Blastospores from Metarhizium anisopliae and Metarhizium rileyi Are Not Always as Virulent as Conidia Are towards Spodoptera frugiperda Caterpillars and Use Different Infection Mechanisms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Isolates and Insect Rearing
2.2. Blastospore and Conidium Production
2.3. Virulence Bioassay
2.4. Microscopy of Propagule Germination and Penetration of Insect Cuticle
2.5. RNA Extraction and Sequencing
2.6. RNA-Seq Reads
2.7. Differential Gene Expression Analysis
3. Results
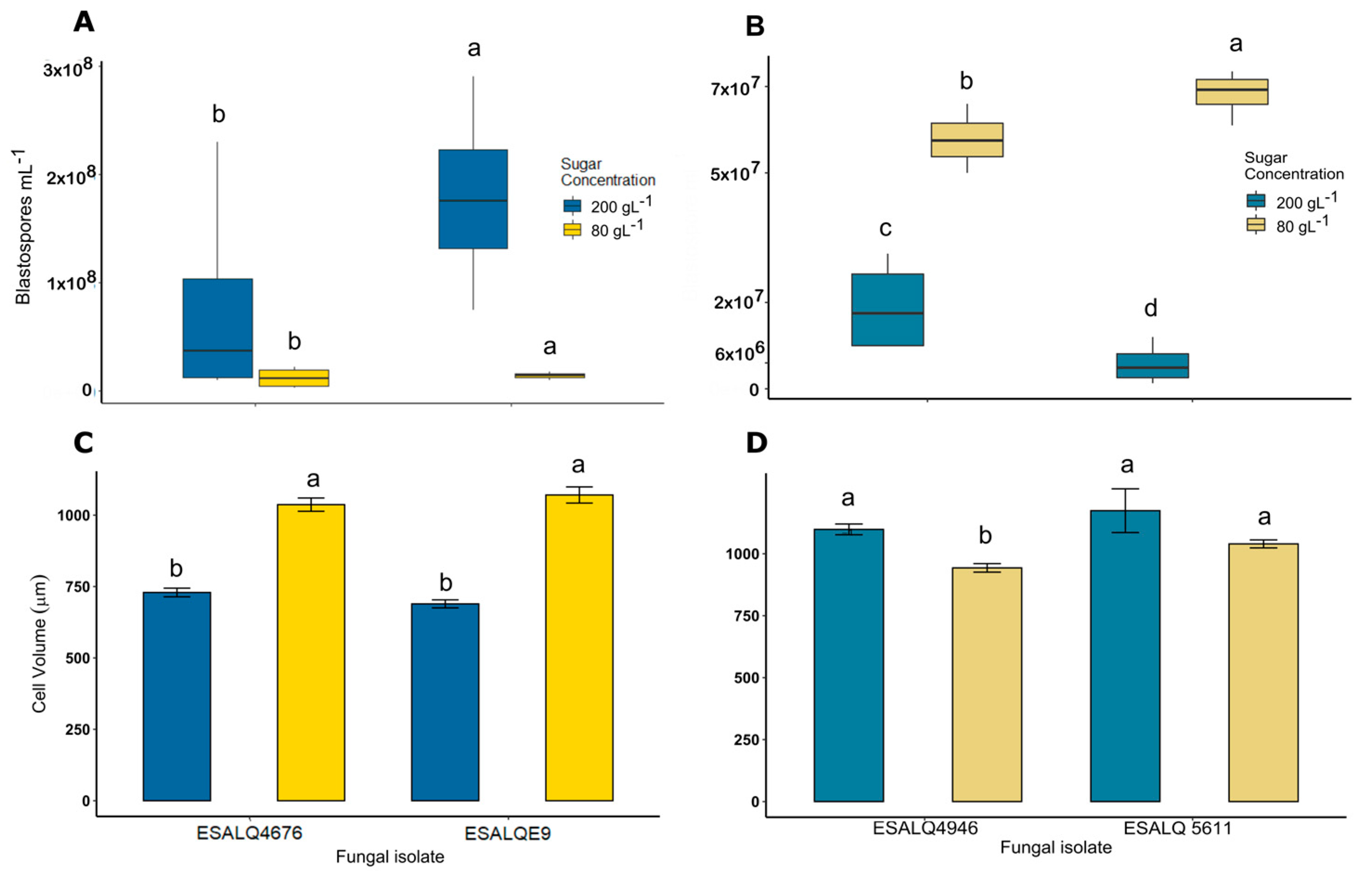
3.1. Blastospore Production under Different Levels of Osmotic Stress
3.2. Virulence Bioassay
3.3. Propagule Germination and Penetration through the Insect Cuticle
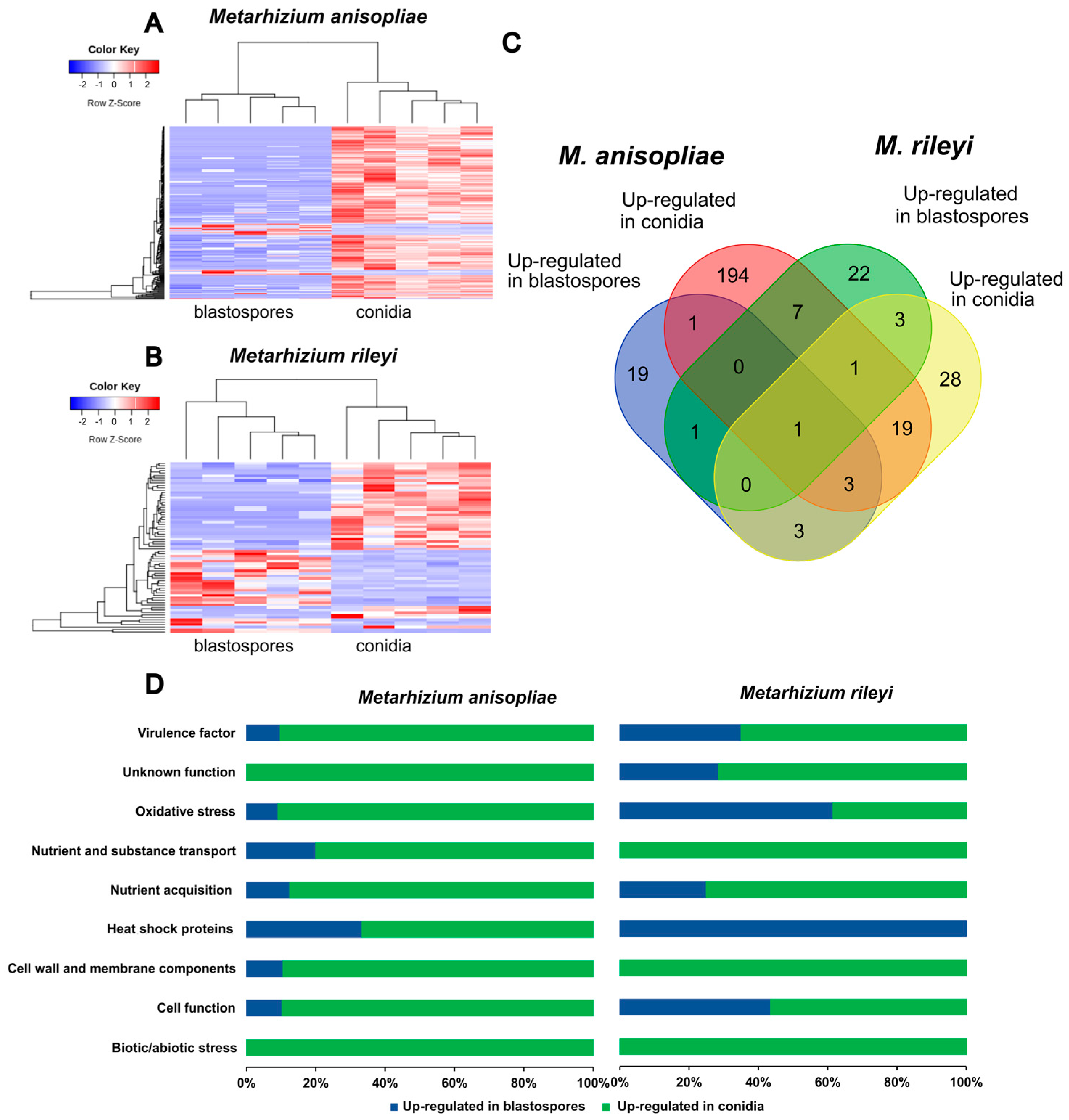
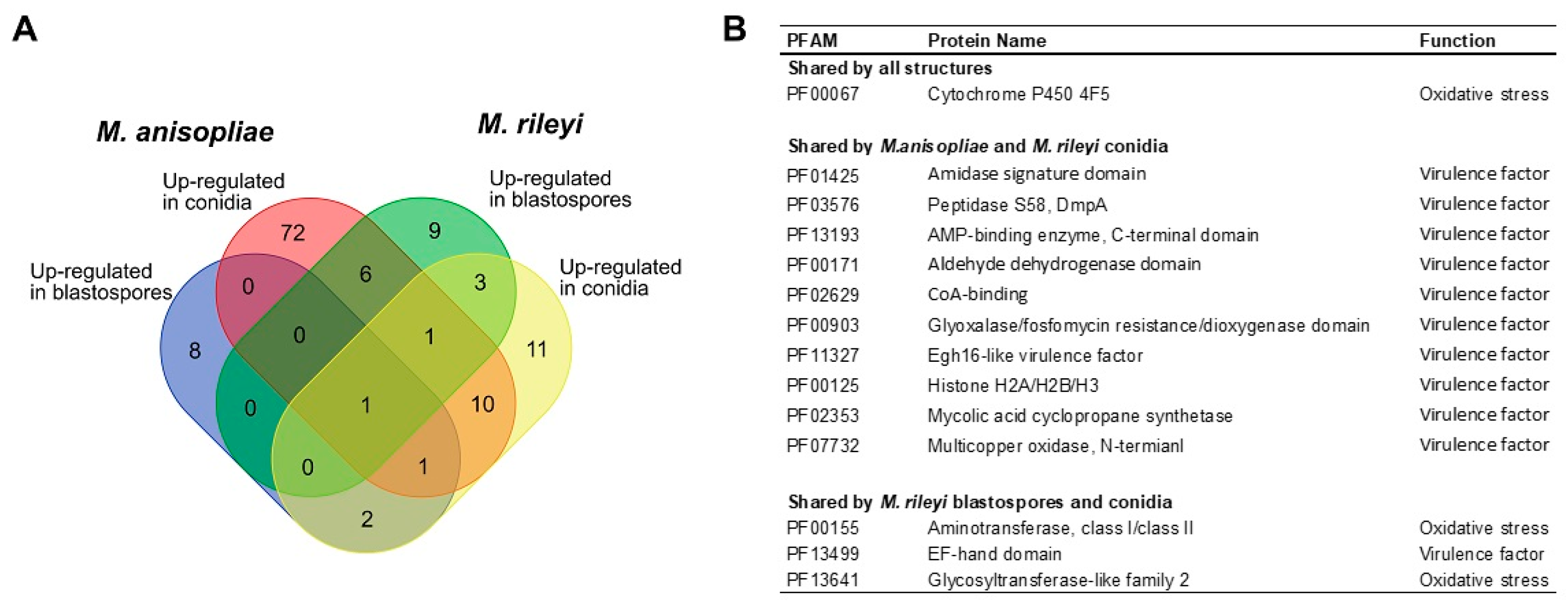
3.4. RNAseq Sequencing and Differential Gene Expression Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ambethgar, V. Potential of entomopathogenic fungi in insecticide resistance management (IRM): A review. J. Biopestic. 2009, 2, 177–193. [Google Scholar]
- St Leger, R.J.; Wang, J.B. Metarhizium: Jack of all trades, master of many. Open Biol. 2020, 10, 200307. [Google Scholar] [CrossRef] [PubMed]
- Sbaraini, N.; Guedes, R.L.M.; Andreis, F.C.; Junges, A.; de Morais, G.L.; Vainstein, M.H.; de Vasconcelos, A.T.R.; Schrank, A. Secondary metabolite gene clusters in the entomopathogen fungus Metarhizium anisopliae: Genome identification and patterns of expression in a cuticle infection model. BMC Genom. 2016, 17, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunner-Mendoza, C.; Reyes-Montes, M.D.; Moonjely, S.; Bidochka, M.J.; Toriello, C. A review on the genus Metarhizium as an entomopathogenic microbial biocontrol agent with emphasis on its use and utility in Mexico. Biocontrol Sci. Technol. 2019, 29, 83–102. [Google Scholar] [CrossRef]
- Parra, J.R.P. Biological Control in Brazil: An overview. Sci. Agric. 2014, 71, 420–429. [Google Scholar] [CrossRef]
- Wang, J.J.; Bai, W.W.; Zhou, W.; Liu, J.; Chen, J.; Liu, X.Y.; Xiang, T.T.; Liu, R.H.; Wang, W.H.; Zhang, B.L.; et al. Transcriptomic analysis of two Beauveria bassiana strains grown on cuticle extracts of the silkworm uncovers their different metabolic response at early infection stage. J. Invertebr. Pathol. 2017, 145, 45–54. [Google Scholar] [CrossRef]
- De Paula, A.R.; Silva, L.E.I.; Ribeiro, A.; Da Silva, G.A.; Silva, C.P.; Butt, T.M.; Samuels, R.I. Metarhizium anisopliae blastospores are highly virulent to adult Aedes aegypti, an important arbovirus vector. Parasites Vectors 2021, 14. [Google Scholar] [CrossRef]
- Mascarin, G.M.; Jackson, M.A.; Kobori, N.N.; Behle, R.W.; Delalibera, I. Liquid culture fermentation for rapid production of desiccation tolerant blastospores of Beauveria bassiana and Isaria fumosorosea strains. J. Invertebr. Pathol. 2015, 127, 11–20. [Google Scholar] [CrossRef]
- Butt, T.M.; Coates, C.J.; Dubovskiy, I.M.; Ratcliffe, N.A. Entomopathogenic Fungi: New Insights into Host-Pathogen Interactions. In Genetics and Molecular Biology of Entomopathogenic Fungi; Lovett, B., StLeger, R.J., Eds.; Advances in Genetics; Elsevier Academic Press Inc.: San Diego, CA, USA, 2016; Volume 94, pp. 307–364. [Google Scholar]
- Mantilla, J.G.; Galeano, N.F.; Gaitan, A.L.; Cristancho, M.A.; Keyhani, N.O.; Gongora, C.E. Transcriptome analysis of the entomopathogenic fungus Beauveria bassiana grown on cuticular extracts of the coffee berry borer (Hypothenemus hampei). Microbiology 2012, 158, 1826–1842. [Google Scholar] [CrossRef] [Green Version]
- Iwanicki, N.S.A.; Mascarin, G.M.; Moreno, S.G.; Eilenberg, J.; Delalibera, I., Jr. Growth kinetic and nitrogen source optimization for liquid culture fermentation of Metarhizium robertsii blastospores and bioefficacy against the corn leafhopper Dalbulus maidis. World J. Microbiol. Biotechnol. 2020, 36, 71. [Google Scholar] [CrossRef]
- Iwanicki, N.S.; Ferreira, B.O.; Mascarin, G.M.; Júnior, Í.D. Modified Adamek’s medium renders high yields of Metarhizium robertsii blastospores that are desiccation tolerant and infective to cattle-tick larvae. Fungal Biol. 2018, 122, 883–890. [Google Scholar] [CrossRef]
- Maldonado-Blanco, M.G.; Gallegos-Sandoval, J.L.; Fernandez-Pena, G.; Sandoval-Coronado, C.F.; Elias-Santos, M. Effect of culture medium on the production and virulence of submerged spores of Metarhizium anisopliae and Beauveria bassiana against larvae and adults of Aedes aegypti (Diptera: Culicidae). Biocontrol Sci. Technol. 2014, 24, 180–189. [Google Scholar] [CrossRef]
- Lohse, R.; Jakobs-Schönwandt, D.; Vidal, S.; Patel, A.V. Evaluation of new fermentation and formulation strategies for a high endophytic establishment of Beauveria bassiana in oilseed rape plants. Biol. Control 2015, 88, 26–36. [Google Scholar] [CrossRef]
- Mascarin, G.M.; Jackson, M.A.; Kobori, N.N.; Behle, R.W.; Dunlap, C.A.; Delalibera, I. Glucose concentration alters dissolved oxygen levels in liquid cultures of Beauveria bassiana and affects formation and bioefficacy of blastospores. Appl. Microbiol. Biotechnol. 2015, 99, 6653–6665. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.B.; St Leger, R.J.; Wang, C. Advances in Genomics of Entomopathogenic Fungi. In Genetics and Molecular Biology of Entomopathogenic Fungi; Lovett, B., StLeger, R.J., Eds.; Advances in Genetics; Elsevier Academic Press Inc: San Diego, CA, USA, 2016; Volume 94, pp. 67–105. [Google Scholar]
- Gao, Q.A.; Jin, K.; Ying, S.H.; Zhang, Y.J.; Xiao, G.H.; Shang, Y.F.; Duan, Z.B.; Hu, X.A.; Xie, X.Q.; Zhou, G.; et al. Genome Sequencing and Comparative Transcriptomics of the Model Entomopathogenic Fungi Metarhizium anisopliae and M. Acridum. PLoS Genet. 2011, 7, e1001264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fronza, E.; Specht, A.; Heinzen, H.; de Barros, N.M. Metarhizium (Nomuraea) rileyi as biological control agent. Biocontrol Sci. Technol. 2017, 27, 1243–1264. [Google Scholar] [CrossRef]
- Grijalba, E.P.; Espinel, C.; Cuartas, P.E.; Chaparro, M.L.; Villamizar, L.F. Metarhizium rileyi biopesticide to control Spodoptera frugiperda: Stability and insecticidal activity under glasshouse conditions. Fungal Biol. 2018, 122, 1069–1076. [Google Scholar] [CrossRef]
- Zhong, K.; Liu, Z.C.; Wang, J.L.; Liu, X.S. The entomopathogenic fungus Nomuraea rileyi impairs cellular immunity of its host Helicoverpa armigera. Arch. Insect Biochem. Physiol. 2017, 96, 10. [Google Scholar] [CrossRef]
- Yan-li, Z.; Hui, D.; Li-sheng, Z.; Zu-min, G.; Jin-cheng, Z. High virulence of a naturally occurring entomopathogenic fungal isolate, Metarhizium (Nomuraea) rileyi, against Spodoptera frugiperda. J. Appl. Entomol. 2022, 146, 659–665. [Google Scholar] [CrossRef]
- Montezano, D.; Specht, A.; Sosa-Gómez, D.; Roque-Specht, V.; Sousa-Silva, J.; Paula-Moraes, S.; Peterson, J.; Hunt, T. Host Plants of Spodoptera frugiperda (Lepidoptera: Noctuidae) in the Americas. Afr. Entomol. 2018, 26, 286–300. [Google Scholar] [CrossRef] [Green Version]
- Jackson, M.A.; Jaronski, S.T. Production of microsclerotia of the fungal entomopathogen Metarhizium anisopliae and their potential for use as a biocontrol agent for soil-inhabiting insects. Mycol. Res. 2009, 113, 842–850. [Google Scholar] [CrossRef]
- Jackson, M.A. Optimizing nutritional conditions for the liquid culture production of effective fungal biological control agents. J. Ind. Microbiol. Biotechnol. 1997, 19, 180–187. [Google Scholar] [CrossRef]
- Hothorn, T.; Bretz, F.; Westfall, P.; Heiberger, R.M.; Schuetzenmeister, A.; Scheibe, S. Package ‘Multcomp’: Simultaneous Inference in General Parametric Models; Texas Tech University: Lubbock, TX, USA, 2023. [Google Scholar]
- Moral, R.d.A.; Hinde, J.; Demetrio, C.G.B. Package ‘hnp’: Half-Normal Plots with Simulation Envelopes. 2022. Available online: https://cran.r-project.org/web/packages/hnp/hnp.pdf (accessed on 17 January 2023).
- Islam, S.M.N.; Chowdhury, M.Z.H.; Mim, M.F.; Momtaz, M.B.; Islam, T. Biocontrol potential of native isolates of Beauveria bassiana against cotton leafworm Spodoptera litura (Fabricius). Sci. Rep. 2023, 13, 8331. [Google Scholar] [CrossRef] [PubMed]
- Pannuti, L.E.R.; Baldin, E.L.L.; Hunt, T.E.; Paula-Moraes, S.V. On-Plant Larval Movement and Feeding Behavior of Fall Armyworm (Lepidoptera: Noctuidae) on Reproductive Corn Stages. Environ. Entomol. 2016, 45, 192–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobson, A.; Wilson, V.; Pileggi, S. Package ‘Parmsurvfit’: Parametric Models for Survival Data. 2022. Available online: https://rdrr.io/cran/parmsurvfit/ (accessed on 17 January 2023).
- Package Ecotox: Lethal Time Probit, Version 1.4.4. Available online: https://cran.r-project.org/web/packages/ecotox/ecotox.pdf (accessed on 25 May 2023).
- Invitrogen TRIzol™ Reagent User Guide. Available online: https://tools.thermofisher.com/content/sfs/manuals/trizol_reagent.pdf (accessed on 8 September 2020).
- Promega Corporation. RQ1 RNase-Free DNase (Cat.# M6101): Usage Information. Available online: https://worldwide.promega.com/products/cloning-and-dna-markers/molecular-biology-enzymes-and-reagents/rq1-rnase_free-dnase/?catNum=M6101 (accessed on 8 September 2020).
- Iwanicki, N.S.; Júnior, I.D.; Eilenberg, J.; De Fine Licht, H.H. Comparative RNAseq Analysis of the Insect-Pathogenic Fungus Metarhizium anisopliae Reveals Specific Transcriptome Signatures of Filamentous and Yeast-Like Development. G3 (Bethesda) 2020, 10, 2141–2157. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; Mcanulla, C.; Mcwilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [Green Version]
- Morales-Reyes, C.; Mascarin, G.M.; Jackson, M.A.; Hall, D.; Sánchez-Peña, S.R.; Arthurs, S.P. Comparison of aerial conidia and blastospores from two entomopathogenic fungi against Diaphorina citri (Hemiptera: Liviidae) under laboratory and greenhouse conditions. Biocontrol Sci. Technol. 2018, 28, 737–749. [Google Scholar] [CrossRef]
- Klein, B.S.; Tebbets, B. Dimorphism and virulence in fungi. Curr. Opin. Microbiol. 2007, 10, 314–319. [Google Scholar] [CrossRef] [Green Version]
- Boucias, D.; Liu, S.; Meagher, R.; Baniszewski, J. Fungal dimorphism in the entomopathogenic fungus Metarhizium rileyi: Detection of an in vivo quorum-sensing system. J. Invertebr. Pathol. 2016, 136, 100–108. [Google Scholar] [CrossRef]
- Albuquerque, P.; Casadevall, A. Quorum sensing in fungi—A review. Med. Mycol. 2012, 50, 337–345. [Google Scholar] [CrossRef] [Green Version]
- Vega, F.E.; Meyling, N.V.; Luangsa-ard, J.J.; Blackwell, M.; Kaya, H.K. Chapter 6—Fungal Entomopathogens. In Insect Pathology, 2nd ed.; Academic Press: San Diego, CA, USA, 2012; pp. 171–220. [Google Scholar]
- Kiruthiga, G.; Jeyarani, S.; Sathiah, N.; Murugan, M.; Sivakumar, U.; Uma, D. Pathogenicity, ultra-structural growth and development of green muscardine fungus, Metarhizium anisopliae (Metschnikoff) Sorokin (Ascomycota: Hypocreales) on maize fall armyworm, Spodoptera frugiperda (J. E. Smith) (Lepidoptera: Noctuidae). Egypt. J. Biol. Pest Control 2022, 32. [Google Scholar] [CrossRef]
- Ortiz-Urquiza, A.; Keyhani, N. Action on the Surface: Entomopathogenic Fungi versus the Insect Cuticle. Insects 2013, 4, 357–374. [Google Scholar] [CrossRef] [Green Version]
- Lovett, B.; St Leger, R.J. Stress is the rule rather than the exception for Metarhizium. Curr. Genet. 2015, 61, 253–261. [Google Scholar] [CrossRef]
- Dubovskiy, I.M.; Whitten, M.M.A.; Kryukov, V.Y.; Yaroslavtseva, O.N.; Grizanova, E.V.; Greig, C.; Mukherjee, K.; Vilcinskas, A.; Mitkovets, P.V.; Glupov, V.V.; et al. More than a colour change: Insect melanism, disease resistance and fecundity. Proc. R. Soc. B Biol. Sci. 2013, 280, 20130584. [Google Scholar] [CrossRef] [Green Version]
- Pedrini, N. Molecular interactions between entomopathogenic fungi (Hypocreales) and their insect host: Perspectives from stressful cuticle and hemolymph battlefields and the potential of dual RNA sequencing for future studies. Fungal Biol. 2018, 122, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Vega, F.; Jackson, M.; McGuire, M. Germination of conidia and blastospores of Paecilomyces fumosoroseus on the cuticle of the silverleaf whitefly, Bemisia Argentifolii. Mycopathol. 1999, 147, 33–35. [Google Scholar] [CrossRef]
- Vilcinskas, A. Coevolution between pathogen-derived proteinases and proteinase inhibitors of host insects. Virulence 2010, 1, 206–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, S.; Sun, Y.; Chen, H.; Wang, C. Suppression of the insect cuticular microbiomes by a fungal defensin to facilitate parasite infection. ISME J. 2023, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Tang, G.; Lu, M.; Wang, C. Host and Environmental Sensing by Entomopathogenic Fungi to Infect Hosts. Curr. Clin. Microbiol. Rep. 2022, 9, 69–74. [Google Scholar] [CrossRef]



| Letal Time 50 (LT50) | Letal Time 95 (LT95) | |||||
|---|---|---|---|---|---|---|
| Fungal Propagule | Estimated | lci * | uci * | Estimated | lci | uci |
| Conidia | 17.5 | 11.6 | 47.8 | 162 | 55.8 | 2306 |
| Blastospores 80 g L−1 | 18.5 | 11.8 | 58.3 | 245 | 71.1 | 6394 |
| Blastospores 200 g L−1 | 20.6 | 12.1 | 99.4 | 530 | 106 | 71405 |
| Letal Time 50 (LT50) | Letal Time 95 (LT95) | |||||
|---|---|---|---|---|---|---|
| Fungal Propagule | Estimated | lci * | uci * | Estimated | lci | uci |
| Conidia | 2.57 | 1.97 | 3.12 | 10.2 | 7.23 | 20.2 |
| Blastospores 80 g L−1 | 6.28 | 5.84 | 6.88 | 18.4 | 15.3 | 27.3 |
| Blastospores 200 g L−1 | 4.84 | 3.99 | 6.12 | 13.3 | 9.06 | 39.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gotti, I.A.; Moreira, C.C.; Delalibera, I., Jr.; De Fine Licht, H.H. Blastospores from Metarhizium anisopliae and Metarhizium rileyi Are Not Always as Virulent as Conidia Are towards Spodoptera frugiperda Caterpillars and Use Different Infection Mechanisms. Microorganisms 2023, 11, 1594. https://doi.org/10.3390/microorganisms11061594
Gotti IA, Moreira CC, Delalibera I Jr., De Fine Licht HH. Blastospores from Metarhizium anisopliae and Metarhizium rileyi Are Not Always as Virulent as Conidia Are towards Spodoptera frugiperda Caterpillars and Use Different Infection Mechanisms. Microorganisms. 2023; 11(6):1594. https://doi.org/10.3390/microorganisms11061594
Chicago/Turabian StyleGotti, Isabella Alice, Camila Costa Moreira, Italo Delalibera, Jr., and Henrik H. De Fine Licht. 2023. "Blastospores from Metarhizium anisopliae and Metarhizium rileyi Are Not Always as Virulent as Conidia Are towards Spodoptera frugiperda Caterpillars and Use Different Infection Mechanisms" Microorganisms 11, no. 6: 1594. https://doi.org/10.3390/microorganisms11061594





