K.B.A.; Kalla, D.; Uppuluri, K.B.; Anbazhagan, V. Green synthesis of silver and gold nanoparticles employing levan, a biopolymer from Acetobacter xylinum NCIM 2526, as a reducing agent and capping agent. Carbohydr. Polym. 2014, 112, 539–545. [Google Scholar] [CrossRef]
- Cinan, E.; Cesur, S.; Erginer Haskoylu, M.; Gunduz, O.; Toksoy Oner, E. Resveratrol-Loaded Levan Nanoparticles Produced by Electrohydrodynamic Atomization Technique. Nanomaterials 2021, 11, 2582. [Google Scholar] [CrossRef]
- Gomes, T.D.; Caridade, S.G.; Sousa, M.P.; Azevedo, S.; Kandur, M.Y.; Öner, E.T.; Alves, N.M.; Mano, J.F. Adhesive free-standing multilayer films containing sulfated levan for biomedical applications. Acta Biomater. 2018, 69, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Erginer, M.; Akcay, A.; Coskunkan, B.; Morova, T.; Rende, D.; Bucak, S.; Baysal, N.; Ozisik, R.; Eroglu, M.S.; Agirbasli, M.; et al. Sulfated levan from Halomonas smyrnensis as a bioactive, heparinmimetic glycan for cardiac tissue engineering. Carbohydr. Polym. 2016, 149, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Avsar, G.; Agirbasli, D.; Agirbasli, M.A.; Gunduz, O.; Oner, E.T. Levan based fibrous scaffolds electrospun via co-axial and, singleneedle techniques for tissue engineering applications. Carbohyd. Polym. 2018, 193, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Koşarsoy Ağçeli, G. A new approach to nanocomposite carbohydrate polymer films: Levan and chia seed mucilage. Int. J. Biol. Macromol. 2022, 218, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Domżał-Kędzia, M.; Lewińska, A.; Jaromin, A.; Weselski, M.; Pluskota, R.; Łukaszewicz, M. Fermentation parameters and conditions affecting levan production and its potential applications in cosmetics. Bioorganic Chem. 2019, 93, 102787. [Google Scholar] [CrossRef] [PubMed]
- Wasilewski, T.; Seweryn, A.; Pannert, D.; Kierul, K.; Domżał-Kędzia, M.; Hordyjewicz-Baran, Z.; Łukaszewicz, M.; Lewińska, A. Application of Levan-Rich Digestate Extract in the Production of Safe-to-Use and Functional Natural Body Wash Cosmetics. Molecules 2022, 27, 2793. [Google Scholar] [CrossRef] [PubMed]
- Revin, V.V.; Novokuptsev, N.V.; Red’kin, N.A. Optimization of Cultivation Conditions for Azotobacter vinelandii D-08, Producer of the Polysaccharide Levan, for Obtaining Biocomposite Materials. Bioresources 2016, 11, 9661–9675. [Google Scholar] [CrossRef]
- Chang, I.; Lee, M.; Tran, A.T.P.; Lee, S.; Kwon, Y.M.; Im, J.; Cho, G.C. Review on biopolymer-based soil treatment (BPST) technology in geotechnical engineering practices. Transp. Geotech. 2020, 24, 100385. [Google Scholar] [CrossRef]
- Mohsin, A.; Akyliyaevna, K.A.; Zaman, W.Q.; Hussain, M.H.; Mohsin, M.Z.; Al-Rashed, S.; Tan, X.; Tian, X.; Aida, K.; Tariq, M.; et al. Kinetically modelled approach of xanthan production using different carbon sources: A study on molecular weight and rheological properties of xanthan. Int. J. Biol. Macromol. 2021, 193 Pt. B, 1226–1236. [Google Scholar] [CrossRef]
- Habibi, H.; Khosravi-Darani, K. Effective variables on production and structure of xanthan gum and its food applications: A review. Biocatal. Agricul. Biotechnol. 2017, 10, 130–140. [Google Scholar] [CrossRef]
- Berninger, T.; Dietz, N.; González López, Ó. Water-soluble polymers in agriculture: Xanthan gum as eco-friendly alternative to synthetics. Microb. Biotechnol. 2021, 14, 1881–1896. [Google Scholar] [CrossRef] [PubMed]
- García-Ochoa, F.; Santos, V.E.; Casas, J.A.; Gómez, E. Xanthan gum: Production, recovery, and properties. Biotechnol. Adv. 2000, 18, 549–579. [Google Scholar] [CrossRef] [PubMed]
- Gbadamosi, A.; Patil, S.; Kamal, M.S.; Adewunmi, A.A.; Yusuff, A.S.; Agi, A.; Oseh, J. Application of Polymers for Chemical Enhanced Oil Recovery: A Review. Polymers 2022, 14, 1433. [Google Scholar] [CrossRef] [PubMed]
- An, S.-Q.; Potnis, N.; Dow, M.; Vorhölter, F.-J.; He, Y.-Q.; Becker, A.; Teper, D.; Li, Y.; Wang, N.; Bleris, L.; et al. Mechanistic insights into host adaptation, virulence and epidemiology of the phytopathogen Xanthomonas. FEMS Microbiol. Rev. 2020, 44, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Vicente, J.G.; Holub, E.B. Xanthomonas campestris pv. campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Mol. Plant. Pathol. 2013, 14, 2–18. [Google Scholar] [CrossRef]
- Li, T.; Li, Y.; Ma, X.; Dan, X.; Huang, X.; Li, Q.; Lei, S.; Zhang, Z.; Huang, S.; Jiang, W.; et al. Comparative Genomic Analysis of Two Xanthomonas oryzae pv. oryzae Strains Isolated from Low Land and High Mountain Paddies in Guangxi, China. Front. Microbiol. 2022, 13, 867633. [Google Scholar] [CrossRef]
- Feng, Y.M.; Long, Z.Q.; Xiang, H.M.; Ran, J.N.; Zhou, X.; Yang, S. Research on Diffusible Signal Factor-Mediated Quorum Sensing in Xanthomonas: A Mini-Review. Molecules 2023, 28, 876. [Google Scholar] [CrossRef]
- Liyanapathiranage, P.; Wagner, N.; Avram, O.; Pupko, T.; Potnis, N. Phylogenetic Distribution and Evolution of Type VI Secretion System in the Genus Xanthomonas. Front. Microbiol. 2022, 13, 840308. [Google Scholar] [CrossRef]
- Cuesta-Morrondo, S.; Redondo, C.; Palacio-Bielsa, A.; Garita-Cambronero, J.; Cubero, J. Complete Genome Sequence Resources of Six Strains of the Most Virulent Pathovars of Xanthomonas arboricola Using Long- and Short-Read Sequencing Approaches. Phytopathology 2022, 112, 1808–1813. [Google Scholar] [CrossRef] [PubMed]
- Bellenot, C.; Carrère, S.; Gris, C.; Noël, L.D.; Arlat, M. Genome Sequences of 17 Strains from Eight Races of Xanthomonas campestris pv. campestris. Microbiol. Resour. Announc. 2022, 11, e0027922. [Google Scholar] [CrossRef] [PubMed]
- Revin, V.V.; Liyas’kina, E.V.; Pokidko, B.V.; Pimenov, N.V.; Mardanov, A.V.; Ravin, N.V. Characteristics of the New Xanthan-Producing Strain Xanthomonas campestris M 28: Study of the Genome, Cultivation Conditions, and Physicochemical and Rheological Properties of the Polysaccharide. Appl. Biochem. Microbiol. 2021, 57, 356–365. [Google Scholar] [CrossRef]
- Wibberg, D.; Alkhateeb, R.S.; Winkler, A.; Albersmeier, A.; Schatschneider, S.; Albaum, S.; Vorholter, F.-J. Draft genome of the xanthan producer Xanthomonas campestris NRRL B-1459 (ATCC 13951). J. Biotechnol. 2015, 204, 45–46. [Google Scholar] [CrossRef]
- Vorholter, F.-J.; Schneiker, S.; Goesmann, A.; Krause, L.; Bekel, T.; Kaiser, O.; Puhler, A. The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J. Biotechnol. 2008, 134, 33–45. [Google Scholar] [CrossRef]
- Tao, F.; Wang, X.; Ma, C.; Yang, C.; Tang, H.; Gai, Z.; Xu, P. Genome Sequence of Xanthomonas campestris JX, an Industrially Productive Strain for Xanthan Gum. J. Bacteriol. 2012, 194, 4755–4756. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.; Horta de Passo, V.; Fang, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Lv, H. Complete Genome Sequence of strain WHRI 3811, race 1 of Xanthomonas campestris pv. campestris, the Causal Agent of Black Rot of Cruciferous Vegetables. Mol. Plant–Microbe Interact. 2019, 32, 1571–1573. [Google Scholar] [CrossRef]
- Oliveira, A.C.; Ferreira, R.M.; Ferro, M.I.T.; Ferro, J.A.; Chandler, M.; Varani, A.M. Transposons and pathogenicity in Xanthomonas: Acquisition of murein lytic transglycosylases by TnXax1 enhances Xanthomonas citri subsp. citri 306 virulence and fitness. Peer J. 2018, 6, 6111. [Google Scholar] [CrossRef]
- Balíková, K.; Farkas, B.; Matúš, P.; Urík, M. Prospects of Biogenic Xanthan and Gellan in Removal of Heavy Metals from Contaminated Waters. Polymers 2022, 14, 5326. [Google Scholar] [CrossRef]
- Rončević, Z.; Zahović, I.; Danilović, N. Potential of different Xanthomonas campestris strains for xanthan biosynthesis on waste glycerol from biodiesel production. J. Process. Energy Agric. 2020, 24, 62–66. [Google Scholar] [CrossRef]
- Niknezhad, S.V.; Asadollahi, M.A.; Zamani, A.; Biria, D. Production of xanthan gum by free and immobilised cells of Xanthomonas campestris and Xanthomonas pelargonii. Int. J. Biol. Macromol. 2016, 82, 751–756. [Google Scholar] [CrossRef] [PubMed]
- Prameela, K.; Murali Mohan, C.; Ramakrishna, C. Biopolymers for food design: Consumer-friendly natural ingredients. In Biopolymers for Food Design; Academic Press: Cambridge, MA, USA, 2018; pp. 1–32. [Google Scholar] [CrossRef]
- Barbosa, R.M.; da Rocha, D.N.; Bombaldi de Souza, R.F.; Santos, J.L.; Ferreira, J.R.M.; Moraes, Â.M. Cell-Friendly Chitosan-Xanthan Gum Membranes Incorporating Hydroxyapatite Designed for Periodontal Tissue Regeneration. Pharmaceutics 2023, 15, 705. [Google Scholar] [CrossRef] [PubMed]
- Jadav, M.; Pooja, D.; Adams, D.J.; Kulhari, H. Advances in Xanthan Gum-Based Systems for the Delivery of Therapeutic Agents. Pharmaceutics 2023, 15, 402. [Google Scholar] [CrossRef] [PubMed]
- Sorze, A.; Valentini, F.; Dorigato, A.; Pegoretti, A. Development of a Xanthan Gum Based Superabsorbent and Water Retaining Composites for Agricultural and Forestry Applications. Molecules 2023, 28, 1952. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, L.; Wu, J.; Zhang, H.; Zhu, L.; Zhan, X. Potential application of a low-viscosity and high-transparency xanthan gum produced from Xanthomonas campestris CCTCC M2015714 in foods. Prep. Biochem. Biotechnol. 2018, 48, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Dzionek, A.; Wojcieszyńska, D.; Guzik, U. Use of xanthan gum for whole cell immobilization and its impact in bioremediation—A review. Bioresour. Technol. 2022, 351, 126918. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, M.; Bhandari, B. Effect of gums on the rheological, microstructural and extrusion printing characteristics of mashed potatoes. Int. J. Biol. Macromol. 2018, 117, 1179–1187. [Google Scholar] [CrossRef] [PubMed]
- Azam, R.S.M.; Zhang, M.; Bhandari, B.; Yang, C. Effect of different gums on features of 3D Printed Object Based on Vitamin-D enriched orange concentrate. Food Biophys. 2018, 13, 250–262. [Google Scholar] [CrossRef]
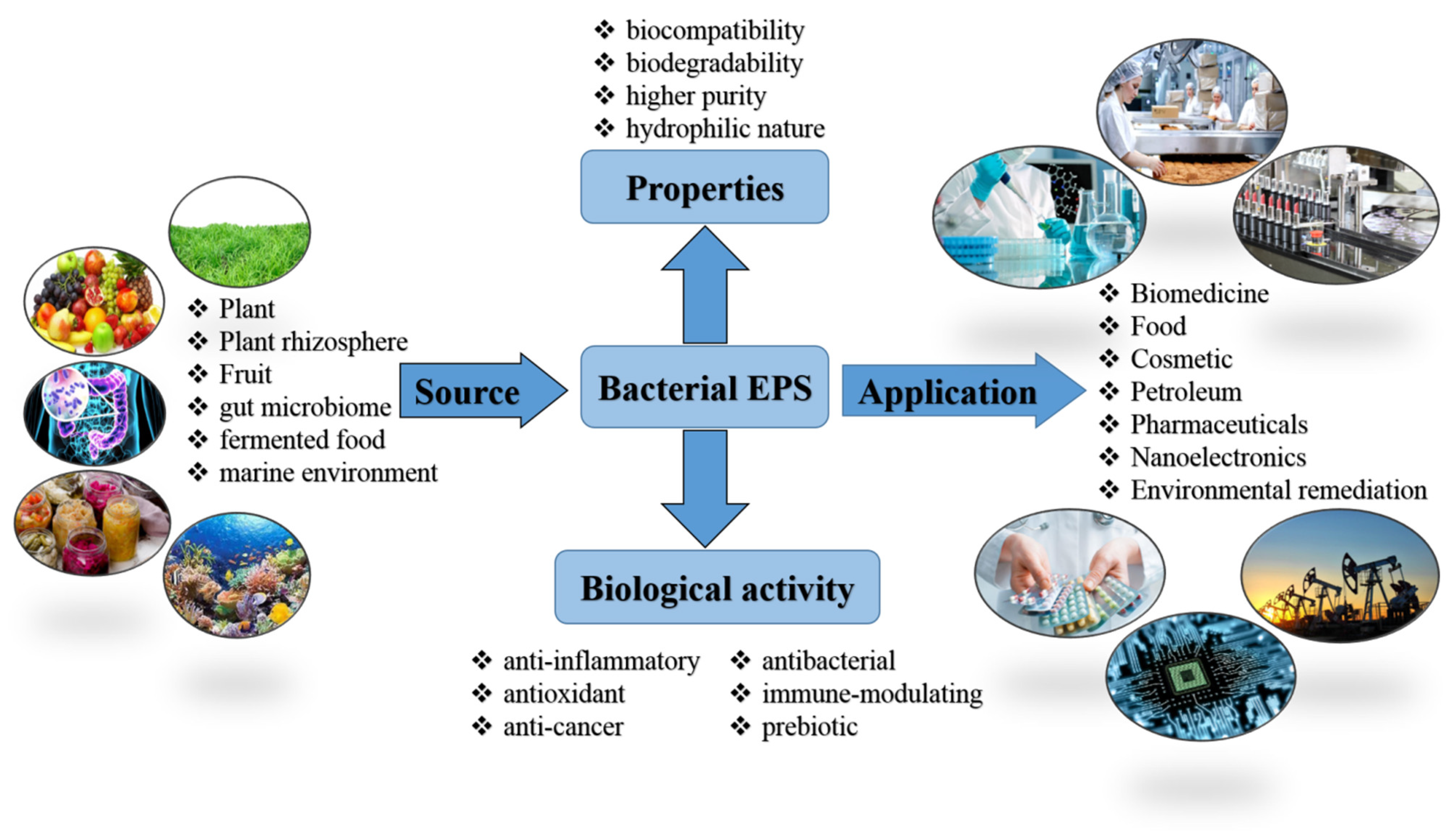



| No | Species | Sources of Isolation | Number of Bases (bp) | DNA G + C Content (mol%) | Ref. |
|---|---|---|---|---|---|
| 1 | K. cocois WE7T (CGMCC 1.15338T; JCM 31140T) | Contaminated coconut milk, China | 3,406,946 | 62.7 | [92] |
| 2 | K. diospyri MSKU 9T (NBRC 113802T; TBRC 9844T) | Persimmon, Thailand | 3,762,373 | 60.4 | [93] |
| 3 | K. europaeus LMG 18890T (ATCC 51845T; DES 11T; DSM 6160T) | Submerged culture vinegar generator, Germany | 4,227,398 | 61.3 | [90] |
| 4 | K. hansenii JCM 7643T (DSM 5602T; ATCC 35959T; BCC 6318 T) | Vinegar, Israel | 3,710,965 | 59.3 | [94] |
| 5 | K. intermedius TF2T (DSM 11804T; BCC 36457T; JCM 16936T; LMG 18909T; BCC 36447T; BCRC 17055T; CIP 105780T) | Kombucha beverage, Switzerland | 3,883,532 | 61.6 | [95] |
| 6 | K. kakiaceti JCM 25156T (DSM 24098T; G5-1T; LMG 26206T; NRIC 798T; BCRC 80743T) | Kaki vinegar, Japan | 3,133,102 | 62.1 | [96] |
| 7 | K. kombuchae LMG 23726T (MTCC 6913T; RG3T) | 3,483,869 | 59.6 | [94] | |
| 8 | K. maltaceti LMG 1529T (IFO 14815T; NBRC 14815T; NCIB 8752T; NCIMB 8752T) | Malt vinegar brewery acetifier | 3,638,012 | 63.2 | [96,97] |
| 9 | K. medellinensis NBRC 3288T (IFO 3288T); Kondo 51T; LMG 1693T) | Vinegar, Japan | 3,136,818 | 60.9 | [96,98] |
| 10 | K. melaceti AV382T (ZIM B1054T; LMG 31303T; CCM 8958T) | Apple vinegar | 3,629,663 | 59.14% | [99] |
| 11 | K. melomenusus AV436T (ZIM B1056T; LMG 31304T; CCM 8959T) | Apple cider Vinegar, Kopivnik, Slovenia | [99] | ||
| 12 | K. nataicola LMG 1536T (BCC 36443T; JCM 25120T; NRIC 616T) | Nata de coco, Philippines | 3,672,972 | 61.5 | [90,98] |
| 13 | K. oboediens LMG 18849T (BCC 36445T; CIP 105763T; DSM 11826T JCM 16937T; NCIMB 13557T; LTH 2460T;BCRC 17057T) | Submerged red wine vinegar, Germany | 3,777,265 | 61.4 | [90] |
| 14 | K. pomaceti T5K1T (CCM 8723T; LMG 30150T; ZIM B1029T) | Apple cider vinegar, Slovenia | 3,449,370 | 62.5 | [94] |
| 15 | K. rhaeticus LMG 22126T (DSM 16663T, BCC 36452T; JCM 17122T; DST GL02T;CIP 109761T) | Apple juice, South Tyrol region, Italy | 3,465,200 | 63.5 | [90,98] |
| 16 | K. saccharivorans LMG 1582T (BCC 36444T; JCM 25121T; NRIC 614; CIP 109786T; CECT 7869T; NCCB 29003T;NRIC 0614T) | Beet juice, Germany | 3,350,941 | 61.6 | [90,98] |
| 17 | K. sucrofermentans LMG 18788T (DSM 5973T; BCC 7227T; JCM 9730T; BPR 2001T; ATCC 700178T; BCRC 80162T; CECT 7291T;CIP 106078T) | Black cherry, Tokyo, Japan | 3,363,922 | 62.3 | [90] |
| 18 | K. swingsii LMG 22125T (DSM 16373T; BCC 36451T; JCM 17123; DST GL01T; CIP 109760T) | Apple juice, South Tyrol region, Italy | 3,732,982 | 62.4 | [90] |
| 19 | K. xylinus LMG 1515T (ATCC 23767T; DSM 6513T; BCC 7226T; JCM 7644T; NBRC 15237T; NCIMB 11664T; BCRC 2952T; CCM 3611T; CCUG 37299T; CECT 7351T; CIP 103107T; IFO 15237T; NCTC 4112T; VTT E-97831T) | Mountains ash berries | 3,660,954 | 66.2 | [90,98] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Netrusov, A.I.; Liyaskina, E.V.; Kurgaeva, I.V.; Liyaskina, A.U.; Yang, G.; Revin, V.V. Exopolysaccharides Producing Bacteria: A Review. Microorganisms 2023, 11, 1541. https://doi.org/10.3390/microorganisms11061541
Netrusov AI, Liyaskina EV, Kurgaeva IV, Liyaskina AU, Yang G, Revin VV. Exopolysaccharides Producing Bacteria: A Review. Microorganisms. 2023; 11(6):1541. https://doi.org/10.3390/microorganisms11061541
Chicago/Turabian StyleNetrusov, Alexander I., Elena V. Liyaskina, Irina V. Kurgaeva, Alexandra U. Liyaskina, Guang Yang, and Viktor V. Revin. 2023. "Exopolysaccharides Producing Bacteria: A Review" Microorganisms 11, no. 6: 1541. https://doi.org/10.3390/microorganisms11061541
APA StyleNetrusov, A. I., Liyaskina, E. V., Kurgaeva, I. V., Liyaskina, A. U., Yang, G., & Revin, V. V. (2023). Exopolysaccharides Producing Bacteria: A Review. Microorganisms, 11(6), 1541. https://doi.org/10.3390/microorganisms11061541









