Abstract
Tomato is the main vegetable cultivated under soilless culture systems (SCSs); production of organic tomato under SCSs has increased due to consumer demands for healthier and environmentally friendly vegetables. However, organic tomato production under SCSs has been associated with low crop performance and fruit quality defects. These agricultural deficiencies could be linked to alterations in tomato plant microbiota; nonetheless, this issue has not been sufficiently addressed. Thus, the main goal of the present study was to characterize the rhizosphere and phyllosphere of tomato plants cultivated under conventional and organic SCSs. To accomplish this goal, tomato plants grown in commercial greenhouses under conventional or organic SCSs were tested at 8, 26, and 44 weeks after seedling transplantation. Substrate (n = 24), root (n = 24), and fruit (n = 24) composite samples were subjected to DNA extraction and high-throughput 16S rRNA gene sequencing. The present study revealed that the tomato core microbiota was predominantly constituted by Proteobacteria, Actinobacteria, and Firmicutes. Remarkably, six bacterial families, Bacillaceae, Microbacteriaceae, Nocardioidaceae, Pseudomonadaceae, Rhodobacteraceae, and Sphingomonadaceae, were shared among all substrate, rhizosphere, and fruit samples. Importantly, it was shown that plants under organic SCSs undergo a dysbiosis characterized by significant changes in the relative abundance of Bradyrhizobiaceae, Caulobacteraceae, Chitinophagaceae, Enterobacteriaceae, Erythrobacteraceae, Flavobacteriaceae, Nocardioidaceae, Rhodobacteraceae, and Streptomycetaceae. These results suggest that microbial alterations in substrates, roots, and fruits could be potential factors in contributing to the crop performance and fruit quality deficiencies observed in organic SCSs.
1. Introduction
Tomato (Solanum lycopersicum L.) fruit is one of the most consumed vegetables around the world, with an estimated production of ∼160 million tons per year [1,2,3]. In recent years, production of tomatoes under soilless culture systems (SCSs), hosted in greenhouses, has emerged as a sustainable and intensive agricultural practice [4,5]. It has been estimated that ∼95% of tomatoes produced in greenhouses from Europe and North America are cultivated under SCSs [5].
In SCSs, soil is replaced by inert substrates (e.g., coconut fiber, peat, perlite, rockwool, and struvite) and nutrients are supplied through irrigation water [4,6]. Tomato production under SCSs provides numerous advantages, for example, efficient use of water and nutrients, increased fruit quality, off season production, reduction of soil-borne pathogens, and low environmental impact [4,5,7]. Importantly, it has been estimated that SCSs generate 30% higher fruit yields than soil-based cultivation systems [4]. Additionally, SCSs require 75% less water (∼50 L/kg of tomato fruits) than soil-based production (∼200 L/kg of tomato fruits) [5]; for these reasons, the SCS is considered a more sustainable intensive production system [4,6].
In the last decade, production of organic tomato fruits has increased considerably due to consumer demands for healthier and safer vegetables [8]. Because certified organic farming strictly prohibits the use of synthetic fertilizers, fungicides, herbicides, and pesticides [9], organic tomatoes are considered as a healthier alternative [10].
Unfortunately, it has been documented that tomato plants under organic farming undergo moderate environmental stress [11], causing lower fruit yields [12,13,14,15,16], a reduction in fruit weight and firmness [14,16], and an increase in phytopathogen infections [9,17], compared to conventional production systems. Interestingly, it has been suggested that a potential cause of the agricultural problems observed in organic production could be associated with alterations of the tomato plant microbiota [18,19,20].
An imbalance in microbial populations (dysbiosis) in roots (rhizosphere) and aerial plant surfaces (phyllosphere) could promote plant disease [21,22,23] and negatively impact tomato plant growth and productivity [19,24]; thus, it could be possible that tomato plants under SCSs experience alterations to their core microbial populations. A core microbiota is defined as microbial taxa that remains stable independently of plant genotype and environmental conditions [23]. Based on this premise, the main goal of the present study was to characterize the rhizosphere and phyllosphere microbiota of tomato plants cultivated under conventional and organic fertilization regimes in SCSs and to gain insights into the potential impact of these agricultural practices on the establishment of potentially beneficial and pathogenic bacterial populations.
2. Materials and Methods
2.1. Tomato Plants Grown in Soilless Culture Systems
The study was conducted in two commercial greenhouses located in Queretaro, Mexico (20°42′22.5″ N 99°56′27.6″ W) during the 2019 growing season. Seeds of Solanum lycopersicum cv. “Merlice” were grown in commercial rockwool substrates; 16 days after germination, seedlings were grafted on Solanum lycopersicum cv. “Maxifort” (DeRuiter, The Netherlands). Two weeks after grafting, two seedlings were transplanted into a commercial soilless grow bag. Grow bags had a dimension of 110 cm × 20 cm × 12 cm and contained growing substrates made of sterile coconut fiber (Galaku International, Vaucluse, NSW, Australia); substrates had a 40% air filled porosity, 55% water holding capacity, 40% water retention efficiency, and a 30 mL 10 min−1 capillary uptake value.
For the conventional and organic SCSs, a drop irrigation system was used to provide plants with nutrients; for the conventional system, nutrient solution contained Ca(NO₃)₂ (123 g L−1), CaCl2 (9 g L−1), Ca-EDTA (2 g L−1), KNO3 (26 g L−1), KCl (5 g L−1), K2SO4 (33 g L−1), MgSO4 (48 g L−1), KH2PO4 (21 g L−1), and a mixture of the commercial fertilizers Quelsel Mix (6.5% Fe, 2.1% Mg, 0.4% Zn, 0.2% Cu, 2% B, 0.1% Mo, 88.7% chelating agents) (3.5 g L−1), Newquel Mn 13% (0.14 g L−1), Newquel Zn 14% (0.405 g L−1), and Quelsel Fe 6% (1 g L−1) (Diosol, Mexico) [1,19].
For the organic SCS, the nutrient solution contained CaSO4 (50 g L−1), MgSO4 (66 g L−1), and K2SO4 (90 g L−1); additionally, an organic fertilizer, Tierra Fertil 5-7-1 (55 mL L−1, Mar y Tierra, Mexico) was added [1,19]. All organic nutrients were obtained from providers registered in the Organic Material Review Institute (OMRI), and production management was in accordance with the National Organic Program (NOP) from the United States Department of Agriculture (USDA) [25].
Nutrient solutions for the conventional and organic SCSs were adjusted to maintain the concentration of the following nutrients: Ca (7.3 mM), Cl (9 mM), K (7 mM), Mg (2.8 mM), NH4 (0.8 mM), NO3 (12.5 mM), PO4 (2 mM), and SO4−2 (3.4 mM); as well as the microelements: B (0.09 mM), Cu (0.001 mM), Fe (0.044 mM), Mn (0.013 mM), Mo (0.001 mM), and Zn (0.009 mM) [1,19]. Additionally, the following commercial pathogen control agents were supplemented in conventional and organic SCSs AgroClean (Koppert, Berkel en Rodenrijs, The Netherlands), Amifol K (Tradecorp, Zapopan, Mexico), Kumulus (BASF, Mexico City, Mexico), MilStop (PHC, Mexico City, Mexico), Serenade (Bayer CropScience, Leverkusen, Germany), and System Cu (Idai Nature, Valencia, Spain) as indicated by the manufacturers. The electrical conductivity and pH of the nutrient solution in the conventional fertilization regime were, on average, 2.6 mS cm−1 and 6.2, respectively; whereas, in the organic fertilization regime, they were 1.84 mS cm−1 and 6.6, respectively. Average day/night temperature and relative humidity inside greenhouses were ~23/17 °C, and ~86/92%, respectively. The tomato fruit production cycle comprised weeks 8 to 44, after seedlings transplantation (AST). During this period, twelve plants from each production system were randomly selected and labeled across the whole greenhouse. Substrate, rhizosphere, and tomato fruit samples from these selected plants were collected for microbial analyses. Overall, tomato yield under organic SCSs was, on average, 19% lower than the conventional fertilization regime (unpublished data).
2.2. Sample Collection and DNA Extraction
Substrate (n = 12), rhizosphere (n = 12), and fruit (n = 12) samples from plants grown in each SCS, conventional and organic, were collected at weeks 8, 26, and 44 AST (Table 1). Substrate samples (~5 g) were collected from root-free zones (>1 cm from roots) as previously described [18,19]. Rhizosphere samples (~2 g) were collected ~10 cm away from the stem; roots were shaken to remove loose substrate particles, and only bacterial communities associated within ~1 mm of the root surface remained [18,26,27]. A tomato sample was composed of three fruits collected from a single cluster harvested at the pink maturity stage and grade 4, according to the USDA color classification requirements [28]. All samples were collected aseptically, using sterile gloves and sampling bags; placed in a cooler (~4 °C); and transported to the laboratory within 3 h of collection.

Table 1.
Sampling scheme for microbial population analyses of substrate, rhizosphere, and fruit samples from conventional and organic SCSs.
To gain insights into the core microbiota, the thirty-six substrate, rhizosphere, and fruit samples collected during the whole production cycle (from weeks 8 to 44 AST) were processed as composite samples (3 individual samples = 1 composite sample) (Table 1). Composite samples (n = 12) were subjected to DNA extraction; briefly, the substrate (1.0 g), rhizosphere (1.0 g), and tomato samples were rinsed with sterile deionized water (1.0, 1.0, and 3.0 mL, respectively) for ~1 min to collect microbial populations associated with the samples [26,29,30]. Sample washes were centrifuged at 8000× g during 1 min and the obtained pellets (~200 mg) were subjected to total DNA extraction using the ZymoBIOMICS® DNA Miniprep kit, (Zymo Research, Irvine, CA, USA) following the manufacturer’s instructions. DNA concentration and quality were measured using a Nanodrop 2000 UV-Vis spectrophotometer (Thermo ScientificTM, Waltham, MA, USA). All DNA samples were diluted at 5 ng/μL and stored at −20 °C.
2.3. High-Throughput Sequencing of 16S rRNA and Data Processing
A total of 72 samples were subjected to next-generation sequencing (NGS) using the ZymoBIOMICS® Targeted Sequencing Service for Microbiome Analysis at the Zymo Research company (https://zymoresearch.eu/pages/16s-its-amplicon-sequencing; Irvine, CA, USA; accessed on 1 April 2023) [31,32,33,34]; briefly, V3–V4 regions of the 16S rRNA gene were PCR amplified and then used for sequence library construction using the Quick-16S™ NGS Library Prep Kit (Zymo Research, Irvine, CA, USA), as described elsewhere [35]. Sequencing via synthesis was performed with the Illumina® MiSeq™ platform (San Diego, CA, USA) [36]. After sequencing, primers and adaptor sequences were removed from the reads, and the sequences were trimmed to the same length (~320 bases, using Illumina Data Analysis Software V2.3) [37]. Low-quality reads and chimeric sequences were removed using the Divisive Amplicon Denoising Algorithm 2 (DADA2) pipeline [38]. Rarefaction curves were generated based on the number of bacterial Amplicon Sequence Variants (ASVs) to confirm that sampling depth was sufficient and that all samples reached a plateau. ASVs were classified at the phylum and family level using UCLUST from QIIME v.1.9.1 [39] using the Zymo Research Database, an internally designed and curated reference database [31,32,33,34,35].
2.4. Analyses of Core Microbiota, Bacterial Abundance, and Diversity
Analyses of core microbiota at each selected niche were performed using relative abundance and occurrence data with cut-off values of ≥1% and ≥50%, respectively, as previously described [40]. The relative abundance of the total bacteria was estimated by means of the 16S rRNA gene copy number generated for each sample using quantitative PCR (qPCR) assays with the absolute quantification method; for this goal, a 16S rRNA gene plasmid-DNA standard curve was used for the analysis. The total gene copy number was calculated using the following equation: number of copies = [amount of DNA (ng) × Avogadro’s number (6.022 × 1023)]/[estimated genome size (4.64 × 106 bp*) × average molecular weight of a DNA bp (660 g/mole/bp)] [41,42,43,44].
Analyses of alpha diversity, richness, Simpson, and evenness indices [45,46,47] were estimated using bacterial relative abundance data at the family level. Beta diversity was estimated using Bray–Curtis and Jaccard indices [48,49]. Comparisons of microbiota composition were carried out using Principal Component Analysis (PCA) [50].
Identification of potential bacterial biomarkers was performed by using linear discriminant analysis (LDA) effect size (LEfSe), an approach highlighting biological consistency and effect size relevance [51]. LDA-LEfSe was performed using the all-against-all mode, calculating the LDA score after 200 bootstrapping iterations and a level of significance ≤ 0.05. The LDA score threshold was set to 4.0 [51,52,53].
2.5. Absolute Quantification of Enterobacteriaceae in Tomato Fruits
An independent set of tomato fruit samples were collected from a different harvest season, six months after the initial experiment. DNA from tomato fruits was extracted as described above; this DNA was subjected to quantification of Enterobacteriaceae by means of quantitative PCR (qPCR) analysis using primers PS1-forward (5′-GGGGATAACYACTGGAAACGGTRGC-3′) and PS1-reverse (5′-GCATGGCTGCATCA GGSTTKC-3′); these primers amplified a ~236 bp segment of the 16S rRNA gene, previously validated [54,55]. Each qPCR reaction included 5 μL of Takara SYBR® Premix Ex Taq™ (Takara Bio Inc., Shiga, Japan), 0.68 μL of each primer (1.0 μM), 0.85 μL of bovine serum albumin (Bioline, London, UK), 10.0 ng of sample DNA, and nuclease-free water for a total volume of 17 μL. The amplification program consisted of 1 min at 95 °C followed by 35 cycles of denaturing at 95 °C for 30 s, annealing at 60 °C for 30 s, an extension at 72 °C for 30 s., and a final extension for 2 min at 72 °C. A qPCR standard curve was generated using DNA extracted from a E. coli reference strain (ATCC 11229).
2.6. Statistical Analyses
Results were also analyzed by means of an unpaired t-test and ANOVA Fisher’s protected least significant difference test using StatView version 5.0.1. Differences were considered significant at p ≤ 0.05. Alpha and beta diversity indices as well as PCA analyses were performed with the PAST software (v. 4.09) [56].
3. Results
3.1. Tomato Core Microbiota
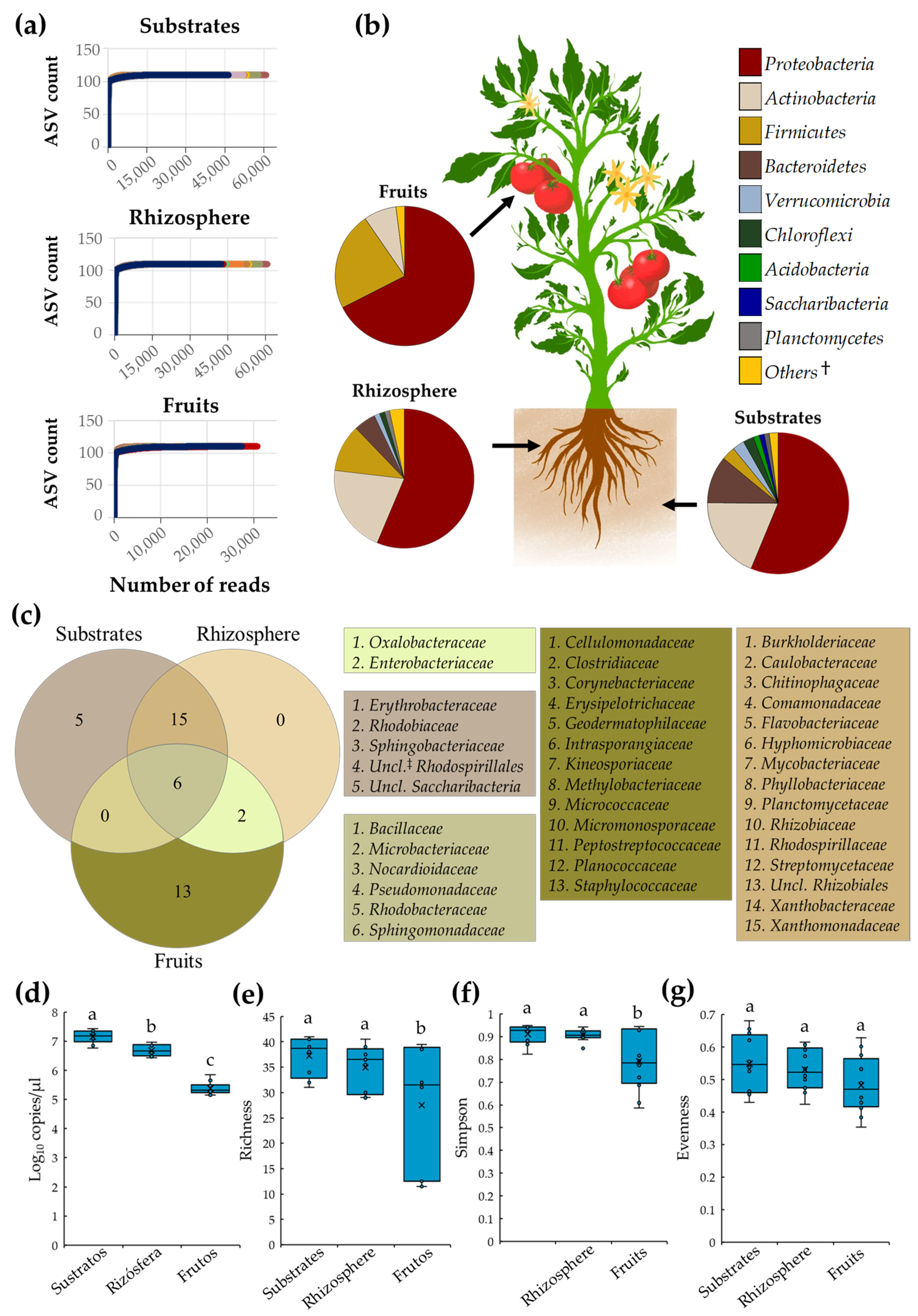
In the present study, tomato plant microbiota grown with conventional and organic SCSs was characterized after three independent samplings, during a whole production cycle. A total of 1,984,561 high-quality reads were derived from the 72 composite samples collected; from these, 613,355, 635,779, and 735,427 reads were obtained from substrate, rhizosphere, and fruit samples, respectively. Rarefaction analyses revealed that sequencing reached a plateau with the number of reads obtained from each niche, suggesting a high coverage of the tomato microbiota (Figure 1a). Microbiota of substrate, rhizosphere, and fruit samples was composed by nine, seven, and three major bacterial phyla (listed in Figure 1b), respectively, from which Proteobacteria was the most abundant microbial member in substrate (56.2%), rhizosphere (56.3%), and tomato fruit samples (67.5%) (Figure 1b).
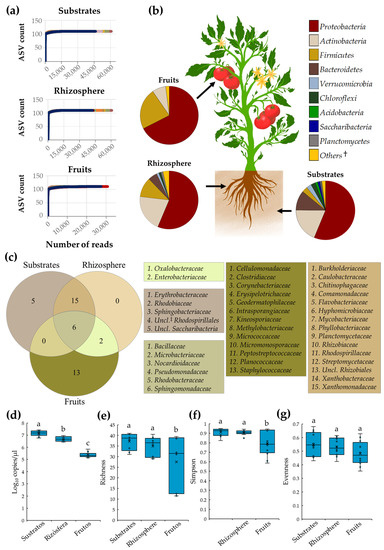
Figure 1.
Diversity and core microbiota of tomato plants cultivated under SCSs. (a) Rarefaction curves for observed ASVs; (b) diversity of core microbiota at the phylum level; (c) Venn diagram depicting bacterial families shared between substrate, root, and fruit samples; boxplots of (d) total bacteria, (e) richness, (f) Simpson, and (g) evenness indices in substrates, rhizosphere, and fruit samples. Boxes indicate the interquartile range of the data; the solid line and the cross inside the box depict the median and average; respectively. Whiskers represent minimum and maximum values; circles within or outside boxes show data distribution. Different letters indicate significant differences (p ≤ 0.05) between samples (n = 24). †: Bacterial groups with relative abundance < 1%. ‡: Unclassified.
Analysis of core microbiota revealed that substrate, rhizosphere, and tomato fruit niches were inhabited by 26, 23, and 21 bacterial families, respectively. Interestingly, six families, Bacillaceae, Microbacteriaceae, Nocardioidaceae, Pseudomonadaceae, Rhodobacteraceae, and Sphingomonadaceae, were shared among all selected niches. As expected, substrates and roots shared the largest number of families (15 taxa), suggesting a close biological interaction between these two niches. In contrast, roots and fruits only shared two families, Enterobacteriaceae and Oxalobacteraceae. Additionally, it was revealed that substrates and fruits were colonized by five and thirteen bacterial families, respectively. Notably, it was found that rhizosphere possesses no unique bacterial families; all members of root-associated microbiota were shared with substrates and fruits (Figure 1c).
Analyses of tomato plant bacterial diversity revealed that substrates had the highest (p ≤ 0.05) relative abundance of total bacteria (average 7.2 log10), measured by 16S rRNA gene copies, followed by rhizosphere (average 6.7 log10), and fruit (average 5.4 log10) samples (Figure 1d). Interestingly, the bacterial diversity between substrate and root samples was comparable (p > 0.05) as indicated by richness, Simpson, and evenness indices, corroborating a close biological interaction between these two niches. In contrast, bacterial diversity indices in tomato fruit samples were lower (p ≤ 0.05) compared to substrate and root samples, suggesting that this niche was colonized by a lessened and uneven number of bacterial families (Figure 1d–g).
3.2. Differences in Tomato Microbiota between Conventional and Organic Soilless Culture Systems
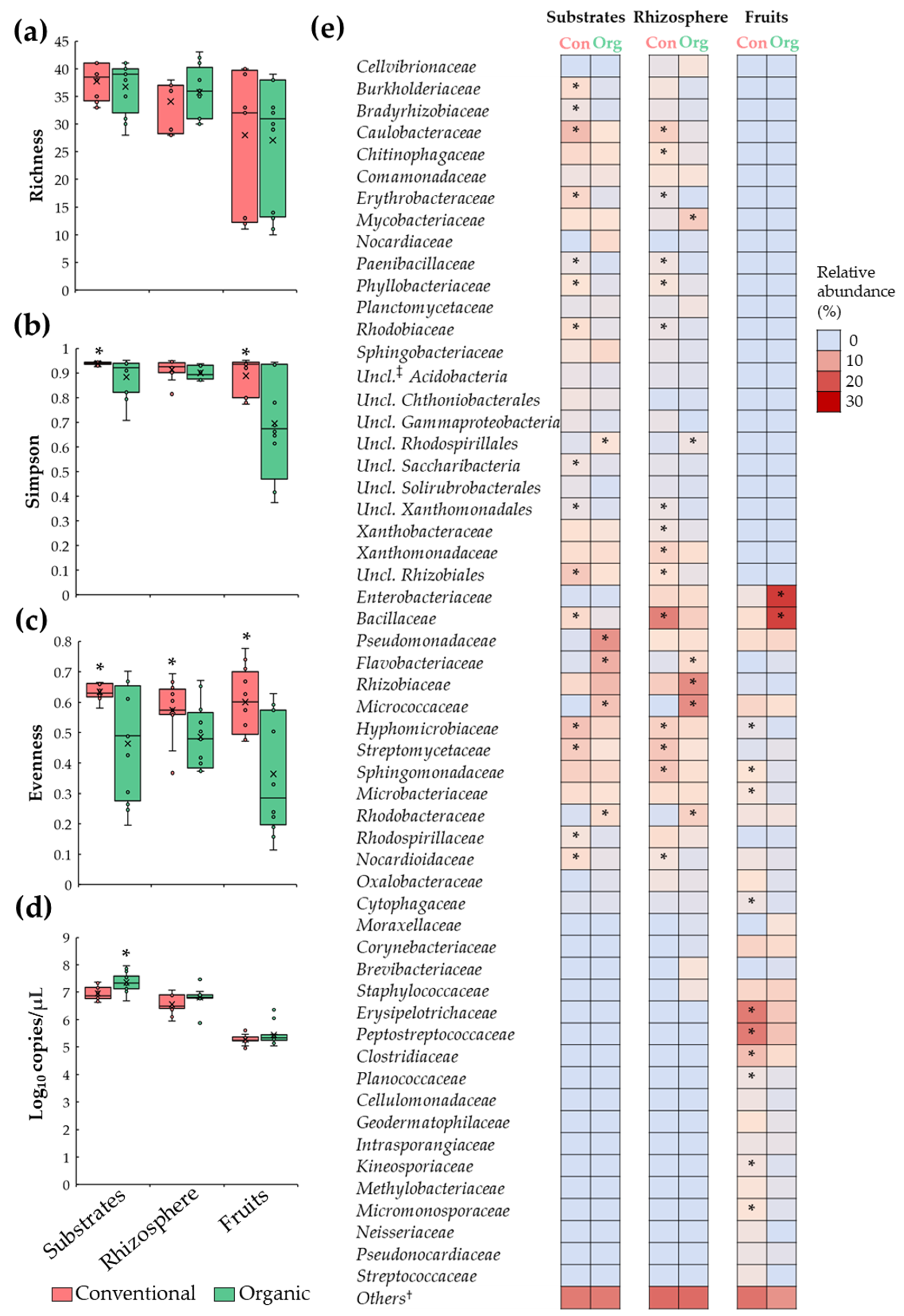
Analyses of microbiota diversity revealed a comparable (p > 0.05) number of bacterial families (richness) in substrate, rhizosphere, and fruit samples between conventional and organic SCS; also, it was observed that the family richness index was more variable among fruit samples (Figure 2a). Moreover, it was revealed that substrates and fruit samples under organic SCSs had a lower Simpson diversity index compared to their conventional counterparts (Figure 2b), suggesting that bacterial populations, in organic samples, are unevenly distributed. This observation was corroborated via evenness index analysis, showing a reduction (p ≤ 0.05) in this parameter in organic samples when compared to conventional samples (Figure 2c). Importantly, analysis of the relative abundance revealed minor (<1 log10) or no differences (p > 0.05) in total bacterial density, measured by 16S rRNA gene copies, between organic and conventional samples (Figure 2d). Together, these data indicate that substrates, rhizosphere, and fruit samples from organic and conventional SCSs are colonized by a comparable number of microbial taxa; however, in organic samples, a few bacterial groups are predominantly abundant.
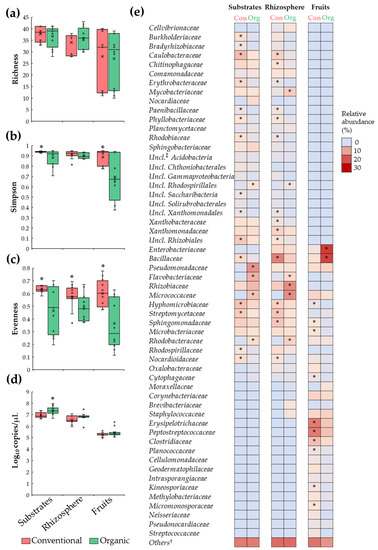
Figure 2.
Comparisons of tomato plant microbiota between conventional (Con) and organic (Org) fertilization regimes in SCSs. Boxplots depicting (a) richness, (b) Simpson, (c) and evenness indices as well as (d) total bacteria. Features of the boxplots are described in Figure 1. (e) Heatmap portraying relative abundance of bacterial families in substrate, rhizosphere, and tomato fruit samples. Asterisks indicate significant differences (p ≤ 0.05) between samples (n = 12). †: Bacterial groups with relative abundance < 1%. ‡: Unclassified.
To corroborate the difference in the relative abundance of bacterial families between conventional and organic systems, inferential and multivariate statistics were performed. These analyses revealed that substrates from the organic system were enriched (p ≤ 0.05) by members of Flavobacteriaceae, Micrococcaceae, Pseudomonadaceae, Rhodobacteraceae, and unclassified Rhodospirillales families; and reduced (p ≤ 0.05) in members of Bacillaceae, Burkholderiaceae, Bradyrhizobiaceae, Caulobacteraceae, Erythrobacteraceae, Hyphomicrobiaceae, Nocardioidaceae, Paenibacillaceae, Phyllobacteriaceae, Rhodobiaceae, Rhodospirillaceae, Streptomycetaceae, unclassified Rhizobiales, unclassified Saccharibacteria, and unclassified Xanthomonadales compared to conventional samples. Additionally, rhizosphere samples from the organic system were enriched (p ≤ 0.05) by members of Flavobacteriaceae, Micrococcaceae, Mycobacteriaceae, Rhizobiaceae, Rhodobacteraceae, and unclassified Rhodospirillales; and reduced in Bacillaceae, Caulobacteraceae, Chitinophagaceae, Erythrobacteraceae, Hyphomicrobiaceae, Nocardioidaceae, Paenibacillaceae, Phyllobacteriaceae, Rhodobiaceae, Sphingomonadaceae, Streptomycetaceae, unclassified Rhizobiales, unclassified Xanthomonadales, Xanthobacteraceae, and Xanthomonadaceae when compared to conventional samples. Moreover, it was shown that tomato fruit samples from organic SCS were enriched with (p ≤ 0.05) Bacillaceae and Enterobacteriaceae; and reduced in Clostridiaceae, Cytophagaceae, Erysipelotrichaceae, Hyphomicrobiaceae, Kineosporiaceae, Microbacteriaceae, Micromonosporaceae, Peptostreptococcaceae, Planococcaceae, and Sphingomonadaceae compared with conventional samples (Figure 2e). Together, these results suggest that the organic fertilization regime induced dysbiosis in substrate, rhizosphere, and fruits microbial populations.
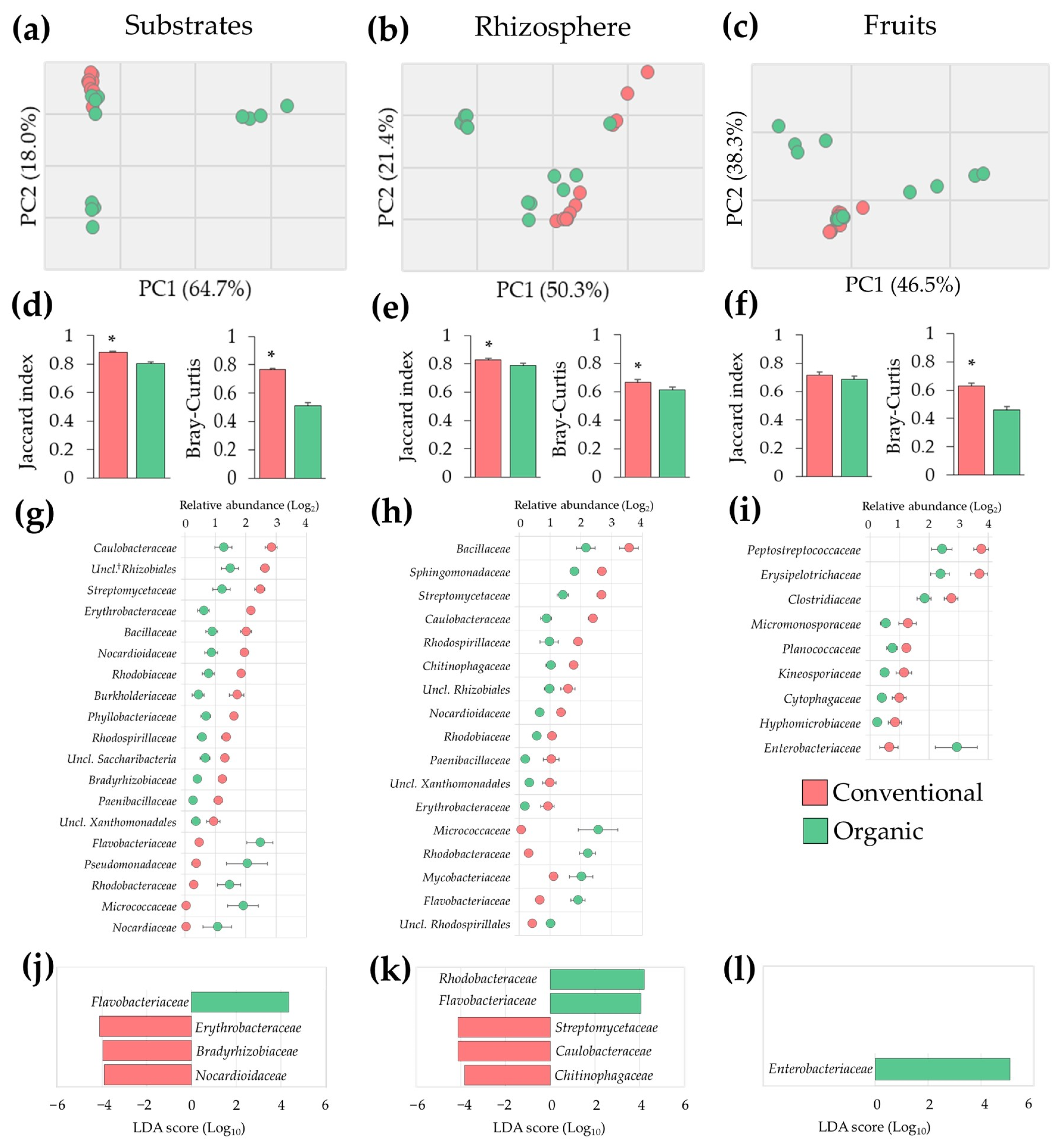
The unbalance in microbial populations observed in samples from organic SCS was corroborated via multivariate analysis. PCA of microbiota profiles revealed a higher variability in substrate, rhizosphere, and fruit samples from the organic system when compared to conventional samples (Figure 3a–c). These results were confirmed via analyses of beta diversity, where samples from the organic system had lower (p ≤ 0.05) Jaccard and Bray–Curtis similarity indices when compared to conventional samples, indicating a higher variability in prevalence and relative abundance of microbial populations (Figure 3d–f). Taken together these results suggest that plants cultivated under organic SCS exhibited microbial dysbiosis in roots and fruits.
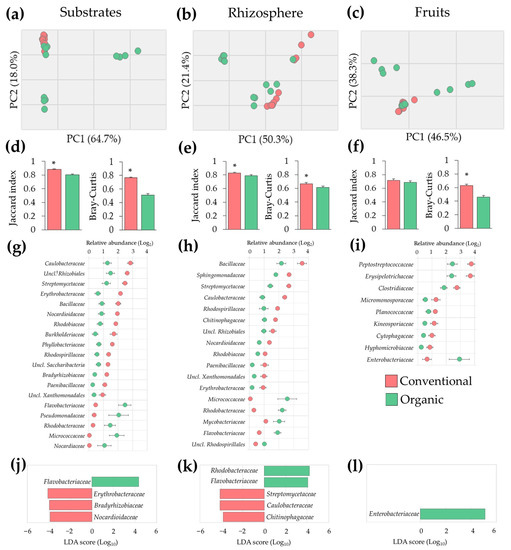
Figure 3.
Tomato plant microbiota alterations in organic compared to conventional fertilization regimes in SCSs. (a–c) Principal Component Analysis (PCA) of microbial populations; (d–f) beta diversity indices; (g–i) altered microbial populations (1.5-fold change and p ≤ 0.05); and (j–l) microbial markers identified via LDA-LEfSe analysis of substrate, rhizosphere, and fruit samples. Asterisks indicate significant differences (p ≤ 0.05) between samples (n = 12). †: Unclassified.
3.3. Microbial Dysbiosis Biomarkers
To gain insights into the nature of the dysbiosis observed in the organic SCS, bacterial densities between conventional and organic samples were compared. These analyses revealed, at least 1.5-fold change (FC) differences (p ≤ 0.05) in nineteen, seventeen, and nine families in substrate, rhizosphere, and fruit samples, respectively. Many of them exhibiting inversely differential abundance; in substrates, the most drastic changes were observed in Micrococcaceae (63.1 FC) and Nocardiaceae (37.1 FC), followed by Pseudomonadaceae (5.7 FC), Flavobacteriaceae (5.3 FC), and Rhodobacteraceae (5.3 FC). In the rhizosphere, major differences were observed in Micrococcaceae (48.6 FC), followed by Rhodobacteraceae (7.5 FC), Paenibacillaceae (5.3 FC), and Erythrobacteraceae (5.3 FC). In fruits, significant differences were observed in Enterobacteriaceae (4.6 FC) and Hyphomicrobiaceae (4 FC) (Figure 3g–i).
Because these microbial communities showed remarkably higher beta diversity variability, analyses of LDA-LEfSe were carried out to identify potential biomarkers explaining most of the microbial effects between conventional and organic SCSs. This approach revealed that four, five, and one families were differentially (p ≤ 0.05) abundant with a LDA score of 4.0 in substrate, rhizosphere, and fruit samples, respectively. Specifically, a significant (p ≤ 0.05) enrichment of Flavobacteriaceae and a marked (p ≤ 0.05) reduction of Erythrobacteraceae, Bradyrhizobiaceae, and Nocardioidaceae were observed in organic substrate samples. Additionally, an increased (p ≤ 0.05) abundance of Rhodobacteraceae and Flavobacteriaceae and a reduction (p ≤ 0.05) of Streptomycetaceae, Caulobacteraceae, and Chitinophagaceae were observed in organic rhizosphere samples. Moreover, a marked (p ≤ 0.05) enrichment of Enterobacteriaceae in organic tomato fruits was observed (Figure 3j–l). Taken together these results indicate that plants under the organic SCS undergo a dysbiosis process in roots and fruits. Importantly, some of these changes in bacterial densities could be used as potential biomarkers to evaluate the health and performance of tomato production systems.
3.4. Absolute Quantification of Enterobacteriaceae in Tomato Fruits
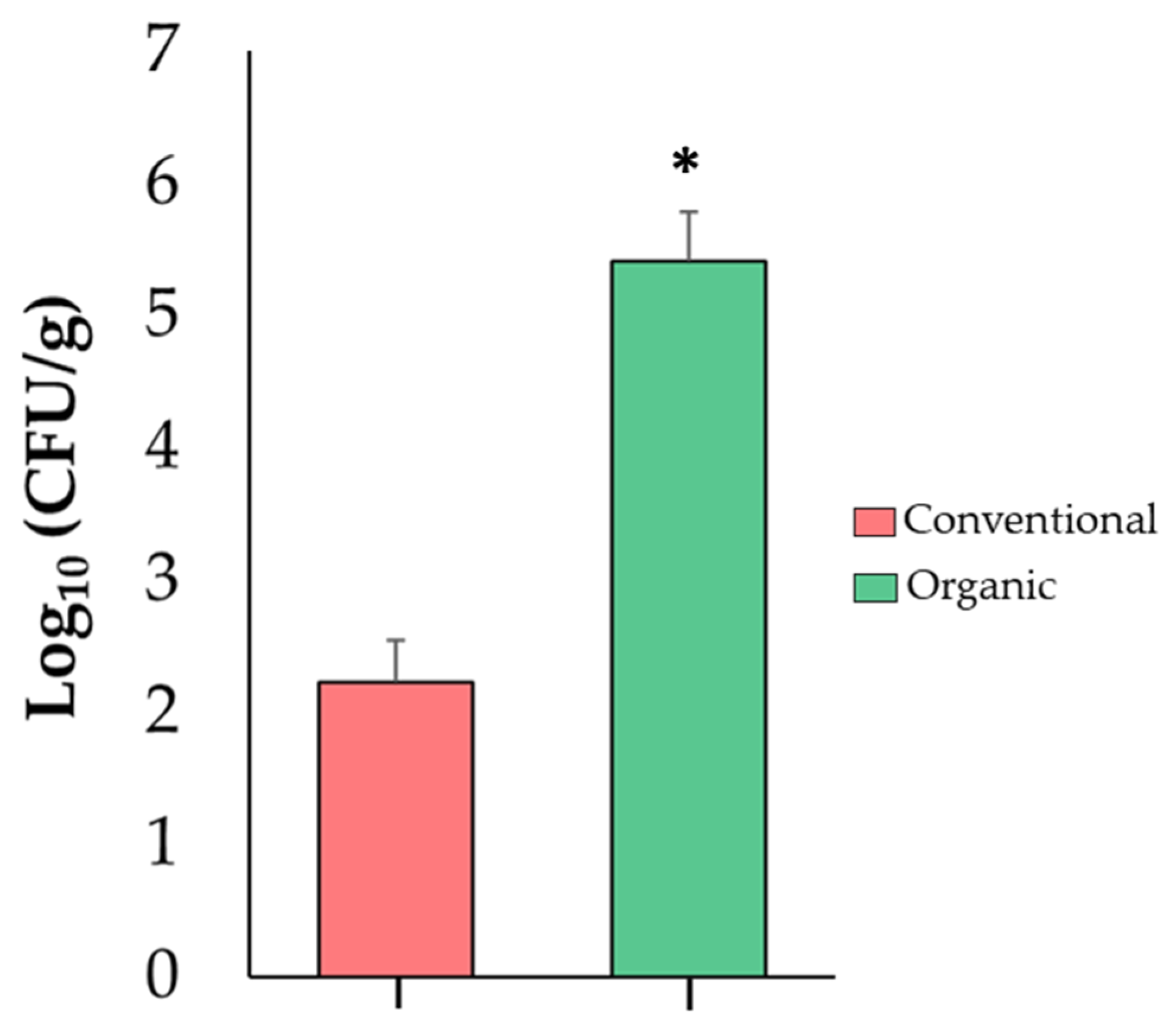
Because the increase in the relative abundance of Enterobacteriaceae in tomato fruits is highly important for food safety, this finding was confirmed via an independent analysis using a qPCR assay. This analysis revealed that organic tomatoes were colonized by higher numbers (p ≤ 0.05) (5.41 log10) of Enterobacteriaceae when compared to conventional fruits (2.23 log10) (Figure 4). These results suggest that an organic SCS favors proliferation of Enterobacteriaceae in tomato fruits and could have an impact on fruit quality and shelf life.
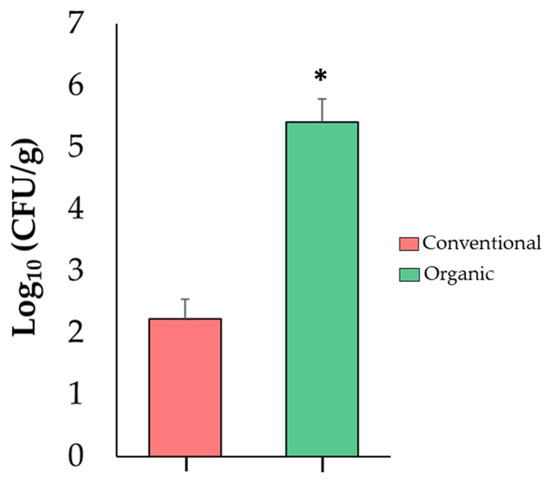
Figure 4.
Abundance of Enterobacteriaceae in tomato fruit samples from conventional and organic SCSs. Asterisk indicates significant differences (p ≤ 0.05) between samples (n = 4).
4. Discussion
Assembly of core microbiota in crop plants is driven by biotic and abiotic factors, including climate conditions, growth stage, and different fertilization regimes [57,58,59]. Numerous studies have shown that microbial populations in soil, the rhizosphere, and the phyllosphere impact crop health, productivity, and safety [60,61,62]; however, there is limited information regarding diversity of microbial populations in SCSs. This topic is of particular importance due to the advantages and opportunities provided by soilless agriculture [5]. Thus, herein we identified the core microbiota associated with tomato plants cultivated under SCSs and microbial differences between conventional and organic fertilization regimes.
4.1. Tomato Core Microbiota in Soilless Culture Systems
Our data revealed that tomato core microbiota was dominated by phyla Proteobacteria, Actinobacteria, and Firmicutes in the substrate, rhizosphere, and fruit samples; comparable bacterial diversity has been reported in numerous studies with tomato plants cultivated in soil-based and SCSs [20,27,63,64,65,66,67,68,69,70]. Together, these findings suggest that, regardless of the cultivation system, tomato plants have evolved a close biological interaction with members of Proteobacteria, Actinobacteria, and Firmicutes. Bacterial groups within Proteobacteria possess a high genomic plasticity, facilitating fast growth and stress adaptation; due to these two features, Proteobacteria successfully colonize plant niches [71]. Furthermore, it has been shown that Proteobacteria harbor genomic traits linked to multiple bacteria–host beneficial processes, such as nitrogen fixation and phosphate solubilization promoted by nitrogenase and pyrroloquinoline quinone-encoding gene expression, respectively [72]. On the other hand, members of Actinobacteria and Firmicutes possess genetic features allowing for production and secretion of bacterial metabolites such as antibiotics, phytohormones, and siderophores capable of promoting plant growth [73,74,75,76]. Collectively, these bacterial groups produce metabolites that induce plant growth and disease resistance [21,72,76,77]. Taken together, these results corroborate that Proteobacteria, Actinobacteria, and Firmicutes are main components of the tomato plant’s core microbiota.
Importantly, the present study revealed that the core microbiota, during the whole production cycle, were integrated by 26, 23, and 21 families (listed in Figure 1c) in the substrate, rhizosphere, and tomato fruits, respectively. The presence of these bacterial families in tomato plants has been observed in other studies [19,26,30,66]. Interestingly, six bacterial families, Bacillaceae, Microbacteriaceae, Nocardioidaceae, Pseudomonadaceae, Rhodobacteraceae, and Sphingomonadaceae, colonized the substrate, rhizosphere, and fruit samples, suggesting the presence of a tomato plant core microbiota. Members of this core microbiota perform important functions linked to plant growth promotion; for example, Bacillaceae and Pseudomonadaceae produce metabolites such as polyketides and pyoverdines, which reduce pathogen colonization and enhance mineral absorption, respectively [75]. Members of Microbacteriaceae produce 1-aminocyclopropane-1-carboxylate (ACC) deaminase, an enzyme that reduces ethylene levels, a plant stress hormone [78]. Sphingomonadaceae synthetize dehydrochlorinases, dehydrogenases, and halidohydrolases, enzymes that metabolize synthetic pesticides such as organochlorines [79,80]. Members of Nocardioidaceae and Rhodobacteraceae secrete phytohormones such as gibberellins and indole-3-acetic acid (IAA) that increase root length and improve nutrient and water absorption [81,82,83]. Taken together, these results suggest that tomato plant core microbiota could influence plant growth, disease resistance, and productivity. Nonetheless, additional studies should be performed to corroborate this notion.
Interestingly, the present work and many other independent studies have identified Burkholderiaceae, Caulobacteraceae, Chitinophagaceae, Flavobacteriaceae, Hyphomicrobiaceae, Rhizobiaceae, Rhodospirillaceae, Streptomycetaceae, and Xanthomonadaceae as constitutive of the core microbiota in soil, substrate, and rhizosphere samples from tomato plants [18,63,84,85,86,87,88]. These results suggest that root tissues have evolved molecular mechanisms to recruit core microbiota populations [89]. Moreover, the co-occurrence of Enterobacteriaceae and Oxalobacteraceae in root and fruit tissues has been reported by other studies [26,30], supporting the idea that rhizosphere microbiota could influence the establishment of bacterial populations in the phyllosphere [90].
Additionally, in the present study, it was revealed that bacterial relative abundance and diversity were higher in substrates, followed by rhizosphere and fruit samples; comparable trends have been reported elsewhere [18,64,91,92]. Together, these results support the long-standing hypothesis [93] that suggests that stress generated by multiple and diverse environmental, biotic, and abiotic factors reduces the diversity and relative abundance of phyllosphere microbiota, when compared with rhizosphere microbiota [93,94].
4.2. Bacterial Relative Abundance Is Influenced by Conventional and Organic Soilless Culture Systems
It has been reported that tomato plants cultivated under organic SCSs have lower fruit yields, higher susceptibility to phytopathogens, as well as lower fruit firmness and weight [9,14,16,95]. It has been proposed that these deficiencies could be linked to dysbiosis in the tomato plant microbiota [20]. Herein, it was shown that conventional and organic plants have comparable bacterial abundance and richness in substrate, rhizosphere, and fruit samples; however, it was revealed that organic plants endured microbiota dysbiosis characterized by an increase in the relative abundance and dominance of some specific bacterial groups. Comparable phenomena have been reported in other studies with tomato, lettuce, and teak plants [67,96,97].
Specifically, the present study revealed that substrate, rhizosphere, and fruit samples under an organic SCS have altered relative abundance of 20, 21, and 12 families (listed in Figure 2e), respectively, with all of them members of Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria phyla. Importantly, the dysbiosis characterized by a remarkable change in the relative abundance of Enterobacteriaceae, Flavobacteriaceae, Hyphomicrobiaceae, Pseudomonadaceae Rhizobiaceae, Rhodobacteraceae, and Xanthomonadaceae in substrate, rhizosphere, and fruit samples from the organic production system has been documented by many other studies [19,69,96,98,99,100], suggesting that these bacterial families could represent a microbial target that could be used to improve tomato plant health, productivity, and quality.
To gain insights into the potential impact of these microbial populations on plant health and productivity, microbial biomarkers (LEfSe analysis) were identified. Particularly, it was revealed that most of the effects in the organic production system could be linked to Flavobacteriaceae, Erythrobacteraceae, Bradyrhizobiaceae, and Nocardioidaceae in substrates; Caulobacteraceae, Chitinophagaceae, Flavobacteriaceae, Rhodobacteraceae, Streptomycetaceae, in the rhizosphere; and Enterobacteriaceae in tomato fruit samples. These potential microbial biomarkers have been highlighted by other studies with tomato and lettuce cultivars [19,67,69,96,99].
Although additional and extensive studies are required to elucidate the effects of these bacterial biomarkers, numerous studies support the importance of these bacterial families on plant health and productivity. For instance, Flavobacteriaceae promotes phosphorus solubilization and pectin degradation; in environments under nutrient limitations, these bacterial groups could improve nutrient uptake and assimilation of organic compounds by plants [101,102]. Bradyrhizobiaceae, Caulobacteraceae, Chitinophagaceae, Erythrobacteraceae, Nocardioidaceae, and Streptomycetaceae promote biological nitrogen fixation by producing enzymes responsible for nitrogen assimilation [103,104,105,106,107,108] and increasing shoot and root biomass growth [109,110,111]. Thus, it could be possible that the reduction in the relative abundance of these bacterial biomarkers could be linked to the poor agricultural performance observed in organic SCSs [112].
Importantly, the present work revealed that Enterobacteriaceae is the most predominant member of tomato fruit microbiota; this finding has been documented in many other studies [26,29,66,113,114,115]. These results are of particular interest because genera within the Enterobacteriaceae family are important pathogens for tomato plants [116] and humans [117] as well as key members of the fruit spoilage microbiota [118,119]. The potential negative effects of Enterobacteriaceae in the organic production system could be related to the ability of this bacterial group to produce adhesins, phytotoxins, and proteases associated with plant pathogenesis [120,121]. Additionally, members of Enterobacteriaceae produce extracellular enzymes such as pectate lyases, polygalaturonases, pectin methylesterases, and pectin acetylesterases involved in fruit cell wall degradation [119]; an increased production of these enzymes has been linked to reductions in fruit firmness and shelf life [116,122].
Moreover, the high abundance of Enterobacteriaceae in tomato fruits is a major food safety problem. First, numerous studies have reported a high prevalence of potential enteropathogens such as Enterobacter spp. (range = 17–28%, [123,124,125]), Escherichia coli (range = 3–18%, [125,126,127]), Klebsiella spp. (range = 2–39%, [123,125]), and Salmonella enterica (range = 8–44%, [128,129]), all of them members of Enterobacteriaceae, in tomato fruits intended for human consumption. Second, in many countries, consumption of tainted tomatoes has been linked to E. coli [130], S. enterica [131,132,133,134,135,136], and Shigella flexneri [137] human outbreaks. For many years, tomato fruits have been described as Enterobacteriaceae pathogen carriers; however, in the last decade, numerous studies have shown that tomato fruits are an alternative host for colonization, replication, and propagation of Enterobacteriaceae [138,139,140]. Importantly, the present study revealed that a major effect of the organic SCS was a twenty-fold increment in the relative abundance of Enterobacteriaceae in tomato fruits. This finding was corroborated via an independent microbial molecular analysis with tomato fruits collected from a different harvest season. Together, these results suggest that the dysbiosis caused by the organic fertilization regime could potentially produce tomato fruits that are more susceptible to pathogen colonization; further studies are in progress to corroborate this idea. Furthermore, we hypothesize that the dysbiosis observed in the organic SCS could be attributed to the organic source of nitrogen, phosphorus, and potassium used in the present study. Numerous studies have shown that the use of organic fertilizers increases the relative abundance of different bacterial groups in soil and soilless cultivation systems [19,100,141,142,143,144]. Additional studies are in progress to evaluate this hypothesis.
5. Conclusions
In the present study, it was revealed that tomato plant microbiota was predominantly colonized by members of the Proteobacteria, Actinobacteria, and Firmicutes phyla families. Additionally, it was found that Bacillaceae, Microbacteriaceae, Nocardioidaceae, Pseudomonadaceae, Rhodobacteraceae, and Sphingomonadaceae colonized substrate, rhizosphere, and phyllosphere samples and are members of the tomato plant core microbiota.
Importantly, it was revealed that tomato plants cultivated under organic SCS endure a dysbiosis in substrates, roots, and fruits, characterized by an increased relative abundance of Flavobacteriaceae, Rhodobacteraceae, and Enterobacteriaceae; and a reduction of Bradyrhizobiaceae, Caulobacteraceae, Chitinophagaceae, Erythrobacteraceae, Nocardioidaceae, and Streptomycetaceae. Altogether, these results suggest that the dysbiosis observed in organic tomato plants could be responsible for the agricultural deficiencies reported for this production system. Importantly, the present study highlights a list of bacterial groups with biotechnological potential to promote plant health and production.
Author Contributions
C.N.R.-N., F.A.-O., H.V.S.-R., A.R.-A., D.M.R.-P., M.J.S., G.M.N. and E.M.M.-S., conceptualized the study; C.N.R.-N. and F.A.-O., performed the experiments; C.N.R.-N., F.A.-O., H.V.S.-R., A.R.-A., D.M.R.-P., M.J.S., G.M.N. and E.M.M.-S., analyzed the data and interpreted results; C.N.R.-N., F.A.-O., H.V.S.-R., A.R.-A., D.M.R.-P., M.J.S., G.M.N. and E.M.M.-S., wrote, edited, and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
C.N.R.-N. was supported by a CONACYT-Mexico graduate scholarship (692848) during the development of this work.
Data Availability Statement
The authors declare that all relevant data supporting the findings of this study are included in this article.
Acknowledgments
The authors thank tomato producers that provided samples for the present study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rodríguez-Ortega, W.M.; Martínez, V.; Nieves, M.; Simón, I.; Lidón, V.; Fernandez-Zapata, J.C.; Martinez-Nicolas, J.J.; Cámara-Zapata, J.M.; García-Sánchez, F. Agricultural and Physiological Responses of Tomato Plants Grown in Different Soilless Culture Systems with Saline Water under Greenhouse Conditions. Sci. Rep. 2019, 9, 6733. [Google Scholar] [CrossRef] [PubMed]
- Araujo, J.C.; Telhado, S.F.P. Organic Food: A Comparative Study of the Effect of Tomato Cultivars and Cultivation Conditions on the Physico-Chemical Properties. Foods 2015, 4, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Floare-Avram, C.V.; Covaciu, F.; Voica, C.; Puscas, R.; Feher, I.; Marincas, O.; Magdas, D.A. Differentiation of Tomatoes Based on Isotopic, Elemental and Organic Markers. J. Food Sci. Technol. 2020, 57, 2222–2232. [Google Scholar] [CrossRef]
- Putra, P.A.; Yuliando, H. Soilless Culture System to Support Water Use Efficiency and Product Quality: A Review. Agric. Agric. Sci. Procedia 2015, 3, 283–288. [Google Scholar] [CrossRef]
- Gruda, N.S. Increasing Sustainability of Growing Media Constituents and Stand-Alone Substrates in Soilless Culture Systems. Agronomy 2019, 9, 298. [Google Scholar] [CrossRef]
- Gonnella, M.; Renna, M. The Evolution of Soilless Systems towards Ecological Sustainability in the Perspective of a Circular Economy. Is It Really the Opposite of Organic Agriculture? Agronomy 2021, 11, 950. [Google Scholar] [CrossRef]
- Maluin, F.N.; Hussein, M.Z.; Nik Ibrahim, N.N.L.; Wayayok, A.; Hashim, N. Some Emerging Opportunities of Nanotechnology Development for Soilless and Microgreen Farming. Agronomy 2021, 11, 1213. [Google Scholar] [CrossRef]
- Vinha, A.F.; Barreira, S.V.P.; Costa, A.S.G.; Alves, R.C.; Oliveira, M.B.P.P. Organic versus Conventional Tomatoes: Influence on Physicochemical Parameters, Bioactive Compounds and Sensorial Attributes. Food Chem. Toxicol. 2014, 67, 139–144. [Google Scholar] [CrossRef]
- Azarbad, H. Conventional vs. Organic Agriculture—Which One Promotes Better Yields and Microbial Resilience in Rapidly Changing Climates? Front. Microbiol. 2022, 13, 903500. [Google Scholar] [CrossRef]
- Vigar, V.; Myers, S.; Oliver, C.; Arellano, J.; Robinson, S.; Leifert, C. A Systematic Review of Organic Versus Conventional Food Consumption: Is There a Measurable Benefit on Human Health? Nutrients 2019, 12, 7. [Google Scholar] [CrossRef]
- Oliveira, A.B.; Moura, C.F.H.; Gomes-Filho, E.; Marco, C.A.; Urban, L.; Miranda, M.R.A. The Impact of Organic Farming on Quality of Tomatoes Is Associated to Increased Oxidative Stress during Fruit Development. PLoS ONE 2013, 8, e56354. [Google Scholar] [CrossRef]
- Ronga, D.; Caradonia, F.; Vitti, A.; Francia, E. Agronomic Comparisons of Heirloom and Modern Processing Tomato Genotypes Cultivated in Organic and Conventional Farming Systems. Agronomy 2021, 11, 349. [Google Scholar] [CrossRef]
- Ronga, D.; Lovelli, S.; Zaccardelli, M.; Perrone, D.; Ulrici, A.; Francia, E.; Milc, J.; Pecchioni, N. Physiological Responses of Processing Tomato in Organic and Conventional Mediterranean Cropping Systems. Sci. Hortic. 2015, 190, 161–172. [Google Scholar] [CrossRef]
- Riahi, A.; Hdider, C.; Sanaa, M.; Tarchoun, N.; Kheder, M.B.; Guezal, I. Effect of Conventional and Organic Production Systems on the Yield and Quality of Field Tomato Cultivars Grown in Tunisia. J. Sci. Food Agric. 2009, 89, 2275–2282. [Google Scholar] [CrossRef]
- Mubarok, S.; Farhah, F.F.; Anas; Suwali, N.; Kurnia, D.; Kusumiyati; Suminar, E.; Ezura, H. Data on the Yield and Quality of Organically Hybrids of Tropical Tomato Fruits at Two Stages of Fruit Maturation. Data Brief 2019, 25, 104031. [Google Scholar] [CrossRef] [PubMed]
- Adekiya, A.O.; Dahunsi, S.O.; Ayeni, J.F.; Aremu, C.; Aboyeji, C.M.; Okunlola, F.; Oyelami, A.E. Organic and In-Organic Fertilizers Effects on the Performance of Tomato (Solanum lycopersicum) and Cucumber (Cucumis sativus) Grown on Soilless Medium. Sci. Rep. 2022, 12, 12212. [Google Scholar] [CrossRef] [PubMed]
- Bettiol, W.; Ghini, R.; Galvão, J.A.H.; Siloto, R.C. Organic and Conventional Tomato Cropping Systems. Sci. Agric. 2004, 61, 253–259. [Google Scholar] [CrossRef]
- Grunert, O.; Robles-Aguilar, A.A.; Hernandez-Sanabria, E.; Schrey, S.D.; Reheul, D.; Van Labeke, M.-C.; Vlaeminck, S.E.; Vandekerckhove, T.G.L.; Mysara, M.; Monsieurs, P.; et al. Tomato Plants Rather than Fertilizers Drive Microbial Community Structure in Horticultural Growing Media. Sci. Rep. 2019, 9, 9561. [Google Scholar] [CrossRef] [PubMed]
- Grunert, O.; Hernandez-Sanabria, E.; Buysens, S.; De Neve, S.; Van Labeke, M.-C.; Reheul, D.; Boon, N. In-Depth Observation on the Microbial and Fungal Community Structure of Four Contrasting Tomato Cultivation Systems in Soil Based and Soilless Culture Systems. Front. Plant Sci. 2020, 11, 520834. [Google Scholar] [CrossRef]
- Readyhough, T.; Neher, D.A.; Andrews, T. Organic Amendments Alter Soil Hydrology and Belowground Microbiome of Tomato (Solanum lycopersicum). Microorganisms 2021, 9, 1561. [Google Scholar] [CrossRef]
- Lee, S.-M.; Kong, H.G.; Song, G.C.; Ryu, C.-M. Disruption of Firmicutes and Actinobacteria Abundance in Tomato Rhizosphere Causes the Incidence of Bacterial Wilt Disease. ISME J. 2021, 15, 330–347. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Elkabetz, D.; Leibman-Markus, M.; Sayas, T.; Schneider, A.; Jami, E.; Kleiman, M.; Bar, M. Cytokinin Drives Assembly of the Phyllosphere Microbiome and Promotes Disease Resistance through Structural and Chemical Cues. ISME J. 2022, 16, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Runge, P.; Ventura, F.; Kemen, E.; Stam, R. Distinct Phyllosphere Microbiome of Wild Tomato Species in Central Peru upon Dysbiosis. Microb. Ecol. 2023, 85, 168–183. [Google Scholar] [CrossRef] [PubMed]
- Mehlferber, E.C.; McCue, K.F.; Ferrel, J.E.; Koskella, B.; Khanna, R. Temporally Selective Modification of the Tomato Rhizosphere and Root Microbiome by Volcanic Ash Fertilizer Containing Micronutrients. Appl. Environ. Microbiol. 2022, 88, e00049-22. [Google Scholar] [CrossRef] [PubMed]
- USDA AMS Guidelines for Organic Crop Certification. Available online: https://www.ams.usda.gov/sites/default/files/media/Crop%20-%20Guidelines.pdf (accessed on 1 April 2023).
- Allard, S.M.; Walsh, C.S.; Wallis, A.E.; Ottesen, A.R.; Brown, E.W.; Micallef, S.A. Solanum lycopersicum (Tomato) Hosts Robust Phyllosphere and Rhizosphere Bacterial Communities When Grown in Soil Amended with Various Organic and Synthetic Fertilizers. Sci. Total Environ. 2016, 573, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Vargas, P.; Bosmans, L.; Van Calenberge, B.; Van Kerckhove, S.; Lievens, B.; Rediers, H. Bacterial Community Dynamics of Tomato Hydroponic Greenhouses Infested with Hairy Root Disease. FEMS Microbiol. Ecol. 2021, 97, fiab153. [Google Scholar] [CrossRef]
- Shinozaki, Y.; Nicolas, P.; Fernandez-Pozo, N.; Ma, Q.; Evanich, D.J.; Shi, Y.; Xu, Y.; Zheng, Y.; Snyder, S.I.; Martin, L.B.B.; et al. High-Resolution Spatiotemporal Transcriptome Mapping of Tomato Fruit Development and Ripening. Nat. Commun. 2018, 9, 364. [Google Scholar] [CrossRef]
- Allard, S.M.; Ottesen, A.R.; Micallef, S.A. Rain Induces Temporary Shifts in Epiphytic Bacterial Communities of Cucumber and Tomato Fruit. Sci. Rep. 2020, 10, 1765. [Google Scholar] [CrossRef]
- Ottesen, A.R.; González Peña, A.; White, J.R.; Pettengill, J.B.; Li, C.; Allard, S.; Rideout, S.; Allard, M.; Hill, T.; Evans, P.; et al. Baseline Survey of the Anatomical Microbial Ecology of an Important Food Plant: Solanum lycopersicum (Tomato). BMC Microbiol. 2013, 13, 114. [Google Scholar] [CrossRef]
- Piraine, R.E.A.; Leite, F.P.L.; Bochman, M.L. Mixed-Culture Metagenomics of the Microbes Making Sour Beer. Fermentation 2021, 7, 174. [Google Scholar] [CrossRef]
- Can-Herrera, L.A.; Gutierrez-Canul, C.D.; Dzul-Cervantes, M.A.A.; Pacheco-Salazar, O.F.; Chi-Cortez, J.D.; Carbonell, L.S. Identification by Molecular Techniques of Halophilic Bacteria Producing Important Enzymes from Pristine Area in Campeche, Mexico. Braz. J. Biol. 2021, 83, e246038. [Google Scholar] [CrossRef] [PubMed]
- Puón-Peláez, X.-H.D.; McEwan, N.R.; Gómez-Soto, J.G.; Álvarez-Martínez, R.C.; Olvera-Ramírez, A.M. Metataxonomic and Histopathological Study of Rabbit Epizootic Enteropathy in Mexico. Animals 2020, 10, 936. [Google Scholar] [CrossRef] [PubMed]
- Çelik, Z.C.; Çakiris, A.; Yanıkoğlu, F.; Abacı, N.; Ekmekçi, S.S.; Ilgın, C.; Çelik, H.; Tağtekin, D. Metagenomic Analysis of Black-Stained Plaques in Permanent Dentition. Arch. Oral Biol. 2021, 128, 105171. [Google Scholar] [CrossRef]
- Gómez-Govea, M.A.; Ramírez-Ahuja, M.d.L.; Contreras-Perera, Y.; Jiménez-Camacho, A.J.; Ruiz-Ayma, G.; Villanueva-Segura, O.K.; Trujillo-Rodríguez, G.d.J.; Delgado-Enciso, I.; Martínez-Fierro, M.L.; Manrique-Saide, P.; et al. Suppression of Midgut Microbiota Impact Pyrethroid Susceptibility in Aedes aegypti. Front. Microbiol. 2022, 13, 2025. [Google Scholar] [CrossRef]
- Ravi, R.K.; Walton, K.; Khosroheidari, M. MiSeq: A Next Generation Sequencing Platform for Genomic Analysis. Methods Mol. Biol. 2018, 1706, 223–232. [Google Scholar] [CrossRef]
- Del Fabbro, C.; Scalabrin, S.; Morgante, M.; Giorgi, F.M. An Extensive Evaluation of Read Trimming Effects on Illumina NGS Data Analysis. PLoS ONE 2013, 8, e85024. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Neu, A.T.; Allen, E.E.; Roy, K. Defining and Quantifying the Core Microbiome: Challenges and Prospects. Proc. Natl. Acad. Sci. USA 2021, 118, e2104429118. [Google Scholar] [CrossRef]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a Prokaryotic Universal Primer for Simultaneous Analysis of Bacteria and Archaea Using Next-Generation Sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef]
- Abellan-Schneyder, I.; Matchado, M.S.; Reitmeier, S.; Sommer, A.; Sewald, Z.; Baumbach, J.; List, M.; Neuhaus, K. Primer, Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing. mSphere 2021, 6, e01202-20. [Google Scholar] [CrossRef] [PubMed]
- Nathani, N.M.; Patel, A.K.; Dhamannapatil, P.S.; Kothari, R.K.; Singh, K.M.; Joshi, C.G. Comparative Evaluation of Rumen Metagenome Community Using qPCR and MG-RAST. AMB Express 2013, 3, 55. [Google Scholar] [CrossRef] [PubMed]
- Větrovský, T.; Baldrian, P. The Variability of the 16S rRNA Gene in Bacterial Genomes and Its Consequences for Bacterial Community Analyses. PLoS ONE 2013, 8, e57923. [Google Scholar] [CrossRef] [PubMed]
- Kleine-Bardenhorst, S.; Vital, M.; Karch, A.; Rübsamen, N. Richness Estimation in Microbiome Data Obtained from Denoising Pipelines. Comput. Struct. Biotechnol. J. 2022, 20, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Li, L.; Gotelli, N.J. Diversity-Disease Relationships and Shared Species Analyses for Human Microbiome-Associated Diseases. ISME J. 2019, 13, 1911–1919. [Google Scholar] [CrossRef]
- Morris, E.K.; Caruso, T.; Buscot, F.; Fischer, M.; Hancock, C.; Maier, T.S.; Meiners, T.; Müller, C.; Obermaier, E.; Prati, D.; et al. Choosing and Using Diversity Indices: Insights for Ecological Applications from the German Biodiversity Exploratories. Ecol. Evol. 2014, 4, 3514–3524. [Google Scholar] [CrossRef]
- Jaccard, P. The Distribution of the Flora in the Alpine Zone. New Phytol. 1912, 11, 37–50. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 326–349. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Vismans, G.; Yu, K.; Song, Y.; de Jonge, R.; Burgman, W.P.; Burmølle, M.; Herschend, J.; Bakker, P.A.H.M.; Pieterse, C.M.J. Disease-Induced Assemblage of a Plant-Beneficial Bacterial Consortium. ISME J. 2018, 12, 1496–1507. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Liu, F.; Hewezi, T.; Lebeis, S.L.; Pantalone, V.; Grewal, P.S.; Staton, M.E. Soil Indigenous Microbiome and Plant Genotypes Cooperatively Modify Soybean Rhizosphere Microbiome Assembly. BMC Microbiol. 2019, 19, 201. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Chen, J.; Xiao, Z.; Zhu, X.; Wang, J.; Wu, H.; Wu, Y.; Zhang, Z.; Lin, W. Barcoded Pyrosequencing Reveals a Shift in the Bacterial Community in the Rhizosphere and Rhizoplane of Rehmannia Glutinosa under Consecutive Monoculture. Int. J. Mol. Sci. 2018, 19, 850. [Google Scholar] [CrossRef] [PubMed]
- Resendiz-Nava, C.N.; Silva-Rojas, H.V.; Rebollar-Alviter, A.; Rivera-Pastrana, D.M.; Mercado-Silva, E.M.; Nava, G.M. A Comprehensive Evaluation of Enterobacteriaceae Primer Sets for Analysis of Host-Associated Microbiota. Pathogens 2022, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Kurina, I.; Popenko, A.; Klimenko, N.; Koshechkin, S.; Chuprikova, L.; Filipenko, M.; Tyakht, A.; Alexeev, D. Development of qPCR Platform with Probes for Quantifying Prevalent and Biomedically Relevant Human Gut Microbial Taxa. Mol. Cell. Probes 2020, 52, 101570. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Jamil, F.; Mukhtar, H.; Fouillaud, M.; Dufossé, L. Rhizosphere Signaling: Insights into Plant–Rhizomicrobiome Interactions for Sustainable Agronomy. Microorganisms 2022, 10, 899. [Google Scholar] [CrossRef]
- Timm, C.M.; Carter, K.R.; Carrell, A.A.; Jun, S.-R.; Jawdy, S.S.; Vélez, J.M.; Gunter, L.E.; Yang, Z.; Nookaew, I.; Engle, N.L.; et al. Abiotic Stresses Shift Belowground Populus-Associated Bacteria Toward a Core Stress Microbiome. mSystems 2018, 3, e00070-17. [Google Scholar] [CrossRef]
- Qi, M.; Berry, J.C.; Veley, K.W.; O’Connor, L.; Finkel, O.M.; Salas-González, I.; Kuhs, M.; Jupe, J.; Holcomb, E.; Glavina del Rio, T.; et al. Identification of Beneficial and Detrimental Bacteria Impacting Sorghum Responses to Drought Using Multi-Scale and Multi-System Microbiome Comparisons. ISME J. 2022, 16, 1957–1969. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A Review on the Plant Microbiome: Ecology, Functions, and Emerging Trends in Microbial Application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–Microbiome Interactions: From Community Assembly to Plant Health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Singh, B.K.; Trivedi, P. Microbiome and the Future for Food and Nutrient Security. Microb. Biotechnol. 2017, 10, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.; Mosca, A.; Dimaria, G.; Nicotra, D.; Tessitori, M.; Privitera, G.F.; Pulvirenti, A.; Leonardi, C.; Catara, V. Soil and Soilless Tomato Cultivation Promote Different Microbial Communities That Provide New Models for Future Crop Interventions. Int. J. Mol. Sci. 2022, 23, 8820. [Google Scholar] [CrossRef] [PubMed]
- Genitsaris, S.; Stefanidou, N.; Leontidou, K.; Matsi, T.; Karamanoli, K.; Mellidou, I. Bacterial Communities in the Rhizosphere and Phyllosphere of Halophytes and Drought-Tolerant Plants in Mediterranean Ecosystems. Microorganisms 2020, 8, 1708. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Du, Y.; Zhu, W.; Pang, X.; Wang, Z. Effects of Organic Materials on Soil Bacterial Community Structure in Long-Term Continuous Cropping of Tomato in Greenhouse. Open Life Sci. 2022, 17, 381–392. [Google Scholar] [CrossRef]
- Escobar-Rodríguez, C.; Novak, J.; Buchholz, F.; Uetz, P.; Bragagna, L.; Gumze, M.; Antonielli, L.; Mitter, B. The Bacterial Microbiome of the Tomato Fruit Is Highly Dependent on the Cultivation Approach and Correlates with Flavor Chemistry. Front. Plant Sci. 2021, 12, 775722. [Google Scholar] [CrossRef]
- Gorrasi, S.; Pasqualetti, M.; Muñoz-Palazon, B.; Novello, G.; Mazzucato, A.; Campiglia, E.; Fenice, M. Comparison of the Peel-Associated Epiphytic Bacteria of Anthocyanin-Rich “Sun Black” and Wild-Type Tomatoes under Organic and Conventional Farming. Microorganisms 2022, 10, 2240. [Google Scholar] [CrossRef]
- Allard, S.M.; Ottesen, A.R.; Brown, E.W.; Micallef, S.A. Insect Exclusion Limits Variation in Bacterial Microbiomes of Tomato Flowers and Fruit. J. Appl. Microbiol. 2018, 125, 1749–1760. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiao, Y.S.; Zhan, Y.; Zhang, Z.; Liu, Y.; Wei, Y.; Xu, T.; Li, J. Tomato Microbiome under Long-Term Organic and Conventional Farming. iMeta 2022, 1, e48. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Zhang, Z.; Liu, Y.; Wei, Y. Tomato Endophytic Bacteria Composition and Mechanism of Suppressiveness of Wilt Disease (Fusarium oxysporum). Front. Microbiol. 2021, 12, 731764. [Google Scholar] [CrossRef]
- Ikeda-Ohtsubo, W.; Brugman, S.; Warden, C.H.; Rebel, J.M.J.; Folkerts, G.; Pieterse, C.M.J. How Can We Define “Optimal Microbiota?”: A Comparative Review of Structure and Functions of Microbiota of Animals, Fish, and Plants in Agriculture. Front. Nutr. 2018, 5, 90. [Google Scholar] [CrossRef]
- Bruto, M.; Prigent-Combaret, C.; Muller, D.; Moënne-Loccoz, Y. Analysis of Genes Contributing to Plant-Beneficial Functions in Plant Growth-Promoting Rhizobacteria and Related Proteobacteria. Sci. Rep. 2014, 4, 6261. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi-Zarandi, M.; Saberi Riseh, R.; Tarkka, M.T. Actinobacteria as Effective Biocontrol Agents against Plant Pathogens, an Overview on Their Role in Eliciting Plant Defense. Microorganisms 2022, 10, 1739. [Google Scholar] [CrossRef] [PubMed]
- Narsing Rao, M.P.; Lohmaneeratana, K.; Bunyoo, C.; Thamchaipenet, A. Actinobacteria–Plant Interactions in Alleviating Abiotic Stress. Plants 2022, 11, 2976. [Google Scholar] [CrossRef] [PubMed]
- Abo-Elyousr, K.A.M.; Khalil Bagy, H.M.M.; Hashem, M.; Alamri, S.A.M.; Mostafa, Y.S. Biological Control of the Tomato Wilt Caused by Clavibacter michiganensis subsp. Michiganensis Using Formulated Plant Growth-Promoting Bacteria. Egypt. J. Biol. Pest Control 2019, 29, 54. [Google Scholar] [CrossRef]
- Hashem, A.; Tabassum, B.; Fathi Abd_Allah, E. Bacillus Subtilis: A Plant-Growth Promoting Rhizobacterium That Also Impacts Biotic Stress. Saudi J. Biol. Sci. 2019, 26, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- López, S.M.Y.; Pastorino, G.N.; Fernández-González, A.J.; Franco, M.E.E.; Fernández-López, M.; Balatti, P.A. The Endosphere Bacteriome of Diseased and Healthy Tomato Plants. Arch. Microbiol. 2020, 202, 2629–2642. [Google Scholar] [CrossRef]
- Samayoa, B.E.; Shen, F.-T.; Lai, W.-A.; Chen, W.-C. Screening and Assessment of Potential Plant Growth-Promoting Bacteria Associated with Allium cepa Linn. Microbes Environ. 2020, 35, ME19147. [Google Scholar] [CrossRef]
- Pearce, S.L.; Oakeshott, J.G.; Pandey, G. Insights into Ongoing Evolution of the Hexachlorocyclohexane Catabolic Pathway from Comparative Genomics of Ten Sphingomonadaceae Strains. G3 2015, 5, 1081–1094. [Google Scholar] [CrossRef]
- Zhao, Q.; Yue, S.; Bilal, M.; Hu, H.; Wang, W.; Zhang, X. Comparative Genomic Analysis of 26 Sphingomonas and Sphingobium Strains: Dissemination of Bioremediation Capabilities, Biodegradation Potential and Horizontal Gene Transfer. Sci. Total Environ. 2017, 609, 1238–1247. [Google Scholar] [CrossRef]
- Wang, Y.; Narayanan, M.; Shi, X.; Chen, X.; Li, Z.; Natarajan, D.; Ma, Y. Plant Growth-Promoting Bacteria in Metal-Contaminated Soil: Current Perspectives on Remediation Mechanisms. Front. Microbiol. 2022, 13, 966226. [Google Scholar] [CrossRef]
- Kang, S.-M.; Imran, M.; Shaffique, S.; Kwon, E.-H.; Park, Y.-S.; Lee, I.-J. Growth and Photosynthetic Characteristics of Sesame Seedlings with Gibberellin-Producing Rhodobacter sphaeroides SIR03 and Biochar. Int. J. Plant Biol. 2022, 13, 257–269. [Google Scholar] [CrossRef]
- Azaroual, S.E.; Kasmi, Y.; Aasfar, A.; El Arroussi, H.; Zeroual, Y.; El Kadiri, Y.; Zrhidri, A.; Elfahime, E.; Sefiani, A.; Meftah Kadmiri, I. Investigation of Bacterial Diversity Using 16S rRNA Sequencing and Prediction of Its Functionalities in Moroccan Phosphate Mine Ecosystem. Sci. Rep. 2022, 12, 3741. [Google Scholar] [CrossRef]
- Antoniou, A.; Tsolakidou, M.-D.; Stringlis, I.A.; Pantelides, I.S. Rhizosphere Microbiome Recruited from a Suppressive Compost Improves Plant Fitness and Increases Protection against Vascular Wilt Pathogens of Tomato. Front. Plant Sci. 2017, 8, 2022. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, J.-T.; Wang, W.-H.; Tsui, C.K.; Cai, L. Changes in Bacterial and Fungal Microbiomes Associated with Tomatoes of Healthy and Infected by Fusarium oxysporum f. sp. Lycopersici. Microb. Ecol. 2021, 81, 1004–1017. [Google Scholar] [CrossRef] [PubMed]
- Nuzzo, A.; Satpute, A.; Albrecht, U.; Strauss, S.L. Impact of Soil Microbial Amendments on Tomato Rhizosphere Microbiome and Plant Growth in Field Soil. Microb. Ecol. 2020, 80, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Choi, J.; Lee, P.A.; Roy, N.; Khan, R.; Lee, H.J.; Weon, H.Y.; Kong, H.G.; Lee, S.-W. Alteration of Bacterial Wilt Resistance in Tomato Plant by Microbiota Transplant. Front. Plant Sci. 2020, 11, 1186. [Google Scholar] [CrossRef]
- Renaut, S.; Masse, J.; Norrie, J.P.; Blal, B.; Hijri, M. A Commercial Seaweed Extract Structured Microbial Communities Associated with Tomato and Pepper Roots and Significantly Increased Crop Yield. Microb. Biotechnol. 2019, 12, 1346–1358. [Google Scholar] [CrossRef]
- Nakayasu, M.; Ohno, K.; Takamatsu, K.; Aoki, Y.; Yamazaki, S.; Takase, H.; Shoji, T.; Yazaki, K.; Sugiyama, A. Tomato Roots Secrete Tomatine to Modulate the Bacterial Assemblage of the Rhizosphere. Plant Physiol. 2021, 186, 270–284. [Google Scholar] [CrossRef]
- Santoyo, G. How Plants Recruit Their Microbiome? New Insights into Beneficial Interactions. J. Adv. Res. 2021, 40, 45–58. [Google Scholar] [CrossRef]
- Bao, L.; Cai, W.; Cao, J.; Zhang, X.; Liu, J.; Chen, H.; Wei, Y.; Zhuang, X.; Zhuang, G.; Bai, Z. Microbial Community Overlap between the Phyllosphere and Rhizosphere of Three Plants from Yongxing Island, South China Sea. Microbiologyopen 2020, 9, e1048. [Google Scholar] [CrossRef]
- Dong, C.-J.; Wang, L.-L.; Li, Q.; Shang, Q.-M. Bacterial Communities in the Rhizosphere, Phyllosphere and Endosphere of Tomato Plants. PLoS ONE 2019, 14, e0223847. [Google Scholar] [CrossRef] [PubMed]
- Vorholt, J.A. Microbial Life in the Phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The Importance of the Microbiome of the Plant Holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Seufert, V.; Ramankutty, N.; Foley, J.A. Comparing the Yields of Organic and Conventional Agriculture. Nature 2012, 485, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Paul Chowdhury, S.; Babin, D.; Sandmann, M.; Jacquiod, S.; Sommermann, L.; Sørensen, S.J.; Fliessbach, A.; Mäder, P.; Geistlinger, J.; Smalla, K.; et al. Effect of Long-term Organic and Mineral Fertilization Strategies on Rhizosphere Microbiota Assemblage and Performance of Lettuce. Environ. Microbiol. 2019, 21, 2426–2439. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhao, W.; Zhou, Z.; Huang, G.; Wang, X.; Han, Q.; Liu, G. The Application of Mixed Organic and Inorganic Fertilizers Drives Soil Nutrient and Bacterial Community Changes in Teak Plantations. Microorganisms 2022, 10, 958. [Google Scholar] [CrossRef]
- Grunert, O.; Hernandez-Sanabria, E.; Vilchez-Vargas, R.; Jauregui, R.; Pieper, D.H.; Perneel, M.; Van Labeke, M.-C.; Reheul, D.; Boon, N. Mineral and Organic Growing Media Have Distinct Community Structure, Stability and Functionality in Soilless Culture Systems. Sci. Rep. 2016, 6, 18837. [Google Scholar] [CrossRef]
- Schmidt, J.E.; Vannette, R.L.; Igwe, A.; Blundell, R.; Casteel, C.L.; Gaudin, A.C.M. Effects of Agricultural Management on Rhizosphere Microbial Structure and Function in Processing Tomato Plants. Appl. Environ. Microbiol. 2019, 85, e01064-19. [Google Scholar] [CrossRef]
- Huang, F.; Mo, C.; Li, L.; Shi, J.; Yang, Y.; Liao, X. Organic Fertilizer Application Mediates Tomato Defense Against Pseudomonas syringae Pv. Tomato, Possibly by Reshaping the Soil Microbiome. Front. Microbiol. 2022, 13, 939911. [Google Scholar] [CrossRef]
- Kraut-Cohen, J.; Shapiro, O.H.; Dror, B.; Cytryn, E. Pectin Induced Colony Expansion of Soil-Derived Flavobacterium Strains. Front. Microbiol. 2021, 12, 651891. [Google Scholar] [CrossRef]
- Lidbury, I.D.E.A.; Scanlan, D.J.; Murphy, A.R.J.; Christie-Oleza, J.A.; Aguilo-Ferretjans, M.M.; Hitchcock, A.; Daniell, T.J. A Widely Distributed Phosphate-Insensitive Phosphatase Presents a Route for Rapid Organophosphorus Remineralization in the Biosphere. Proc. Natl. Acad. Sci. USA 2022, 119, e2118122119. [Google Scholar] [CrossRef]
- Rilling, J.I.; Acuña, J.J.; Sadowsky, M.J.; Jorquera, M.A. Putative Nitrogen-Fixing Bacteria Associated with the Rhizosphere and Root Endosphere of Wheat Plants Grown in an Andisol from Southern Chile. Front. Microbiol. 2018, 9, 2710. [Google Scholar] [CrossRef] [PubMed]
- Zuluaga, M.Y.A.; Milani, K.M.L.; Gonçalves, L.S.A.; de Oliveira, A.L.M. Diversity and Plant Growth-Promoting Functions of Diazotrophic/N-Scavenging Bacteria Isolated from the Soils and Rhizospheres of Two Species of Solanum. PLoS ONE 2020, 15, e0227422. [Google Scholar] [CrossRef] [PubMed]
- Nonthakaew, N.; Panbangred, W.; Songnuan, W.; Intra, B. Plant Growth-Promoting Properties of Streptomyces spp. Isolates and Their Impact on Mung Bean Plantlets’ Rhizosphere Microbiome. Front. Microbiol. 2022, 13, 967415. [Google Scholar] [CrossRef]
- Tang, T.; Sun, X.; Dong, Y.; Liu, Q. Erythrobacter aureus sp. Nov., a Plant Growth-Promoting Bacterium Isolated from Sediment in the Yellow Sea, China. 3 Biotech 2019, 9, 430. [Google Scholar] [CrossRef] [PubMed]
- Favero, V.O.; Carvalho, R.H.; Motta, V.M.; Leite, A.B.C.; Coelho, M.R.R.; Xavier, G.R.; Rumjanek, N.G.; Urquiaga, S. Bradyrhizobium as the Only Rhizobial Inhabitant of Mung Bean (Vigna radiata) Nodules in Tropical Soils: A Strategy Based on Microbiome for Improving Biological Nitrogen Fixation Using Bio-Products. Front. Plant Sci. 2021, 11, 602645. [Google Scholar] [CrossRef]
- Passari, A.K.; Upadhyaya, K.; Singh, G.; Abdel-Azeem, A.M.; Thankappan, S.; Uthandi, S.; Hashem, A.; Abd_Allah, E.F.; Malik, J.A.; A.S.A.; et al. Enhancement of Disease Resistance, Growth Potential, and Photosynthesis in Tomato (Solanum lycopersicum) by Inoculation with an Endophytic Actinobacterium, Streptomyces thermocarboxydus Strain BPSAC147. PLoS ONE 2019, 14, e0219014. [Google Scholar] [CrossRef]
- Soumare, A.; Diedhiou, A.G.; Thuita, M.; Hafidi, M.; Ouhdouch, Y.; Gopalakrishnan, S.; Kouisni, L. Exploiting Biological Nitrogen Fixation: A Route Towards a Sustainable Agriculture. Plants 2020, 9, 1011. [Google Scholar] [CrossRef]
- Goyal, R.K.; Schmidt, M.A.; Hynes, M.F. Molecular Biology in the Improvement of Biological Nitrogen Fixation by Rhizobia and Extending the Scope to Cereals. Microorganisms 2021, 9, 125. [Google Scholar] [CrossRef]
- Mus, F.; Crook, M.B.; Garcia, K.; Garcia Costas, A.; Geddes, B.A.; Kouri, E.D.; Paramasivan, P.; Ryu, M.-H.; Oldroyd, G.E.D.; Poole, P.S.; et al. Symbiotic Nitrogen Fixation and the Challenges to Its Extension to Nonlegumes. Appl. Environ. Microbiol. 2016, 82, 3698–3710. [Google Scholar] [CrossRef]
- de Ponti, T.; Rijk, B.; van Ittersum, M.K. The Crop Yield Gap between Organic and Conventional Agriculture. Agric. Syst. 2012, 108, 1–9. [Google Scholar] [CrossRef]
- Ottesen, A.R.; Gorham, S.; Pettengill, J.B.; Rideout, S.; Evans, P.; Brown, E. The Impact of Systemic and Copper Pesticide Applications on the Phyllosphere Microflora of Tomatoes. J. Sci. Food Agric. 2015, 95, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Telias, A.; White, J.R.; Pahl, D.M.; Ottesen, A.R.; Walsh, C.S. Bacterial Community Diversity and Variation in Spray Water Sources and the Tomato Fruit Surface. BMC Microbiol. 2011, 11, 81. [Google Scholar] [CrossRef] [PubMed]
- Ottesen, A.; Ramachandran, P.; Reed, E.; Gu, G.; Gorham, S.; Ducharme, D.; Newell, M.; Rideout, S.; Turini, T.; Hill, T.; et al. Metagenome Tracking Biogeographic Agroecology: Phytobiota of Tomatoes from Virginia, Maryland, North Carolina and California. Food Microbiol. 2019, 79, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. The Changing Face of the Family Enterobacteriaceae (Order: “Enterobacterales”): New Members, Taxonomic Issues, Geographic Expansion, and New Diseases and Disease Syndromes. Clin. Microbiol. Rev. 2021, 34, e00174-20. [Google Scholar] [CrossRef]
- Kang, E.; Crouse, A.; Chevallier, L.; Pontier, S.M.; Alzahrani, A.; Silué, N.; Campbell-Valois, F.-X.; Montagutelli, X.; Gruenheid, S.; Malo, D. Enterobacteria and Host Resistance to Infection. Mamm. Genome 2018, 29, 558–576. [Google Scholar] [CrossRef]
- Balali, G.I.; Yar, D.D.; Afua Dela, V.G.; Adjei-Kusi, P. Microbial Contamination, an Increasing Threat to the Consumption of Fresh Fruits and Vegetables in Today’s World. Int. J. Microbiol. 2020, 2020, 3029295. [Google Scholar] [CrossRef]
- Abbott, D.W.; Boraston, A.B. Structural Biology of Pectin Degradation by Enterobacteriaceae. Microbiol. Mol. Biol. Rev. 2008, 72, 301–316. [Google Scholar] [CrossRef]
- Palacio-Bielsa, A.; Roselló, M.; Llop, P.; López, M.M. Erwinia spp. from Pome Fruit Trees: Similarities and Differences among Pathogenic and Non-Pathogenic Species. Trees 2012, 26, 13–29. [Google Scholar] [CrossRef]
- Charkowski, A.; Blanco, C.; Condemine, G.; Expert, D.; Franza, T.; Hayes, C.; Hugouvieux-Cotte-Pattat, N.; Solanilla, E.L.; Low, D.; Moleleki, L.; et al. The Role of Secretion Systems and Small Molecules in Soft-Rot Enterobacteriaceae Pathogenicity. Annu. Rev. Phytopathol. 2012, 50, 425–449. [Google Scholar] [CrossRef]
- Rasch, M.; Andersen, J.B.; Nielsen, K.F.; Flodgaard, L.R.; Christensen, H.; Givskov, M.; Gram, L. Involvement of Bacterial Quorum-Sensing Signals in Spoilage of Bean Sprouts. Appl. Environ. Microbiol. 2005, 71, 3321–3330. [Google Scholar] [CrossRef] [PubMed]
- Al-Kharousi, Z.S.; Guizani, N.; Al-Sadi, A.M.; Al-Bulushi, I.M.; Shaharoona, B. Hiding in Fresh Fruits and Vegetables: Opportunistic Pathogens May Cross Geographical Barriers. Int. J. Microbiol. 2016, 2016, 4292417. [Google Scholar] [CrossRef] [PubMed]
- Boehme, S.; Werner, G.; Klare, I.; Reissbrodt, R.; Witte, W. Occurrence of Antibiotic-Resistant Enterobacteria in Agricultural Foodstuffs. Mol. Nutr. Food Res. 2004, 48, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Saksena, R.; Malik, M.; Gaind, R. Bacterial Contamination and Prevalence of Antimicrobial Resistance Phenotypes in Raw Fruits and Vegetables Sold in Delhi, India. J. Food Saf. 2020, 40, e12739. [Google Scholar] [CrossRef]
- Waturangi, D.E.; Hudiono, F.; Aliwarga, E. Prevalence of Pathogenic Escherichia Coli from Salad Vegetable and Fruits Sold in Jakarta. BMC Res. Notes 2019, 12, 247. [Google Scholar] [CrossRef]
- Feng, P.C.H.; Reddy, S. Prevalences of Shiga Toxin Subtypes and Selected Other Virulence Factors among Shiga-Toxigenic Escherichia Coli Strains Isolated from Fresh Produce. Appl. Environ. Microbiol. 2013, 79, 6917–6923. [Google Scholar] [CrossRef]
- Bell, R.L.; Zheng, J.; Burrows, E.; Allard, S.; Wang, C.Y.; Keys, C.E.; Melka, D.C.; Strain, E.; Luo, Y.; Allard, M.W.; et al. Ecological Prevalence, Genetic Diversity, and Epidemiological Aspects of Salmonella Isolated from Tomato Agricultural Regions of the Virginia Eastern Shore. Front. Microbiol. 2015, 6, 415. [Google Scholar] [CrossRef]
- Gurtler, J.B.; Harlee, N.A.; Smelser, A.M.; Schneider, K.R. Salmonella Enterica Contamination of Market Fresh Tomatoes: A Review. J. Food Prot. 2018, 81, 1193–1213. [Google Scholar] [CrossRef]
- Santos, R.F.C.; Nascimento, J.D.S.; Geimba, M.P.; Hessel, C.T.; Tondo, E.C. First Report of Human Gastroenteritis Caused by Escherichia Coli O157:NM in Brazil. Foodborne Pathog. Dis. 2017, 14, 665–666. [Google Scholar] [CrossRef]
- Cummings, K.; Barrett, E.; Mohle-Boetani, J.C.; Brooks, J.T.; Farrar, J.; Hunt, T.; Fiore, A.; Komatsu, K.; Werner, S.B.; Slutsker, L. A Multistate Outbreak of Salmonella enterica Serotype Baildon Associated with Domestic Raw Tomatoes. Emerg. Infect. Dis. 2001, 7, 1046–1048. [Google Scholar] [CrossRef]
- Gupta, S.K.; Nalluswami, K.; Snider, C.; Perch, M.; Balasegaram, M.; Burmeister, D.; Lockett, J.; Sandt, C.; Hoekstra, R.M.; Montgomery, S. Outbreak of Salmonella Braenderup Infections Associated with Roma Tomatoes, Northeastern United States, 2004: A Useful Method for Subtyping Exposures in Field Investigations. Epidemiol. Infect. 2007, 135, 1165–1173. [Google Scholar] [CrossRef]
- Hedberg, C.W.; Angulo, F.J.; White, K.E.; Langkop, C.W.; Schell, W.L.; Stobierski, M.G.; Schuchat, A.; Besser, J.M.; Dietrich, S.; Helsel, L.; et al. Outbreaks of Salmonellosis Associated with Eating Uncooked Tomatoes: Implications for Public Health. The Investigation Team. Epidemiol. Infect. 1999, 122, 385–393. [Google Scholar] [CrossRef]
- Behravesh, C.B.; Blaney, D.; Medus, C.; Bidol, S.A.; Phan, Q.; Soliva, S.; Daly, E.R.; Smith, K.; Miller, B.; Taylor, T.; et al. Multistate Outbreak of Salmonella serotype Typhimurium Infections Associated with Consumption of Restaurant Tomatoes, USA, 2006: Hypothesis Generation through Case Exposures in Multiple Restaurant Clusters. Epidemiol. Infect. 2012, 140, 2053–2061. [Google Scholar] [CrossRef]
- Heaton, J.C.; Jones, K. Microbial Contamination of Fruit and Vegetables and the Behaviour of Enteropathogens in the Phyllosphere: A Review. J. Appl. Microbiol. 2008, 104, 613–626. [Google Scholar] [CrossRef]
- Müller, L.; Kjelsø, C.; Frank, C.; Jensen, T.; Torpdahl, M.; Søborg, B.; Dorleans, F.; Rabsch, W.; Prager, R.; Gossner, C.M.; et al. Outbreak of Salmonella Strathcona Caused by Datterino Tomatoes, Denmark, 2011. Epidemiol. Infect. 2016, 144, 2802–2811. [Google Scholar] [CrossRef]
- Reller, M.E.; Nelson, J.M.; Mølbak, K.; Ackman, D.M.; Schoonmaker-Bopp, D.J.; Root, T.P.; Mintz, E.D. A Large, Multiple-Restaurant Outbreak of Infection with Shigella flexneri Serotype 2a Traced to Tomatoes. Clin. Infect. Dis. 2006, 42, 163–169. [Google Scholar] [CrossRef]
- Holden, N.; Pritchard, L.; Toth, I. Colonization Outwith the Colon: Plants as an Alternative Environmental Reservoir for Human Pathogenic Enterobacteria. FEMS Microbiol. Rev. 2009, 33, 689–703. [Google Scholar] [CrossRef]
- Gu, G.; Hu, J.; Cevallos-Cevallos, J.M.; Richardson, S.M.; Bartz, J.A.; van Bruggen, A.H.C. Internal Colonization of Salmonella enterica Serovar Typhimurium in Tomato Plants. PLoS ONE 2011, 6, e27340. [Google Scholar] [CrossRef]
- de Moraes, M.H.; Desai, P.; Porwollik, S.; Canals, R.; Perez, D.R.; Chu, W.; McClelland, M.; Teplitski, M. Salmonella Persistence in Tomatoes Requires a Distinct Set of Metabolic Functions Identified by Transposon Insertion Sequencing. Appl. Environ. Microbiol. 2017, 83, e03028-16. [Google Scholar] [CrossRef]
- Bonanomi, G.; Alioto, D.; Minutolo, M.; Marra, R.; Cesarano, G.; Vinale, F. Organic Amendments Modulate Soil Microbiota and Reduce Virus Disease Incidence in the TSWV-Tomato Pathosystem. Pathogens 2020, 9, 379. [Google Scholar] [CrossRef]
- Caradonia, F.; Ronga, D.; Catellani, M.; Giaretta Azevedo, C.V.; Terrazas, R.A.; Robertson-Albertyn, S.; Francia, E.; Bulgarelli, D. Nitrogen Fertilizers Shape the Composition and Predicted Functions of the Microbiota of Field-Grown Tomato Plants. Phytobiomes J. 2019, 3, 315–325. [Google Scholar] [CrossRef]
- Jiang, S.-Q.; Yu, Y.-N.; Gao, R.-W.; Wang, H.; Zhang, J.; Li, R.; Long, X.-H.; Shen, Q.-R.; Chen, W.; Cai, F. High-Throughput Absolute Quantification Sequencing Reveals the Effect of Different Fertilizer Applications on Bacterial Community in a Tomato Cultivated Coastal Saline Soil. Sci. Total Environ. 2019, 687, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xiong, W.; Zhang, R.; Hang, X.; Wang, D.; Li, R.; Shen, Q. Continuous Application of Different Organic Additives Can Suppress Tomato Disease by Inducing the Healthy Rhizospheric Microbiota through Alterations to the Bulk Soil Microflora. Plant Soil 2018, 423, 229–240. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).