Diversification of Bourbon Virus in New York State
Abstract
:1. Introduction
2. Materials and Methods
2.1. Viruses
2.2. Sequencing
2.3. In Vitro Growth Kinetics
2.4. Synchronous Infection of Ticks and Viral Detection
3. Results
3.1. NYS BRBV Strains Are Genetically Distinct from Midwestern US BRBV Strains
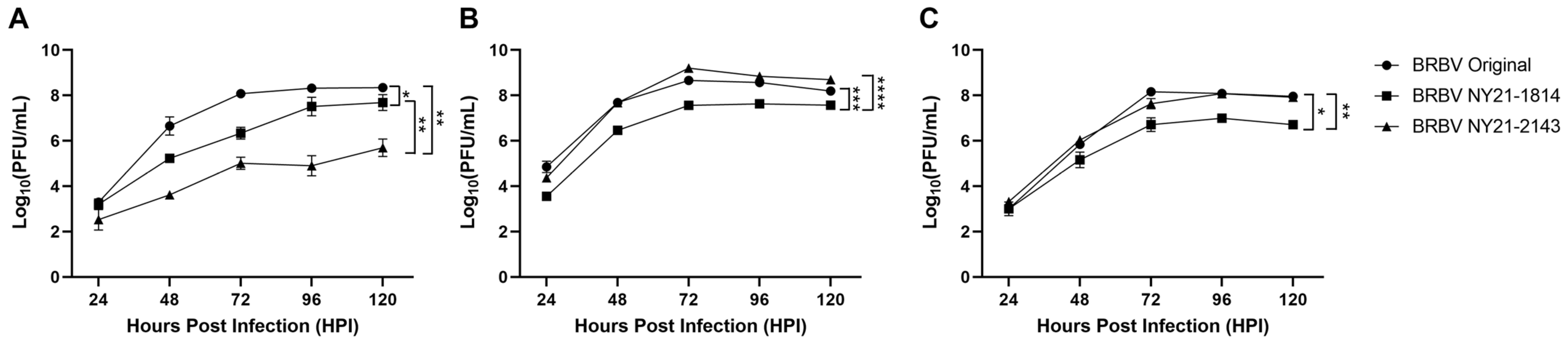
3.2. NYS BRBV Strains Display Phenotypic Variability in Mammalian Cells
3.3. Fitness Advantage of NYS BRBV Strain BRBV NY21-2143 in Experimentally Infected Amblyomma americanum
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hao, S.; Ning, K.; Kuz, C.A.; McFarlin, S.; Cheng, F.; Qiu, J. Eight Years of Research Advances in Bourbon Virus, a Tick-borne Thogotovirus of the Orthomyxovirus Family. Zoonoses 2022, 2. [Google Scholar] [CrossRef]
- Anderson, C.R.; Casals, J. Dhori virus, a new agent isolated from Hyalomma dromedarii in India. Indian J. Med. Res. 1973, 61, 1416–1420. [Google Scholar]
- Briese, T.; Chowdhary, R.; da Rosa, A.T.; Hutchison, S.K.; Popov, V.; Street, C.; Tesh, R.B.; Lipkin, W.I. Upolu virus and Aransas Bay virus, two presumptive bunyaviruses, are novel members of the family Orthomyxoviridae. Virol. J. 2014, 88, 5298–5309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ejiri, H.; Lim, C.K.; Isawa, H.; Fujita, R.; Murota, K.; Sato, T.; Kobayashi, D.; Kan, M.; Hattori, M.; Kimura, T.; et al. Characterization of a novel thogotovirus isolated from Amblyomma testudinarium ticks in Ehime, Japan: A significant phylogenetic relationship to Bourbon virus. Virus Res. 2018, 249, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Yunker, C.E.; Clifford, C.M.; Keirans, J.E.; Thomas, L.A.; Rice, R.C.A. Aransas Bay virus, a new arbovirus of the Upolu serogroup from Ornithodoros capensis (Acari: Argasidae) in coastal Texas. J. Med. Entomol. 1979, 16, 453–460. [Google Scholar] [CrossRef]
- Savage, H.M.; Burkhalter, K.L.; Godsey, M.S., Jr.; Panella, N.A.; Ashley, D.C.; Nicholson, W.L.; Lambert, A.J. Bourbon virus in field-collected ticks, Missouri, USA. Emerg. Infect. Dis. 2017, 23, 2017–2022. [Google Scholar] [CrossRef] [PubMed]
- Savage, H.M.; Godsey, M.S., Jr.; Panella, N.A.; Burkhalter, K.L.; Manford, J.; Trevino-Garrison, I.C.; Straily, A.; Wilson, S.; Bowen, J.; Raghavan, R.K. Surveillance for tick-borne viruses near the location of a fatal human case of bourbon virus (Family Orthomyxoviridae: Genus Thogotovirus) in Eastern Kansas, 2015. J. Med. Entomol. 2018, 55, 701–705. [Google Scholar] [CrossRef]
- Godsey, M.S., Jr.; Rose, D.; Burkhalter, K.L.; Breuner, N.; Bosco-Lauth, A.M.; Kosoy, O.I.; Savage, H.M. Experimental infection of Amblyomma americanum (Acari: Ixodidae) with bourbon virus (Orthomyxoviridae: Thogotovirus). J. Med. Entomol. 2021, 58, 873–879. [Google Scholar] [CrossRef]
- Cumbie, A.N.; Trimble, R.N.; Eastwood, G. Pathogen spillover to an invasive tick species: First detection of Bourbon virus in Haemaphysalis longicornis in the United States. Pathogens 2022, 11, 454. [Google Scholar] [CrossRef]
- Jackson, K.C.; Gidlewski, T.; Root, J.J.; Bosco-Lauth, A.M.; Lash, R.R.; Harmon, J.R.; Brault, A.C.; Panella, N.A.; Nicholson, W.L.; Komar, N. Bourbon virus in wild and domestic animals, Missouri, USA, 2012–2013. Emerg. Infect. Dis. 2019, 25, 1752. [Google Scholar] [CrossRef]
- Dupuis, I.I.; Alan, P.; Prusinski, M.A.; O’Connor, C.; Maffei, J.G.; Koetzner, C.A.; Zembsch, T.E.; Zink, S.D.; White, A.L.; Santoriello, M.P.; et al. Bourbon Virus Transmission, New York, USA. Emerg. Infect. Dis. 2023, 29, 145–148. [Google Scholar] [CrossRef]
- Bamunuarachchi, G.; Harastani, H.; Rothlauf, P.W.; Dai, Y.N.; Ellebedy, A.; Fremont, D.; Whelan, S.P.J.; Wang, D.; Boon, A.C.M. Detection of Bourbon virus-specific serum neutralizing antibodies in human serum in Missouri, USA. Msphere 2022, 7, e00164-22. [Google Scholar] [CrossRef]
- Kosoy, O.I.; Lambert, A.J.; Hawkinson, D.J.; Pastula, D.M.; Goldsmith, C.S.; Hunt, D.C.; Staples, J.E. Novel thogotovirus associated with febrile illness and death, United States, 2014. Emerg. Infect. Dis. 2015, 21, 760. [Google Scholar] [CrossRef] [PubMed]
- Miller, M. Bourbon Virus Linked to Death of Park Official—CDC Testing Tick Samples. The Missourian. 8 March 2018. Available online: https://www.emissourian.com/local_news/county/bourbon-virus-linked-to-deathof-park-official—Cdc-testing-tick-samples/article_057f0f0a-bddd-5f7a-aa0d-735187e4c379.html (accessed on 31 October 2021).
- Bricker, T.L.; Shafiuddin, M.; Gounder, A.P.; Janowski, A.B.; Zhao, G.; Williams, G.D.; Boon, A.C. Therapeutic efficacy of favipiravir against Bourbon virus in mice. PLoS Pathog. 2019, 15, e1007790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roe, M.K.; Huffman, E.R.; Batista, Y.S.; Papadeas, G.G.; Kastelitz, S.R.; Restivo, A.M.; Stobart, C.C. Comprehensive Review of Emergence and Virology of Tickborne Bourbon Virus in the United States. Emerg. Infect. Dis. 2023, 29, 1–7. [Google Scholar] [CrossRef]
- Aziati, I.D.; McFarland, D., Jr.; Antia, A.; Joshi, A.; Aviles-Gamboa, A.; Lee, P.; Harastani, H.; Wang, D.; Adalsteinsson, S.A.; Boon, A.C. Prevalence of Bourbon and Heartland viruses in field collected ticks at an environmental field station in St. Louis County, Missouri, USA. Ticks Tick Borne Dis. 2023, 14, 102080. [Google Scholar] [CrossRef] [PubMed]
- Brault, A.C.; Savage, H.M.; Duggal, N.K.; Eisen, R.J.; Staples, J.E. Heartland virus epidemiology, vector association, and disease potential. Viruses 2018, 10, 498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, S.; Ning, K.; Wang, X.; Wang, J.; Cheng, F.; Ganaie, S.S.; Tavis, J.E.; Qiu, J. Establishment of a Replicon Reporter of the Emerging Tick-Borne Bourbon Virus and Use It for Evaluation of Antivirals. Front. Microbiol. 2020, 11, 572631. [Google Scholar] [CrossRef]
- Tokarz, R.; Williams, S.H.; Sameroff, S.; Leon, M.S.; Jain, K.; Lipkin, W.I. Virome analysis of Amblyomma americanum, Dermacentor variabilis, and Ixodes scapularis ticks reveals novel highly divergent vertebrate and invertebrate viruses. Virol. J. 2014, 88, 11480–11492. [Google Scholar] [CrossRef] [Green Version]
- Sagurova, I.; Ludwig, A.; Ogden, N.H.; Pelcat, Y.; Dueymes, G.; Gachon, P. Predicted Northward Expansion of the Geographic Range of the Tick Vector Amblyomma americanum in North America under Future Climate Conditions. Environ. Health Perspect. 2019, 127, 107014. [Google Scholar] [CrossRef] [Green Version]
- Komar, N.; Hamby, N.; Palamar, M.B.; Staples, J.E.; Williams, C. Indirect evidence of Bourbon virus (Thogotovirus, Orthomyxoviridae) infection in North Carolina. N. C. Med. J. 2020, 81, 214–215. [Google Scholar] [CrossRef]
- Egizi, A.; Wagner, N.E.; Jordan, R.A.; Price, D.C. Lone star ticks (Acari: Ixodidae) infected with Bourbon virus in New Jersey, USA. J. Med. Entomol. 2023, tjad052. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, J.; Lamkiewicz, K.; Kolesnikova, L.; Hölzer, M.; Marz, M.; Kochs, G. Comparative study of ten thogotovirus isolates and their distinct in vivo characteristics. Virol. J. 2022, 96, e01556-21. [Google Scholar] [CrossRef]
- Lambert, A.J.; Velez, J.O.; Brault, A.C.; Calvert, A.E.; Bell-Sakyi, L.; Bosco-Lauth, A.M.; Staples, J.E.; Kosoy, O.I. Molecular, serological and in vitro culture-based characterization of Bourbon virus, a newly described human pathogen of the genus Thogotovirus. J. Clin. Virol. 2015, 73, 127–132. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lange, R.E.; Dupuis, A.P., II.; Prusinski, M.A.; Maffei, J.G.; Koetzner, C.A.; Ngo, K.; Backenson, B.; Oliver, J.; Vogels, C.B.F.; Grubaugh, N.D.; et al. Identification and characterization of novel lineage 1 Powassan virus strains in New York State. Emerg. Microbes Infect. 2023, 12, 2155585. [Google Scholar] [CrossRef]
- Policastro, P.F.; Schwan, T.G. Experimental infection of Ixodes scapularis larvae (Acari: Ixodidae) by immersion in low passage cultures of Borrelia burgdorferi. J. Med. Entomol. 2003, 40, 364–370. [Google Scholar] [CrossRef] [Green Version]
- Mitzel, D.N.; Wolfinbarger, J.B.; Long, R.D.; Masnick, M.; Best, S.M.; Bloom, M.E. Tick-borne flavivirus infection in Ixodes scapularis larvae: Development of a novel method for synchronous viral infection of ticks. Virology 2007, 365, 410–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bondaryuk, A.N.; Kulakova, N.V.; Belykh, O.I.; Bukin, Y.S. Dates and Rates of Tick-Borne Encephalitis Virus-The Slowest Changing Tick-Borne Flavivirus. Int. J. Mol. Sci. 2023, 24, 2921. [Google Scholar] [CrossRef] [PubMed]
- Diuk-Wasser, M.A.; VanAcker, M.C.; Fernandez, M.P. Impact of Land Use Changes and Habitat Fragmentation on the Eco-epidemiology of Tick-Borne Diseases. J. Med. Entomol. 2021, 58, 1546–1564. [Google Scholar] [CrossRef]
- Rochlin, I.; Egizi, A.; Ginsberg, H.S. Modeling of historical and current distributions of lone star tick, Amblyomma americanum (Acari: Ixodidae), is consistent with ancestral range recovery. Exp. Appl. Acarol. 2023, 89, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Paddock, C.D.; Yabsley, M.J. Ecological havoc, the rise of white-tailed deer, and the emergence of Amblyomma americanum—Associated zoonoses in the United States. Curr. Top. Microbiol. 2007, 315, 289–324. [Google Scholar]
- Sonenshine, D.E. Range Expansion of Tick Disease Vectors in North America: Implications for Spread of Tick-Borne Disease. Int. J. Environ. Res. Public Health 2018, 15, 478. [Google Scholar] [CrossRef] [Green Version]
- Ogden, N.H.; Ben Beard, C.; Ginsberg, H.S.; Tsao, J.I. Possible Effects of Climate Change on Ixodid Ticks and the Pathogens They Transmit: Predictions and Observations. J. Med. Entomol. 2021, 58, 1536–1545. [Google Scholar] [CrossRef] [PubMed]
- Tsao, J.I.; Hamer, S.A.; Han, S.; Sidge, J.L.; Hickling, G.J. The Contribution of Wildlife Hosts to the Rise of Ticks and Tick-Borne Diseases in North America. J. Med. Entomol. 2021, 58, 1565–1587. [Google Scholar] [CrossRef]
- Warren, R.J. Deer overabundance in the USA: Recent advances in population control. Anim. Prod. Sci. 2011, 51, 259–266. [Google Scholar] [CrossRef]
- Loss, S.R.; Noden, B.H.; Hamer, G.L.; Hamer, S.A. A quantitative synthesis of the role of birds in carrying ticks and tick-borne pathogens in North America. Oecologia 2016, 182, 947–959. [Google Scholar] [CrossRef] [PubMed]
- Buczek, A.M.; Buczek, W.; Buczek, A.; Bartosik, K. The potential role of migratory birds in the rapid spread of ticks and tick-borne pathogens in the changing climatic and environmental conditions in Europe. Int. J. Environ. 2020, 17, 2117. [Google Scholar] [CrossRef] [Green Version]
- Ogden, N.H.; Lindsay, L.R.; Hanincová, K.; Barker, I.K.; Bigras-Poulin, M.; Charron, D.F.; Heagy, A.; Francis, C.M.; O’Callaghan, C.J.; Schwartz, I.; et al. Role of migratory birds in introduction and range expansion of Ixodes scapularis ticks and of Borrelia burgdorferi and Anaplasma phagocytophilum in Canada. Appl. Environ. Microbiol. 2008, 74, 1780–1790. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, N.; Beati, L.; Sellers, M.; Burton, L.; Adamson, S.; Robbins, R.G.; Moore, F.; Karim, S. Importation of exotic ticks and tick-borne spotted fever group rickettsiae into the United States by migrating songbirds. Ticks Tick Borne Dis. 2014, 5, 127–134. [Google Scholar] [CrossRef] [Green Version]
- Schneider, S.C.; Parker, C.M.; Miller, J.R.; Fredericks, L.P.; Allan, B.F. Assessing the contribution of songbirds to the movement of ticks and Borrelia burgdorferi in the Midwestern United States during fall migration. Ecohealth 2015, 12, 164–173. [Google Scholar] [CrossRef]
- Brinkerhoff, R.J.; Folsom-O’Keefe, C.M.; Streby, H.M.; Bent, S.J.; Tsao, K.; Diuk-Wasser, M.A. Regional variation in immature Ixodes scapularis parasitism on North American songbirds: Implications for transmission of the Lyme pathogen, Borrelia burgdorferi. J. Med. Entomol. 2011, 48, 422–428. [Google Scholar] [CrossRef]
- Scott, J.D.; Anderson, J.F.; Durden, L.A. Widespread dispersal of Borrelia burgdorferi-infected ticks collected from songbirds across Canada. J. Parasitol. 2012, 98, 49–59. [Google Scholar] [CrossRef]
- Lauring, A.S.; Andino, R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010, 6, e1001005. [Google Scholar] [CrossRef] [Green Version]
- Wagner, V.; Sabachvili, M.; Bendl, E.; Fuchs, J.; Kochs, G. The Antiviral Activity of Equine Mx1 against Thogoto Virus Is Determined by the Molecular Structure of Its Viral Specificity Region. Virol. J. 2023, 97, e01938-22. [Google Scholar] [CrossRef] [PubMed]
- Vogt, C.; Preuss, E.; Mayer, D.; Weber, F.; Schwemmle, M.; Kochs, G. The interferon antagonist ML protein of thogoto virus targets general transcription factor IIB. Virol. J. 2008, 82, 11446–11453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jennings, S.; Martínez-Sobrido, L.; García-Sastre, A.; Weber, F.; Kochs, G. Thogoto virus ML protein suppresses IRF3 function. Virology 2005, 331, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Hagmaier, K.; Jennings, S.; Buse, J.; Weber, F.; Kochs, G. Novel gene product of Thogoto virus segment 6 codes for an interferon antagonist. Virol. J. 2003, 77, 2747–2752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buettner, N.; Vogt, C.; Martínez-Sobrido, L.; Weber, F.; Waibler, Z.; Kochs, G. Thogoto virus ML protein is a potent inhibitor of the interferon regulatory factor-7 transcription factor. J. Gen. Virol. 2010, 91, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Stanwick, T.L.; Hallum, J.V. Role of interferon in six cell lines persistently infected with rubella virus. Infect. Immun. 1974, 10, 810–815. [Google Scholar] [CrossRef] [Green Version]
- Desmyter, J.A.N.; Melnick, J.L.; Rawls, W.E. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). Virol. J. 1968, 2, 955–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.; Chen, Z.; Kato, N.; Gale, M.; Lemon, S.M. Distinct poly (IC) and virus-activated signaling pathways leading to interferon-β production in hepatocytes. J. Biol. Chem. 2005, 280, 16739–16747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, E.H.; Smith, D.K.; Rabadan, R.; Peiris, M.; Poon, L.L. Codon usage bias and the evolution of influenza A viruses. Codon Usage Biases of Influenza Virus. BMC Evol. Biol. 2010, 10, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.E.; Yoon, H.; Lee, H.J.; Lee, J.H.; Chang, B.J.; Song, C.S.; Nahm, S.S. Expression patterns of influenza virus receptors in the respiratory tracts of four species of poultry. J. Vet. Sci. 2011, 12, 7–13. [Google Scholar] [CrossRef] [Green Version]
- Kim, A.S.; Zimmerman, O.; Fox, J.M.; Nelson, C.A.; Basore, K.; Zhang, R.; Durnell, L.; Desai, C.; Bullock, C.; Deem, S.L.; et al. An evolutionary insertion in the Mxra8 receptor-binding site confers resistance to alphavirus infection and pathogenesis. Cell Host Microbe 2020, 27, 428–440. [Google Scholar] [CrossRef]
- Deardorff, E.R.; Fitzpatrick, K.A.; Jerzak, G.V.; Shi, P.Y.; Kramer, L.D.; Ebel, G.D. West Nile virus experimental evolution in vivo and the trade-off hypothesis. PLoS Pathog. 2011, 7, e1002335. [Google Scholar] [CrossRef]
- Ciota, A.T.; Payne, A.F.; Kramer, L.D. West Nile virus adaptation to ixodid tick cells is associated with phenotypic trade-offs in primary hosts. Virology 2015, 482, 128–132. [Google Scholar] [CrossRef] [Green Version]
- Morse, M.A.; Marriott, A.C.; Nuttall, P.A. The glycoprotein of Thogoto virus (a tick-borne orthomyxo-like virus) is related to the baculovirus glycoprotein GP64. Virology 1992, 186, 640–646. [Google Scholar] [CrossRef]
- Bai, C.; Qi, J.; Wu, Y.; Wang, X.; Gao, G.F.; Peng, R.; Gao, F. Postfusion structure of human-infecting Bourbon virus envelope glycoprotein. J. Struct. Biol. 2019, 208, 99–106. [Google Scholar] [CrossRef]
- Kataoka, C.; Kaname, Y.; Taguwa, S.; Abe, T.; Fukuhara, T.; Tani, H.; Moriishi, K.; Matsuura, Y. Baculovirus GP64-mediated entry into mammalian cells. Virol. J. 2012, 86, 2610–2620. [Google Scholar] [CrossRef] [Green Version]
- Katou, Y.; Yamada, H.; Ikeda, M.; Kobayashi, M. A single amino acid substitution modulates low-pH-triggered membrane fusion of GP64 protein in Autographa californica and Bombyx mori nucleopolyhedroviruses. Virology 2010, 404, 204–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.P.; Gu, L.Z.; Lou, Y.H.; Cheng, R.L.; Xu, H.J.; Wang, W.B.; Zhang, C.X. A baculovirus isolated from wild silkworm encompasses the host ranges of Bombyx mori nucleopolyhedrosis virus and Autographa californica multiple nucleopolyhedrovirus in cultured cells. J. Gen. Virol. 2012, 93, 2480–2489. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Zhang, S.; Cui, Y.; Shi, Y.; Gao, G.F.; Qi, J. Structures of human-infecting Thogotovirus fusogens support a common ancestor with insect baculovirus. Proc. Natl. Acad. Sci. USA 2017, 114, E8905–E8912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portela, A.; Jones, L.D.; Nuttall, P. Identification of viral structural polypeptides of Thogoto virus (a tick-borne orthomyxo-like virus) and functions associated with the glycoprotein. J. Gen. Virol. 1992, 73, 2823–2830. [Google Scholar] [CrossRef] [PubMed]


| BRBV-1814 | |||
|---|---|---|---|
| Segment | BRBV-Original | BRBV-MO2013 | BRBV-STL |
| 1 (PB2) | 94.83% | 94.85% | 94.85% |
| 2 (PB1) | 95.67% | 94.44% | 96.02% |
| 3 (PA) | 93.56% | 93.77% | 93.83% |
| 4 (GP) | 94.88% | 95.08% | 95.32% |
| 5 (NP) | 94.43% | 94.65% | 94.72% |
| 6 (M) | 97.91% | 97.91% | 97.79% |
| BRBV-2143 | |||
| 1 (PB2) | 99.17% | 99.12% | 99.34% |
| 2 (PB1) | 99.07% | 97.31% | 99.57% |
| 3 (PA) | 99.06% | 98.85% | 99.42% |
| 4 (GP) | 98.54% | 98.47% | 98.99% |
| 5 (NP) | 99.21% | 99.49% | 99.49% |
| 6 (M) | 99.69% | 99.69% | 99.57% |
| BRBV-2666 | |||
| 1 (PB2) | 94.79% | 94.81% | 94.81% |
| 2 (PB1) | 95.55% | 94.33% | 95.90% |
| 3 (PA) | 93.38% | 93.59% | 93.64% |
| 4 (GP) | 94.95% | 94.88% | 95.40% |
| 5 (NP) | 94.87% | 95.08% | 95.16% |
| 6 (M) | 97.91% | 97.91% | 97.79% |
| Segment | Isolate | NT Change | AA Change |
|---|---|---|---|
| PB2 | 1814 | A1125G | I375M |
| PB1 | 2666 | A1069G | S357G |
| PA | 1814 | A1152G | I384M |
| PA | 1814 | A1439G | K480R |
| PA | 2143 | A893G | Q298R |
| PA | 2666 | C413A | P138Q |
| PA | 2666 | A1333G | I445V |
| GP | 2143 | A190G | I64V |
| GP | 2143 | A851G, T852C | N284S |
| GP | 2666 | A1379G | K460R |
| PB2 | 1814, 2666 | A32G | K11R |
| PB2 | 1814, 2666 | A1032G | I344M |
| PB2 | 1814, 2666 | G1247A | R416K |
| PB2 | 1814, 2666 | C1358T | A453V |
| PB2 | 1814, 2666 | T1359G | A453V |
| PB2 | 1814, 2666 | G1406A | S469N |
| PB1 | 1814, 2666 | G167A | R56K |
| PB1 | 1814, 2666 | A278G | H93R |
| PB1 | 1814, 2666 | A1669T | I557L |
| PA | 1814, 2666 | A70G | I24V |
| PA | 1814, 2666 | G214A | V72I |
| PA | 1814, 2666 | G493A | A165T |
| PA | 1814, 2666 | A538G | T180A |
| PA | 1814, 2666 | A547G | T183A |
| PA | 1814, 2143, 2666 | A670G | I224V |
| PA | 1814, 2666 | G677A | R226K |
| PA | 1814, 2666 | A809G | H270R |
| PA | 1814, 2666 | A916G | I306V |
| PA | 1814, 2666 | A939C | K313N |
| PA | 1814, 2666 | G956A | R319K |
| PA | 1814, 2666 | A957G | R319K |
| PA | 1814, 2666 | T1086A | D362E |
| PA | 1814, 2666 | G1378A | V460I |
| PA | 1814, 2666 | A1397G | E466G |
| PA | 1814, 2666 | T1426C | F476L |
| PA | 1814, 2666 | A1445C | N482T |
| PA | 1814, 2143, 2666 | G1908A | M636I |
| GP | 1814, 2666 | C29T | A10V |
| GP | 1814, 2666 | T43G/C | S15P |
| GP | 1814, 2666 | A304G | T102A |
| GP | 1814, 2666 | A1219G | I407V |
| GP | 1814, 2666 | A1259G | L420R |
| NP | 1814, 2666 | G899A | S300N |
| M | 1814, 2666 | A428G | N143S |
| M | 1814, 2666 | A647G | K216R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lange, R.E.; Dupuis, A.P., II; Ciota, A.T. Diversification of Bourbon Virus in New York State. Microorganisms 2023, 11, 1590. https://doi.org/10.3390/microorganisms11061590
Lange RE, Dupuis AP II, Ciota AT. Diversification of Bourbon Virus in New York State. Microorganisms. 2023; 11(6):1590. https://doi.org/10.3390/microorganisms11061590
Chicago/Turabian StyleLange, Rachel E., Alan P. Dupuis, II, and Alexander T. Ciota. 2023. "Diversification of Bourbon Virus in New York State" Microorganisms 11, no. 6: 1590. https://doi.org/10.3390/microorganisms11061590
APA StyleLange, R. E., Dupuis, A. P., II, & Ciota, A. T. (2023). Diversification of Bourbon Virus in New York State. Microorganisms, 11(6), 1590. https://doi.org/10.3390/microorganisms11061590






