The Mystery of Piezophiles: Understudied Microorganisms from the Deep, Dark Subsurface
Abstract
1. Introduction
2. Provenance and Description of Piezophiles
2.1. Geological Provenance of Microorganisms
2.1.1. Deep Marine Environments
2.1.2. Deep Terrestrial Environments
2.2. Microorganisms Adapted to Pressurized Environments
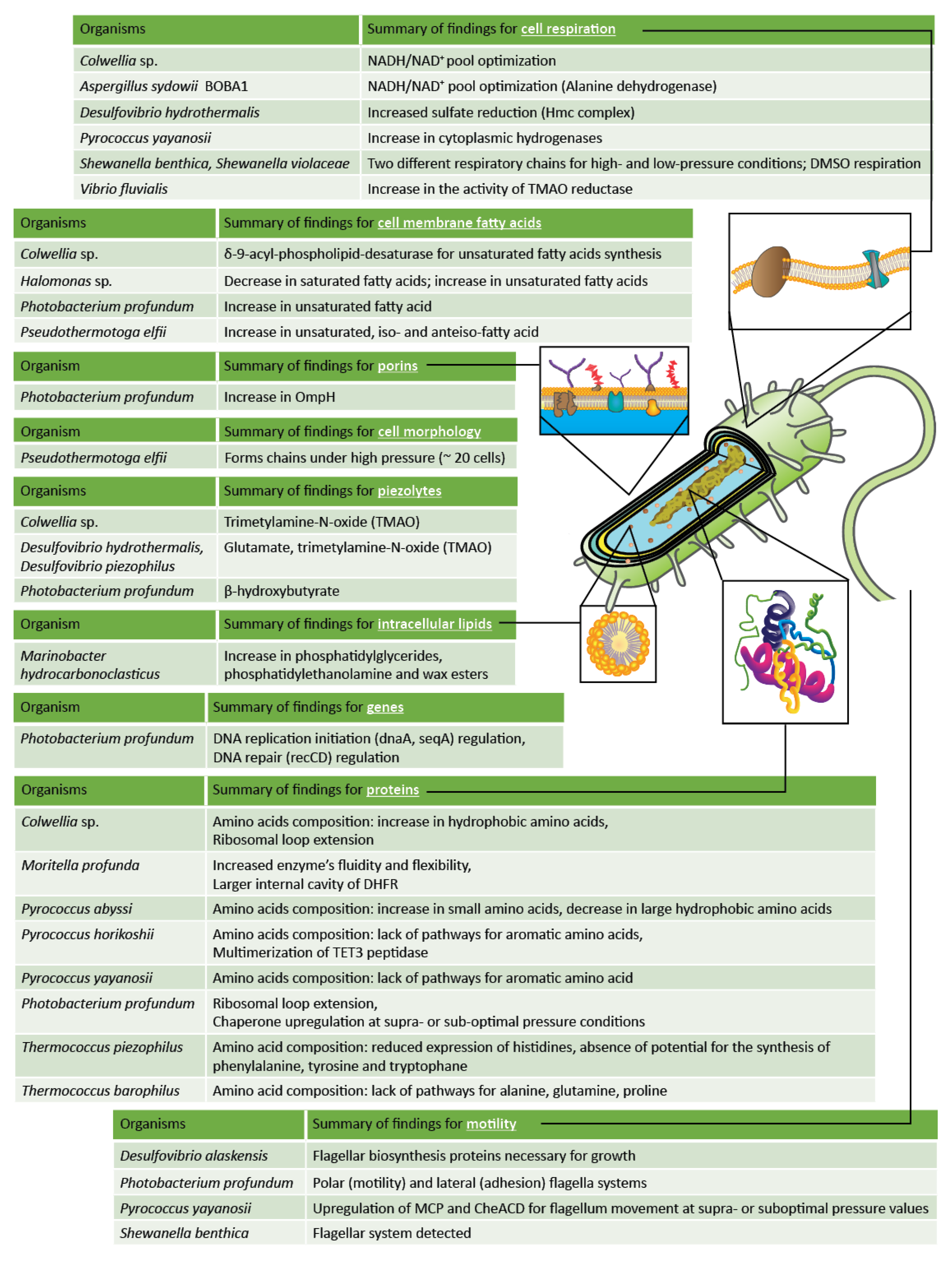
3. Current Knowledge of High-Pressure Cellular Adaptations in the Deep Biosphere
3.1. Motility, Chemotaxis, and Biofilm Adaptations
3.1.1. Adaptations in Microorganisms Retrieved from the Deep Marine Subsurface
3.1.2. Adaptations in Microorganisms Retrieved from the Deep Terrestrial Subsurface
3.2. Cell Morphology Adaptations
Adaptations in Microorganisms Retrieved from the Deep Terrestrial Subsurface
3.3. Cell Membrane Adaptations
3.3.1. Adaptations in Microorganisms Retrieved from the Deep Marine Subsurface
3.3.2. Adaptations in Microorganisms Retrieved from the Deep Terrestrial Subsurface
3.4. Energetic and Respiration Adaptations
3.4.1. Adaptations in Microorganisms Retrieved from the Deep Marine Subsurface
3.4.2. Adaptations in Microorganisms Retrieved from the Deep Terrestrial Subsurface
3.5. Piezolytes as Adaptations
Adaptations in Microorganisms Retrieved from the Deep Marine Subsurface
3.6. Intracellular Lipid Adaptations
Adaptations in Microorganisms Retrieved from the Deep Marine Subsurface
3.7. Protein Adaptations
3.7.1. Adaptations in Amino Acid Composition
Adaptations in Microorganisms Retrieved from the Deep Marine Subsurface
3.7.2. Adaptations in Multimerization
Adaptations in Microorganisms Retrieved from the Deep Marine Subsurface
3.7.3. Adaptations in Protein Structure and Conformation
Adaptations in Microorganisms Retrieved from the Deep Marine Subsurface
3.7.4. Ribosome Adaptations
Adaptations in Microorganisms Retrieved from the Deep Marine Subsurface
3.7.5. Chaperone Adaptations
Adaptations in Microorganisms Retrieved from the Deep Marine Subsurface
Adaptations in Microorganisms Retrieved from the Deep Terrestrial Subsurface
3.8. DNA Adaptations
3.8.1. Adaptations in Microorganisms Retrieved from the Deep Marine Subsurface
3.8.2. Adaptations in Microorganisms Retrieved from the Deep Terrestrial Subsurface
3.9. Overall Summary of High-Pressure Adaptations in Microorganisms
4. Microbial Diversity in the Deep Biosphere
5. Methodological Challenges in Studying Piezophiles
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Merino, N.; Aronson, H.S.; Bojanova, D.P.; Feyhl-Buska, J.; Wong, M.L.; Zhang, S.; Giovannelli, D. Living at the extremes: Extremophiles and the limits of life in a planetary context. Front. Microbiol. 2019, 10, 780. [Google Scholar] [CrossRef] [PubMed]
- Picard, A.; Daniel, I. Pressure as an environmental parameter for microbial life—A review. Biophys. Chem. 2013, 183, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Lauro, F.M.; Bartlett, D.H. Prokaryotic lifestyles in deep sea habitats. Extremophiles 2008, 12, 15–25. [Google Scholar] [CrossRef]
- Dolan, J. The origins of oceanography in France: The scientific expeditions of Travailleur and Talisman (1880–1883). Oceanography 2020, 33, 126–133. [Google Scholar] [CrossRef]
- ZoBell, C.E.; Johnson, F.H. The influence of hydrostatic pressure on the growth and viability of terrestrial and marine bacteria. J. Bacteriol. 1949, 57, 179–189. [Google Scholar] [CrossRef]
- Kim, S.-J.; Kato, C. Sampling, isolation, cultivation, and characterization of piezophilic microbes. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 3869–3881. ISBN 978-3-540-77587-4. [Google Scholar]
- Yayanos, A.A.; Dietz, A.S.; Boxtel, R. Isolation of a deep-sea bacterium and some of its growth characteristics. Science 1979, 205, 808–810. [Google Scholar] [CrossRef]
- Jebbar, M.; Franzetti, B.; Girard, E.; Oger, P. Microbial diversity and adaptation to high hydrostatic pressure in deep-sea hydrothermal vents prokaryotes. Extremophiles 2015, 19, 721–740. [Google Scholar] [CrossRef]
- Michoud, G.; Jebbar, M. High hydrostatic pressure adaptive strategies in an obligate piezophile Pyrococcus yayanosii. Sci. Rep. 2016, 6, 27289. [Google Scholar] [CrossRef] [PubMed]
- Gunbin, K.V.; Afonnikov, D.A.; Kolchanov, N.A. Molecular evolution of the hyperthermophilic archaea of the Pyrococcus genus: Analysis of adaptation to different environmental conditions. BMC Genom. 2009, 10, 639–651. [Google Scholar] [CrossRef]
- Di Giulio, M. A comparison of proteins from Pyrococcus furiosus and Pyrococcus abyssi: Barophily in the physicochemical properties of amino acids and in the genetic code. Gene 2005, 346, 1–6. [Google Scholar] [CrossRef]
- Martins, L.O.; Santos, H. Accumulation of mannosylglycerate and di-myo-inositol-phosphate by Pyrococcus furiosus in response to salinity and temperature. Appl. Environ. Microbiol. 1995, 61, 3299–3303. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, E.; Gabel, F.; Dura, M.A.; Finet, S.; Barraud, C.C.; Masson, P.; Franzetti, B. Effects of hydrostatic pressure on the quaternary structure and enzymatic activity of a large peptidase complex from Pyrococcus horikoshii. Arch. Biochem. Biophys. 2012, 517, 104–110. [Google Scholar] [CrossRef]
- Kaye, J.Z.; Baross, J.A. Synchronous effects of temperature, hydrostatic pressure, and salinity on growth, phospholipid profiles, and protein patterns of four Halomonas species isolated from deep-sea hydrothermal-vent and sea surface environments. Appl. Environ. Microbiol. 2004, 70, 6220–6229. [Google Scholar] [CrossRef] [PubMed]
- Smedile, F.; Foustoukos, D.I.; Patwardhan, S.; Mullane, K.; Schlegel, I.; Adams, M.W.; Schut, G.J.; Giovannelli, D.; Vetriani, C. Adaptations to high pressure of Nautilia sp. strain PV-1, a piezophilic Campylobacterium (aka Epsilonproteobacterium) isolated from a deep-sea hydrothermal vent. Environ. Microbiol. 2022, 24, 6164–6183. [Google Scholar] [CrossRef] [PubMed]
- Amrani, A.; Bergon, A.; Holota, H.; Tamburini, C.; Garel, M.; Ollivier, B.; Imbert, J.; Dolla, A.; Pradel, N. Transcriptomics reveal several gene expression patterns in the piezophile Desulfovibrio hydrothermalis in response to hydrostatic pressure. PLoS ONE 2014, 9, e106831. [Google Scholar] [CrossRef]
- Hervé, G.; Evans, H.G.; Fernado, R.; Patel, C.; Hachem, F.; Evans, D.R. Activation of latent dihydroorotase from Aquifex aeolicus by pressure. J. Biol. Chem. 2017, 292, 629–637. [Google Scholar] [CrossRef]
- Dalmasso, C.; Oger, P.; Selva, G.; Courtine, D.; L’Haridon, S.; Garlaschelli, A.; Roussel, E.; Miyazaki, J.; Reveillaud, J.; Jebbar, M.; et al. Thermococcus piezophilus, sp. nov., a novel hypothermophilic and piezophilic archaeaon with a broad pressure range for growth, isolated from a deepest hydrothermal vent at the Mid-Cayman Rise. Sys. Appl. Microbiol. 2016, 39, 440–444. [Google Scholar] [CrossRef]
- Moalic, Y.; Hartunians, J.; Dalmasso, C.; Courtine, D.; Georges, M.; Oger, P.; Shao, Z.; Jebbar, M.; Alain, K. The piezo-hyperthermophilic archaeon Thermococcus piezophilus regulates its energy efficiency system to cope with large hydrostatic pressure variations. Front. Microbiol. 2021, 12, 20. [Google Scholar] [CrossRef]
- Cario, A.; Jebbar, M.; Thiel, A.; Kervarec, N.; Oger, P.M. Molecular chaperone accumulation as a function of stress evidences adaptation to high hydrostatic pressure in piezophilic archaeon Thermococcus barophilus. Sci. Rep. 2016, 6, 29483. [Google Scholar] [CrossRef]
- Grossi, V.; Yakimov, M.M.; Al Ali, B.; Tapilatu, Y.; Cuny, P.; Goutx, M.; La Cono, V.; Giuliano, L.; Tamburini, C. Hydrostatic pressure affects membrane and storage lipid compositions of the piezotolerant hydrocarbon-degrading Marinobacter hydrocarbonoclasticus strain #5. Environ. Microbiol. 2010, 12, 2020–2033. [Google Scholar] [CrossRef]
- Yin, Q.J.; Zhang, W.J.; Qi, X.Q.; Zhang, S.D.; Jiang, T.; Li, L.F.; Chen, Y.; Santini, C.L.; Zhou, H.; Chou, I.M.; et al. High hydrostatic pressure inducible trimethylamine N-oxide reductase improves the pressure tolerance of piezosensitive bacteria Vibio fluvialis. Front. Microbiol. 2018, 8, 2646. [Google Scholar] [CrossRef] [PubMed]
- Pradel, N.; Ji, B.; Gimenez, G.; Talla, E.; Lenoble, P.; Garel, M.; Tamburini, C.; Fourquet, P.; Lebrun, R.; Bertin, P.; et al. The first genomic and proteomic characterization of a deep-sea sulfate reducer: Insights into the piezophilic lifestyle of Desulfovibrio piezophilus. PLoS ONE 2013, 8, e55130. [Google Scholar] [CrossRef] [PubMed]
- Peoples, L.M.; Kyaw, T.S.; Ugalde, J.A.; Mullane, K.K.; Chastain, R.A.; Yayanos, A.A.; Kusube, M.; Methé, B.A.; Bartlett, D.H. Distinctive gene and protein characteristics of extremely piezophilic Colwellia. BMC Genom. 2020, 21, 692. [Google Scholar] [CrossRef]
- Ohmae, E.; Murakami, C.; Tate, S.; Gekko, K.; Hata, K.; Akasaka, K.; Kato, C. Pressure dependence of activity and stability of dihydrofolate reductases of the deep-sea bacterium Moritella profunda and Escherichia coli. Biochim. Biophys. Acta 2012, 1824, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Penhallurick, R.W.; Ichiye, T. Pressure adaptations in deep-sea Moritella dihydrofolate reductases: Compressibility versus stability. Biology 2021, 10, 1211. [Google Scholar] [CrossRef]
- Yamada, M.; Nakasone, K.; Tamegai, H.; Kato, C.; Usami, R.; Horikoshi, K. Pressure regulation of soluble cytochromes c in a deep-sea piezophilic bacterium, Shewanella violacea. J. Bacteriol. 2000, 182, 2945–2952. [Google Scholar] [CrossRef]
- Zhang, W.-J.; Cui, X.-H.; Chen, L.-H.; Yang, J.; Li, X.-G.; Zhang, C.; Barbe, V.; Mangenot, S.; Fouteau, S.; Guerin, T.; et al. Complete genome sequence of Shewanella benthica DB21MT-2, an obligate piezophilic bacterium isolated from the deepest Mariana Trench sediment. Mar. Genom. 2019, 44, 52–56. [Google Scholar] [CrossRef]
- Bartlett, D.; Wright, M.; Yayanos, A.A.; Silverman, M. Isolation of a gene regulated by hydrostatic pressure in a deep-sea bacterium. Nature 1989, 342, 572–574. [Google Scholar] [CrossRef]
- Campanaro, S.; Vezzi, A.; Vitulo, N.; Lauro, F.M.; D’Angelo, M.; Simonato, F.; Cestaro, A.; Malacrida, G.; Bertoloni, G.; Valle, G.; et al. Laterally transferred elements and high pressure adaptation in Photobacterium profundum strains. BMC Genom. 2005, 6, 122–135. [Google Scholar] [CrossRef]
- Campanaro, S.; Pascale, F.D.; Telatin, A.; Schiavon, R.; Bartlett, D.H.; Valle, G. The transcriptional landscape of the deep-sea bacterium Photobacterium profundum in both a ToxR mutant and its parental strain. BMC Genom. 2012, 13, 567. [Google Scholar] [CrossRef]
- El-Hajj, Z.W.; Tryfona, T.; Allcock, D.J.; Hasan, F.; Lauro, F.M.; Sawyer, L.; Bartlett, D.H.; Ferguson, G.P. Importance of proteins controlling initiation of DNA replication in the growth of the high-pressure-loving bacterium Photobacterium profundum SS9. J. Bacteriol. 2009, 191, 6383–6393. [Google Scholar] [CrossRef] [PubMed]
- Eloe, E.A.; Lauro, F.M.; Vogel, R.F.; Bartlett, D.H. The deep-sea bacterium Photobacterium profundum SS9 utilizes separate flagellar systems for swimming and swarming under high-pressure conditions. Appl. Environ. Microbiol. 2008, 74, 6298–6305. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.; Bartlett, D.H.; Roberts, M.F. Solute accumulation in the deep-sea bacterium Photobacterium profundum. Extremophiles 2002, 6, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Welch, T.J.; Bartlett, D.H. Isolation and characterization of the structural gene for OmpL, a pressure-regulated porin-like protein from the deep-sea bacterium Photobacterium species strain SS9. J. Bacteriol. 1996, 178, 5027–5031. [Google Scholar] [CrossRef]
- Ganesh Kumar, A.; Manisha, D.; Sujitha, K.; Magesh Peter, D.; Kirubagaran, R.; Dharani, G. Genome sequence analysis of deep sea Aspergillus sydowii BOBA1 and effect of high pressure on biodegradation of spent engine oil. Sci. Rep. 2021, 11, 9347. [Google Scholar] [CrossRef]
- Xiong, L.; Jian, H.; Zhang, Y.; Xiao, X. The two sets of DMSO respiratory systems of Shewanella piezotolerans WP3 are involved in deep sea environmental adaptation. Front. Microbiol. 2016, 7, 1418. [Google Scholar] [CrossRef]
- Roumagnac, M.; Pradel, N.; Bartoli, M.; Garel, M.; Jones, A.A.; Armougom, F.; Fenouil, R.; Tamburini, C.; Ollivier, B.; Summers, Z.M.; et al. Responses to the hydrostatic pressure of surface and subsurface strains of Pseudothermotoga elfii revealing the piezophilic nature of the strain originating from an oil-producing well. Front. Microbiol. 2020, 11, 558771. [Google Scholar] [CrossRef]
- Williamson, A.J.; Carlson, H.K.; Kuehl, J.V.; Huang, L.L.; Iavarone, A.T.; Deutschbauer, A.; Coates, J.D. Dissimilatory sulfate reduction under high pressure by Desulfovibrio alaskensis G20. Front. Microbiol. 2018, 9, 1465. [Google Scholar] [CrossRef]
- Dutta, A.; Peoples, L.M.; Gupta, A.; Bartlett, D.H.; Sar, P. Exploring the piezotolerant/piezophilic microbial community and genomic basis of piezotolerance within the deep subsurface Deccan Traps. Extremophiles 2019, 23, 421–433. [Google Scholar] [CrossRef]
- Dutta, A.; Dutta Gupta, S.; Gupta, A.; Sarkar, J.; Roy, S.; Mukherjee, A.; Sar, P. Exploration of deep terrestrial subsurface microbiome in late cretaceous Deccan traps and underlying Archean basement, India. Sci. Rep. 2018, 8, 17459. [Google Scholar] [CrossRef]
- Kalwasińska, A.; Krawiec, A.; Deja-Sikora, E.; Gołębiewski, M.; Kosobucki, P.; Swiontek Brzezinska, M.; Walczak, M. Microbial diversity in deep-subsurface hot brines of northwest Poland: From community structure to isolate characteristics. Appl. Environ. Microbiol. 2020, 86, e00252-20. [Google Scholar] [CrossRef] [PubMed]
- Takai, K.; Moser, D.P.; DeFlaun, M.; Onstott, T.C.; Fredrickson, J.K. Archaeal diversity in waters from deep South African gold mines. Appl. Environ. Microbiol. 2001, 67, 5750–5760. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, H.; Kietäväinen, R.; Sohlberg, E.; Numminen, M.; Ahonen, L.; Itävaara, M. Microbiome composition and geochemical characteristics of deep subsurface high-pressure environment, Pyhäsalmi mine Finland. Front. Microbiol. 2015, 6, 1203. [Google Scholar] [CrossRef] [PubMed]
- Oger, P.M.; Jebbar, M. The many ways of coping with pressure. Res. Microbiol. 2010, 161, 799–809. [Google Scholar] [CrossRef]
- Chu, E.W.; Karr, J.R. Environmental impact: Concept, consequences, measurement. In Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2017; pp. 278–296. [Google Scholar] [CrossRef]
- Tiab, D.; Donaldson, E.C. Chapter 2-Introduction to petroleum geology. In Petrophysics, 3rd ed.; Tiab, D., Donaldson, E.C., Eds.; Gulf Professional Publishing: Boston, MA, USA, 2012; pp. 27–83. ISBN 978-0-12-383848-3. [Google Scholar]
- Lopez-Fernandez, M.; Broman, E.; Turner, S.; Wu, X.; Bertilsson, S.; Dopson, M. Investigation of viable taxa in the deep terrestrial biosphere suggests high rates of nutrient recycling. FEMS Microbiol. Ecol. 2018, 94, fiy121. [Google Scholar] [CrossRef]
- Yamamoto, K.; Hackley, K.C.; Kelly, W.R.; Panno, S.V.; Sekiguchi, Y.; Sanford, R.A.; Liu, W.-T.; Kamagata, Y.; Tamaki, H. Diversity and geochemical community assembly processes of the living rare biosphere in a sand-and-gravel aquifer ecosystem in the midwestern United States. Sci. Rep. 2019, 9, 13484. [Google Scholar] [CrossRef]
- Scoma, A. Functional groups in microbial ecology: Updated definitions of piezophiles as suggested by hydrostatic pressure dependence on temperature. ISME J. 2021, 15, 1871–1878. [Google Scholar] [CrossRef]
- Allan, E.E.; Bartlett, D.H. Piezophiles: Microbial adaptations to the deep-sea environment. In Extremophiles; Eolss Publishers Co. Ltd.: Oxford, UK, 2011; Volume 10, pp. 1–4. [Google Scholar]
- Grossart, H.-P.; Riemann, L.; Azam, F. Bacterial motility in the sea and its ecological implications. Aquat. Microb. Ecol. 2001, 25, 247–258. [Google Scholar] [CrossRef]
- Matz, C.; Boenigk, J.; Arndt, H.; Jürgens, K. Role of bacterial phenotypic traits in seletive feeding of the heterotrophic nanoflagellate Spumella sp. Aquat. Microb. Ecol. 2002, 27, 137–148. [Google Scholar] [CrossRef]
- Szurmant, H.; Ordal, G.W. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 2004, 68, 301–319. [Google Scholar] [CrossRef] [PubMed]
- Stewart, B.J.; McCarter, L.L. Lateral flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 2003, 185, 4508–4518. [Google Scholar] [CrossRef] [PubMed]
- Shinoda, S.; Okamoto, K. Formation and function of Vibrio parahaemolyticus lateral flagella. J. Bacteriol. 1977, 129, 1266–1271. [Google Scholar] [CrossRef] [PubMed]
- Das, C.; Mokashi, C.; Mande, S.S.; Saini, S. Dynamics and control of flagella assembly in Salmonella typhimurium. Front. Cell. Inf. Microbiol. 2018, 8, 36. [Google Scholar] [CrossRef]
- Bardy, S.L.; Mori, T.; Komoriya, K.; Aizawa, S.; Jarrell, K.F. Identification and localization of flagellins FlaA and FlaB3 within flagella of Methanococcus voltae. J. Bacteriol. 2002, 19, 5223–5233. [Google Scholar] [CrossRef]
- Vanlint, D.; Mitchell, R.; Bailey, E.; Meersman, F.; McMillan, P.F.; Michiels, C.W.; Aertsen, A. Rapid acquisition of gigapascal-high-pressure resistance by Escherichia coli. mBio 2011, 2, e00130-10. [Google Scholar] [CrossRef]
- Chi, E.; Bartlett, D.H. Use of a reporter gene to follow high-pressure signal transduction in the deep-sea bacterium Photobacterium sp. strain SS9. J. Bacteriol. 1993, 175, 7533–7540. [Google Scholar] [CrossRef]
- Kato, C.; Qureshi, M.H. Pressure response in deep-sea piezophilic bacteria. J. Mol. Microbiol. Biotech. 1999, 1, 87–92. [Google Scholar]
- Gralnick, J.A.; Vali, H.; Lies, D.P.; Newman, D.K. Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc. Natl. Acad. Sci. USA 2006, 103, 4669–4674. [Google Scholar] [CrossRef]
- Wu, C.-H.; Haja, D.K.; Adams, M.W.W. Cytoplasmic and membrane-bound hydrogenases from Pyrococcus furiosus. Methods Enzymol. 2018, 613, 153–168. [Google Scholar] [CrossRef]
- Robinson, C.R.; Sligar, S.G. Hydrostatic and osmotic pressure as tools to study macromolecular recognition. Methods Enzymol. 1995, 259, 395–427. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Greening, C.; Rattray, J.E.; Chakraborty, A.; Chuvochina, M.; Mayumi, D.; Dolfing, J.; Li, C.; Brooks, J.M.; Bernard, B.B.; et al. Metabolic potential of uncultured bacteria and archaea associated with petroleum seepage in deep-sea sediments. Nat. Commun. 2019, 10, 1816. [Google Scholar] [CrossRef] [PubMed]
- Gaussier, H.; Nouailler, M.; Champaud, E.; Garcin, E.B.; Sebban-Kreuzer, C.; Bornet, O.; Garel, M.; Tamburini, C.; Pieulle, L.; Dolla, A.; et al. Glutamate optimizes enzymatic activity under high hydrostatic pressure in Desulfovibrio species: Effects on the ubiquitous thioredoxin system. Extremophiles 2021, 25, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Martins, L.O.; Carreto, L.S.; Da Costa, M.S.; Santos, H. New compatible solutes related to di-myo-inositol-phosphate in members of the order Thermotogales. J. Bacteriol. 1996, 178, 5644–5651. [Google Scholar] [CrossRef] [PubMed]
- Santos, H.; da Costa, M.S. Organic solutes from thermophiles and hyperthermophiles. Methods Enzymol. 2001, 15, 302–334. [Google Scholar]
- Gunde-Cimerman, N.; Plemenitaš, A.; Oren, A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 2018, 42, 353–375. [Google Scholar] [CrossRef]
- Reed, C.J.; Lewis, H.; Trejo, E.; Winston, V.; Evilia, C. Protein adaptations in archaeal extremophiles. Archaea 2013, 2013, 373275. [Google Scholar] [CrossRef]
- Ichiye, T. Enzymes from piezophiles. Semin. Cell. Dev. Biol. 2018, 84, 138–146. [Google Scholar] [CrossRef]
- Hammes, G.G.; Benkovic, S.J.; Hammes-Schiffer, S. Flexibility, diversity, and cooperativity: Pillars of enzyme catalysis. Biochemistry 2011, 50, 10422–10430. [Google Scholar] [CrossRef]
- Loveridge, E.J.; Tey, L.H.; Behiry, E.M.; Dawson, W.M.; Evans, R.M.; Whittaker, S.B.M.; Gunther, U.L.; Williams, C.; Crump, M.P.; Allemann, R.K. The role of large-scale motions in catalysis by dihydrofolate reductase. J. Am. Chem. Soc. 2011, 50, 20561–20570. [Google Scholar] [CrossRef]
- D’Amico, S.; Collins, T.; Marx, J.-C.; Feller, G.; Gerday, C. Psychrophilic microorganisms: Challenges for life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef]
- Lauro, F.M.; Chastain, R.A.; Blankenship, L.E.; Yayanos, A.A.; Bartlett, D. The unique 16S rRNA genes of piezophiles reflect both phylogeny and adaptation. Appl. Environ. Microbiol. 2007, 73, 838–845. [Google Scholar] [CrossRef]
- Landau, J.V.; Smith, W.P.; Pope, D.H. Role of the 30S ribosomal subunit, initiation factors, and specific ion concentration in barotolerant protein synthesis in Pseudomonas bathycetes. J. Bacteriol. 1977, 130, 154–159. [Google Scholar] [CrossRef]
- Saibil, H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell. Biol. 2013, 14, 630–642. [Google Scholar] [CrossRef]
- Lu, M.; Campbell, J.L.; Boye, E.; Kleckner, N. SeqA: A negative modulator of replication initiation in E. coli. Cell 1994, 77, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Akimitsu, N.; Kashioka, T.; Hatano, M.; Kubota, T.; Ogata, Y.; Sekimizu, K.; Katayama, T. DiaA, a novel DnaA-binding protein, ensures the timely initiation of Escherichia coli chromosome replication. J. Biol. Chem. 2004, 279, 45546–45555. [Google Scholar] [CrossRef]
- Pavankumar, T.L.; Sinha, A.K.; Ray, M.K. All three subunits of RecBCD enzyme are essential for DNA repair and low-temperature growth in the antarctic Pseudomonas syringae Lz4W. PLoS ONE 2010, 5, e9412. [Google Scholar] [CrossRef] [PubMed]
- Fowler, R.G.; Schaaper, R.M. The role of the mutT gene of Escherichia coli in maintaining replication fidelity. FEMS Microbiol. Rev. 1997, 21, 43–54. [Google Scholar] [CrossRef]
- Crowley, D.J.; Boubriak, I.; Berquist, B.R.; Clark, M.; Richard, E.; Sullivan, L.; DasSarma, S.; McCready, S. The uvrA, uvrB and uvrC genes are required for repair of ultraviolet light induced DNA photoproducts in Halobacterium sp. NRC-1. Saline Syst. 2006, 2, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Wit, R.D.; Bouvier, T. ‘Everything is everywhere, but, the environment selects’; what did Baas Becking and Beijerinck really say? Environ. Microbiol. 2006, 8, 755–758. [Google Scholar] [CrossRef]
- Vellend, M. Conceptual synthesis in community ecology. Q. Rev. Biol. 2010, 85, 183–206. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Papke, R.T.; Ward, D.M. The importance of physical isolation to microbial diversification. FEMS Microbiol. Ecol. 2004, 48, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N. Microbial biogeography: Patterns in microbial diversity across space and time. In Accessing Uncultivated Microorganisms; ASM Press: Washington, DC, USA, 2008; pp. 95–115. ISBN 978-1-68367-146-6. [Google Scholar]
- Pholchan, M.K.; Baptista, J.; Davenport, R.J.; Sloan, W.T.; Curtis, T.P. Microbial community assembly, theory and rare functions. Front. Microbiol. 2013, 4, 68. [Google Scholar] [CrossRef] [PubMed]
- Mayol, E.; Arrieta, J.M.; Jiménez, M.A.; Martínez-Asensio, A.; Garcias-Bonet, N.; Dachs, J.; González-Gaya, B.; Royer, S.-J.; Benítez-Barrios, V.M.; Fraile-Nuez, E. Long-range transport of airborne microbes over the global tropical and subtropical ocean. Nat. Commun. 2017, 8, 201. [Google Scholar] [CrossRef]
- Luijendijk, E.; Gleeson, T.; Moosdorf, N. Fresh groundwater discharge insignificant for the world’s oceans but important for coastal ecosystems. Nat. Commun. 2020, 11, 1260–1273. [Google Scholar] [CrossRef] [PubMed]
- Filippidou, S.; Wunderlin, T.; Junier, T.; Jeanneret, N.; Dorador, C.; Molina, V.; Johnson, D.R.; Junier, P. A combination of extreme environmental conditions favor the prevalence of endospore-forming Firmicutes. Front. Microbiol. 2016, 7, 1707. [Google Scholar] [CrossRef]
- Mouser, P.J.; Borton, M.; Darrah, T.H.; Hartsock, A.; Wrighton, K.C. Hydraulic fracturing offers view of microbial life in the deep terrestrial subsurface. FEMS Microbiol. Ecol. 2016, 92, fiw166. [Google Scholar] [CrossRef]
- Soares, A.; Edwards, A.; An, D.; Bagnoud, A.; Bomberg, M.; Budwill, K.; Caffrey, S.M.; Fields, M.; Gralnick, J.; Kadnikov, V.; et al. A global perspective on bacterial diversity in the terrestrial deep subsurface. Microbiology 2023, 169, 001172. [Google Scholar] [CrossRef]
- Wilkins, M.J.; Daly, R.; Mouser, P.; Trexler, R.; Sharma, S.; Cole, D.R.; Wrighton, K.; Biddle, J.F.; Denis, E.H.; Fredrickson, J.K.; et al. Trends and future challenges in sampling in the deep terrestrial biosphere. Front. Microbiol. 2014, 5, 481. [Google Scholar] [CrossRef]
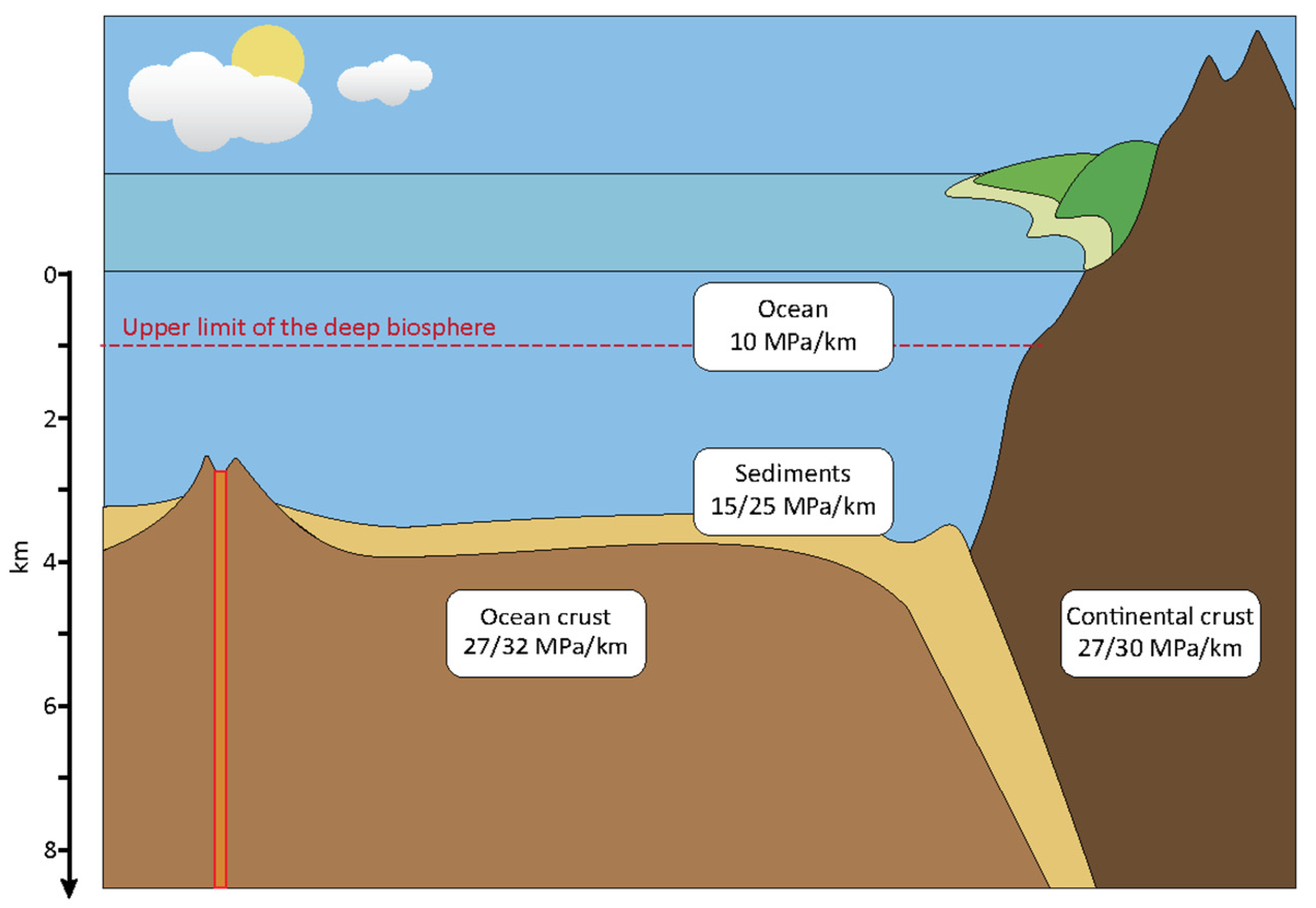
| Term | Definition |
|---|---|
| Non-piezophile | Microorganism that has optimal growth under atmospheric pressure and cannot grow under higher pressures |
| Piezosensitive | Microorganism that has optimal growth under atmospheric pressure but can still grow under higher pressures |
| Piezophile | Microorganism that has optimal growth under high pressures |
| Marine or Terrestrial Environment? | Environment | Studied High- Pressure Adaptations or Diversity? | Organisms Retrieved | Reference(s) |
|---|---|---|---|---|
| Deep marine subsurface | Hydrothermal vents (Mid-Atlantic ridge) | Adaptations | Pyrococcus yayanosii | Michoud and Jebbar (2016) [9] |
| Adaptations | Pyrococcus furiosus, Pyrococcus abyssi, Pyrococcus horikoshii | Gunbin et al. [10], Di Giulio [11], Martins and Santos [12], Rosenbaum et al. [13] | ||
| Hydrothermal vents (Location not specified) | Adaptations | Halomonas sp. | Kaye and Baross [14] | |
| Hydrothermal vents (East Pacific rise) | Adaptations | Nautilia sp. PV-1 | Smedile et al. [15] | |
| Adaptations | Desulfovibrio hydrothermalis | Amrani et al. [16] | ||
| Hydrothermal vents (Italian islands) | Adaptations | Aquifex aeolicus | Hervé et al. [17] | |
| Hydrothermal vents (Mid-Cayman Rise) | Diversity | - | Dalmasso et al. [18] | |
| Adaptations | Thermococcus piezophilus | Moalic et al. [19] | ||
| Hydrothermal vents (Snake Pit) | Adaptations | Thermococcus barophilus | Cario et al. [20] | |
| Deep sea (Mediterranean seawater) | Adaptations | Marinobacter hydrocarbonoclasticus | Grossi et al. [21] | |
| Deep sea (South China Sea) | Adaptations | Vibrio fluvialis | Yin et al. [22] | |
| Deep sea (Mediterranean Sea) | Adaptations | Desulfovibrio piezophilus | Pradel et al. [23] | |
| Deep sea (Mariana Trench) | Adaptations | Colwellia sp. | Peoples et al. [24] | |
| Marine sediment (West African Coast) | Adaptations | Moritella profunda, Moritella yayanosii | Ohmae et al. [25], Penhallurick and Ichiye [26] | |
| Marine sediments (Ryukyu Trench) | Adaptations | Shewanella benthica, Shewanella violaceae | Yamada et al. [27], Zhang et al. [28] | |
| Adaptations | Photobacterium profundum SS9 | Bartlett et al. [29], Campanaro et al. [30], Campanaro et al. [31], El-Hajj et al. [32], Eloe et al. [33], Martin et al. [34], Welch and Bartlett [35] | ||
| Marine sediments (India) | Adaptations | Aspergillus sydowii BOBA1 | Ganesh Kumar et al. [36] | |
| Marine sediments (West Pacific) | Adaptations | Shewanella piezotolerans | Xiong et al. [37] | |
| Deep terrestrial subsurface | Oil reservoir (Location not specified) | Adaptations | Pseudothermotoga elfii DMS9442 | Roumagnac et al. [38] |
| Oil reservoir (USA) | Adaptations | Desulfovibrio alaskensis | Williamson et al. [39] | |
| Deccan Traps (India) | Adaptations | Methylotenera sp., Caulobacter sp., Alcanivorax sp. | Dutta et al. [40] | |
| Diversity | - | Dutta et al. [41] | ||
| Hot brines (Poland) | Diversity | - | Kalwasińska et al. [42] | |
| Gold mine (Africa) | Diversity | - | Takai et al. [43] | |
| Pyhäsalmi mines (Finland) | Diversity | - | Miettinen et al. [44] |
| Term | Definition |
|---|---|
| Dispersal | The transport of microorganisms by wind, water, or other macro-organisms |
| Diversification | The evolution and adaptation of microorganisms through horizontal gene transfer or mutations, for example |
| Drift | The changes in the relative abundance distribution of a microbial community |
| Selection | The effect of abiotic factors (pH, temperature, salinity, pressure, availability of carbon) selecting for the microbial community structure |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scheffer, G.; Gieg, L.M. The Mystery of Piezophiles: Understudied Microorganisms from the Deep, Dark Subsurface. Microorganisms 2023, 11, 1629. https://doi.org/10.3390/microorganisms11071629
Scheffer G, Gieg LM. The Mystery of Piezophiles: Understudied Microorganisms from the Deep, Dark Subsurface. Microorganisms. 2023; 11(7):1629. https://doi.org/10.3390/microorganisms11071629
Chicago/Turabian StyleScheffer, Gabrielle, and Lisa M. Gieg. 2023. "The Mystery of Piezophiles: Understudied Microorganisms from the Deep, Dark Subsurface" Microorganisms 11, no. 7: 1629. https://doi.org/10.3390/microorganisms11071629
APA StyleScheffer, G., & Gieg, L. M. (2023). The Mystery of Piezophiles: Understudied Microorganisms from the Deep, Dark Subsurface. Microorganisms, 11(7), 1629. https://doi.org/10.3390/microorganisms11071629







