Potential Oral Microbial Markers for Differential Diagnosis of Crohn’s Disease and Ulcerative Colitis Using Machine Learning Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population and Sample Collection
2.2. DNA Extraction, PCR Amplification, and 16S rRNA Gene Sequencing
2.3. 16S rRNA Gene Sequencing Data Analysis
2.4. Machine Learning for Diagnosis Model
3. Results
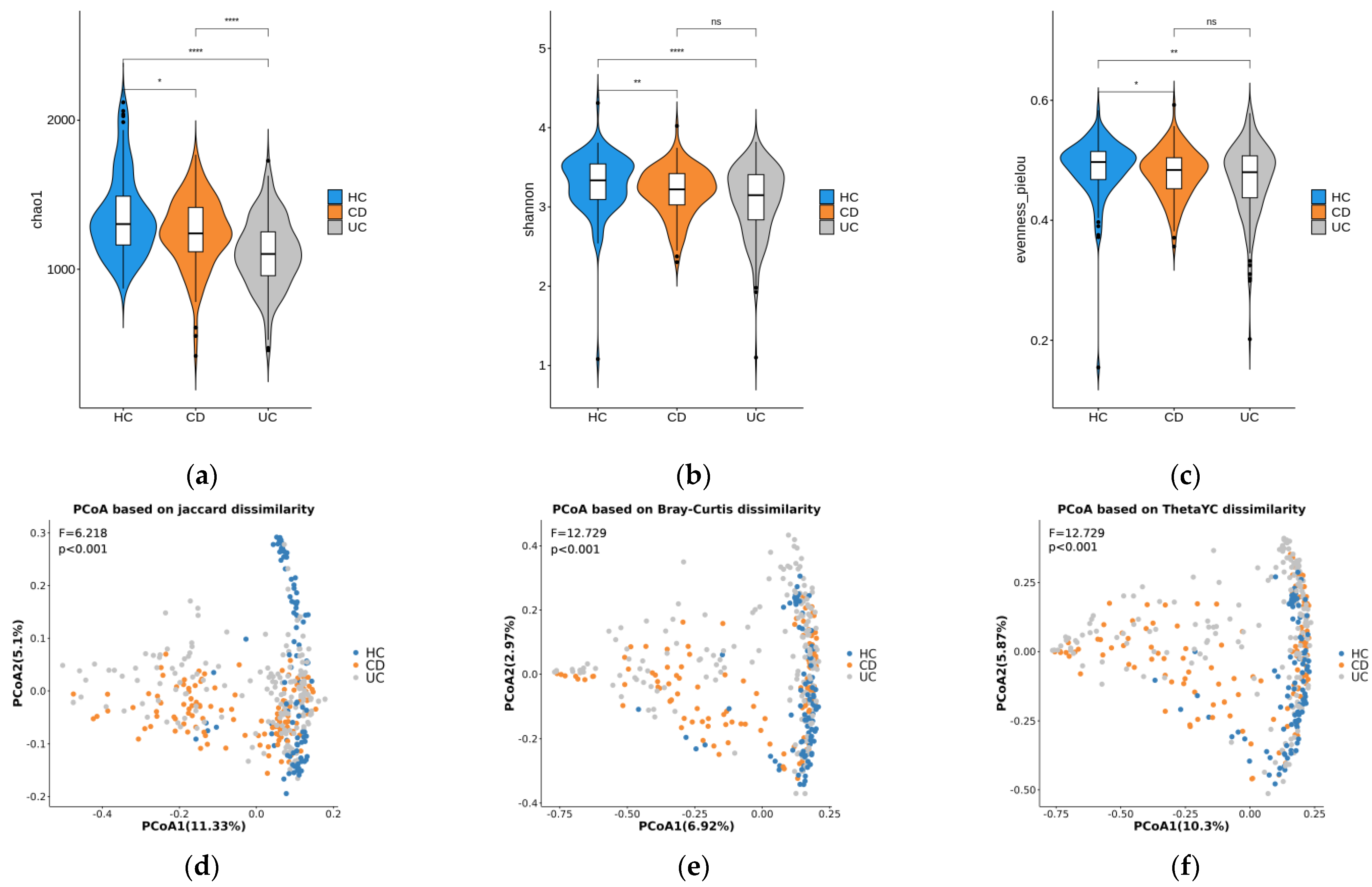
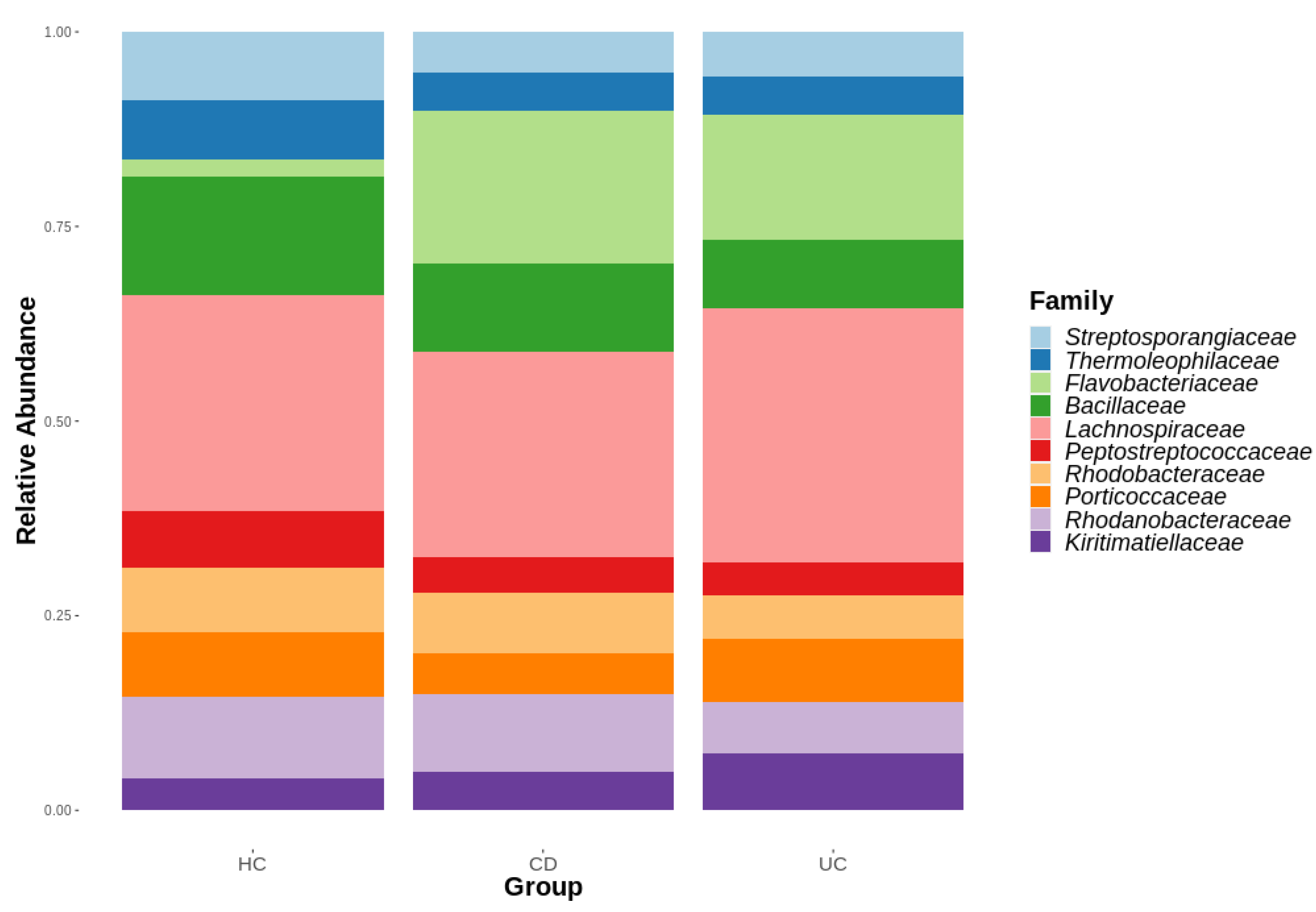
3.1. Diversity Analysis
3.2. Multi-Class Machine Learning Model Based on PLS-DA
3.3. Hierarchical Diagnosis Models Based on PLS-DA and sPLS-DA
3.3.1. Development of a Prediction Model That Classifies IBD vs. HC
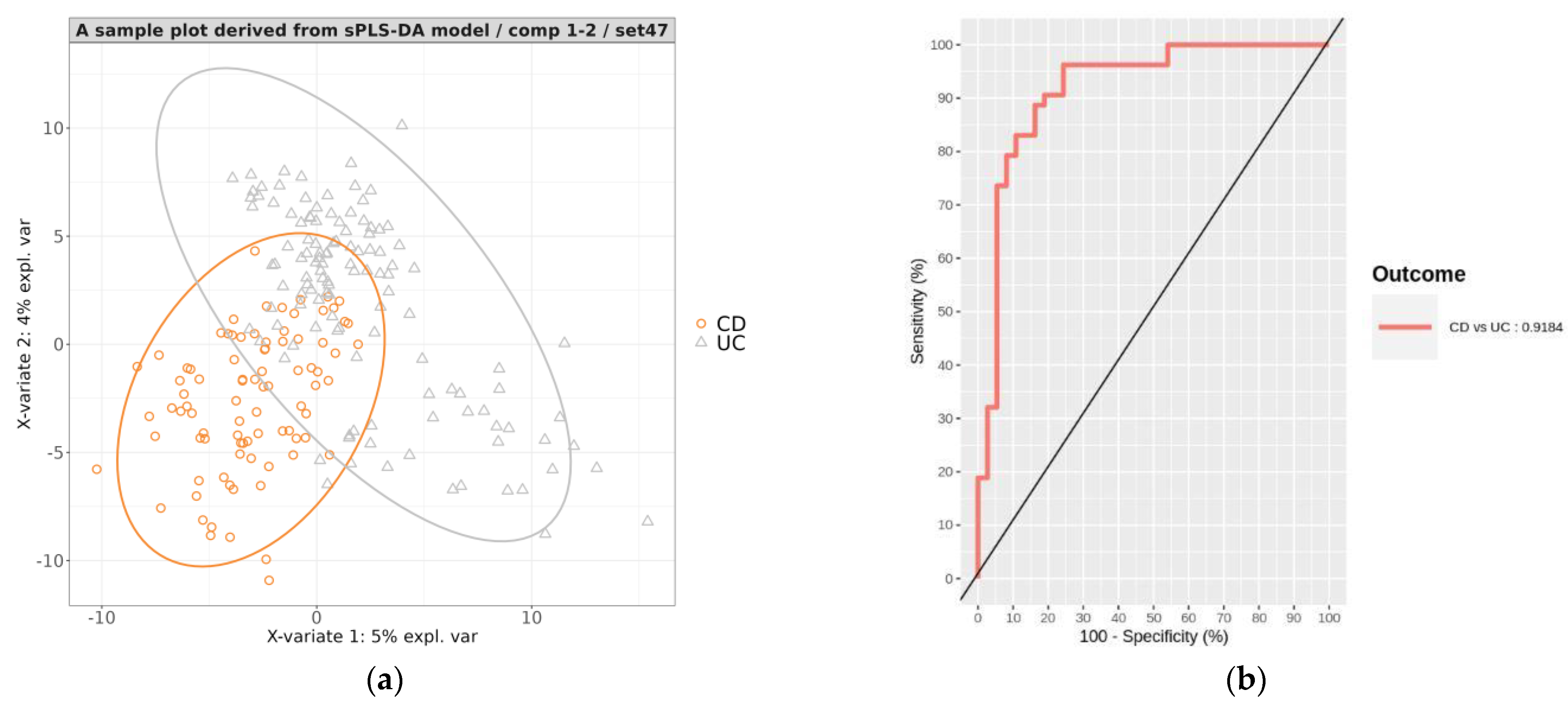
3.3.2. Development of a Prediction Model That Classifies CD vs. UC
3.3.3. Evaluation of the Hierarchical Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Podolsky, D.K. Inflammatory Bowel Disease. N. Engl. J. Med. 2002, 347, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Abreu, M.T. The pathogenesis of inflammatory bowel disease: Translational implications for clinicians. Curr. Gastroenterol. Rep. 2002, 4, 481–489. [Google Scholar] [CrossRef]
- Korzenik, J.R.; Podolsky, D.K. Evolving knowledge and therapy of inflammatory bowel disease. Nat. Rev. Drug Discov. 2006, 5, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Noor, N.M.; Sousa, P.; Paul, S.; Roblin, X. Early Diagnosis, Early Stratification, and Early Intervention to Deliver Precision Medicine in IBD. Inflamm. Bowel Dis. 2021, 28, 1254–1264. [Google Scholar] [CrossRef] [PubMed]
- Moazzami, B.; Moazzami, K.; Rezaei, N. Early onset inflammatory bowel disease: Manifestations, genetics and diagnosis. Turk. J. Pediatr. 2019, 61, 637–647. [Google Scholar] [CrossRef]
- Na, S.-Y.; Moon, W. Recent advances in surveillance colonoscopy for dysplasia in inflammatory bowel disease. Clin. Endosc. 2022, 55, 726–735. [Google Scholar] [CrossRef]
- Khor, B.; Gardet, A.; Xavier, R.J. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011, 474, 307–317. [Google Scholar] [CrossRef] [Green Version]
- Loftus, E.V.; Sandborn, W.J. Epidemiology of inflammatory bowel disease. Gastroenterol. Clin. N. Am. 2002, 31, 1–20. [Google Scholar] [CrossRef]
- Andres, P.G.; Friedman, L.S. Epidemiology and the Natural Course of Inflammatory Bowel Disease. Gastroenterol. Clin. N. Am. 1999, 28, 255–281. [Google Scholar] [CrossRef]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef] [Green Version]
- Elmaghrawy, K.; Hussey, S.; Moran, G.P. The Oral Microbiome in Pediatric IBD: A Source of Pathobionts or Biomarkers? Front. Pediatr. 2021, 8, 620254. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Wu, H.-M.; Yang, Z.; Zhou, Y.-F.; Jin, L.; Yang, M.-F.; Wang, F.-Y. New Insights into the Role of Oral Microbiota Dysbiosis in the Pathogenesis of Inflammatory Bowel Disease. Dig. Dis. Sci. 2021, 67, 42–55. [Google Scholar] [CrossRef]
- Xun, Z.; Zhang, Q.; Xu, T.; Chen, N.; Chen, F. Dysbiosis and Ecotypes of the Salivary Microbiome Associated With Inflammatory Bowel Diseases and the Assistance in Diagnosis of Diseases Using Oral Bacterial Profiles. Front. Microbiol. 2018, 9, 1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Docktor, M.J.; Paster, B.J.; Abramowicz, S.; Ingram, J.; Wang, Y.E.; Correll, M.; Jiang, H.; Cotton, S.L.; Kokaras, A.S.; Bousvaros, A. Alterations in diversity of the oral microbiome in pediatric inflammatory bowel disease. Inflamm. Bowel Dis. 2012, 18, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Somineni, H.K.; Weitzner, J.H.; Venkateswaran, S.; Dodd, A.; Prince, J.; Karikaran, A.; Sauer, C.G.; Abramowicz, S.; Zwick, M.; Cutler, D.J.; et al. Site- and Taxa-Specific Disease-Associated Oral Microbial Structures Distinguish Inflammatory Bowel Diseases. Inflamm. Bowel Dis. 2021, 27, 1889–1900. [Google Scholar] [CrossRef] [PubMed]
- Lam, G.; Albarrak, H.; McColl, C.J.; Pizarro, A.; Sanaka, H.; Gomez-Nguyen, A.; Cominelli, F.; da Silva, A.P.B. The Oral-Gut Axis: Periodontal Diseases and Gastrointestinal Disorders. Inflamm. Bowel Dis. 2022, izac241. [Google Scholar] [CrossRef]
- Manandhar, I.; Alimadadi, A.; Aryal, S.; Munroe, P.B.; Joe, B.; Cheng, X. Gut microbiome-based supervised machine learning for clinical diagnosis of inflammatory bowel diseases. Am. J. Physiol. Liver Physiol. 2021, 320, G328–G337. [Google Scholar] [CrossRef]
- Gubatan, J.; Levitte, S.; Patel, A.; Balabanis, T.; Wei, M.T.; Sinha, S.R. Artificial intelligence applications in inflammatory bowel disease: Emerging technologies and future directions. World J. Gastroenterol. 2021, 27, 1920–1935. [Google Scholar] [CrossRef]
- Kubinski, R.; Djamen-Kepaou, J.-Y.; Zhanabaev, T.; Hernandez-Garcia, A.; Bauer, S.; Hildebrand, F.; Korcsmaros, T.; Karam, S.; Jantchou, P.; Kafi, K.; et al. Benchmark of Data Processing Methods and Machine Learning Models for Gut Microbiome-Based Diagnosis of Inflammatory Bowel Disease. Front. Genet. 2022, 13, 784397. [Google Scholar] [CrossRef]
- Lê Cao, K.-A.; Boitard, S.; Besse, P. Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinform. 2011, 12, 253. [Google Scholar] [CrossRef] [Green Version]
- Park, S.-K.; Kim, S.; Lee, G.-Y.; Kim, S.-Y.; Kim, W.; Lee, C.-W.; Park, J.-L.; Choi, C.-H.; Kang, S.-B.; Kim, T.-O.; et al. Development of a Machine Learning Model to Distinguish between Ulcerative Colitis and Crohn’s Disease Using RNA Sequencing Data. Diagnostics 2021, 11, 2365. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.Y.; Kim, S.; Choi, J.W.; Kang, S.-B.; Kim, T.O.; Seo, G.S.; Cha, J.M.; Chun, J.; Jung, Y.; Im, J.P.; et al. The Common and Unique Pattern of Microbiome Profiles among Saliva, Tissue, and Stool Samples in Patients with Crohn’s Disease. Microorganisms 2022, 10, 1467. [Google Scholar] [CrossRef] [PubMed]
- Molinero, N.; Taladrid, D.; Zorraquín-Peña, I.; de Celis, M.; Belda, I.; Mira, A.; Bartolomé, B.; Moreno-Arribas, M.V. Ulcerative Colitis Seems to Imply Oral Microbiome Dysbiosis. Curr. Issues Mol. Biol. 2022, 44, 1513–1527. [Google Scholar] [CrossRef] [PubMed]
- Ribaldone, D.G.; Brigo, S.; Mangia, M.; Saracco, G.M.; Astegiano, M.; Pellicano, R. Oral Manifestations of Inflammatory Bowel Disease and the Role of Non-Invasive Surrogate Markers of Disease Activity. Medicines 2020, 7, 33. [Google Scholar] [CrossRef]
- Amos, G.C.A.; Sergaki, C.; Logan, A.; Iriarte, R.; Bannaga, A.; Chandrapalan, S.; Wellington, E.M.H.; Rijpkema, S.; Arasaradnam, R.P. Exploring how microbiome signatures change across inflammatory bowel disease conditions and disease locations. Sci. Rep. 2021, 11, 18699. [Google Scholar] [CrossRef]
- Lin, H.; Das Peddada, S. Analysis of microbial compositions: A review of normalization and differential abundance analysis. npj Biofilms Microbiomes 2020, 6, 60. [Google Scholar] [CrossRef]
- López, R.L.; Burgos, M.J.G.; Gálvez, A.; Perez-Pulido, R. The human gastrointestinal tract and oral microbiota in inflammatory bowel disease: A state of the science review. Apmis 2016, 125, 3–10. [Google Scholar] [CrossRef]
- Abdelbary, M.M.H.; Hatting, M.; Bott, A.; Dahlhausen, A.; Keller, D.; Trautwein, C.; Conrads, G. The oral-gut axis: Salivary and fecal microbiome dysbiosis in patients with inflammatory bowel disease. Front. Cell. Infect. Microbiol. 2022, 12, 1010853. [Google Scholar] [CrossRef]
- Ahmad, H.A.; East, J.E.; Panaccione, R.; Travis, S.; Canavan, J.B.; Usiskin, K.; Byrne, M.F. Artificial intelligence in inflammatory bowel disease: Implications for clinical practice and future directions. Intest. Res. 2023. epub ahead of print. [Google Scholar] [CrossRef]
- Liñares-Blanco, J.; Fernandez-Lozano, C.; Seoane, J.A.; López-Campos, G. Machine Learning Based Microbiome Signature to Predict Inflammatory Bowel Disease Subtypes. Front. Microbiol. 2022, 13, 872671. [Google Scholar] [CrossRef]
- Zuo, W.; Wang, B.; Bai, X.; Luan, Y.; Fan, Y.; Michail, S.; Sun, F. 16S rRNA and metagenomic shotgun sequencing data revealed consistent patterns of gut microbiome signature in pediatric ulcerative colitis. Sci. Rep. 2022, 12, 6421. [Google Scholar] [CrossRef]
- Ma, S.; Shungin, D.; Mallick, H.; Schirmer, M.; Nguyen, L.H.; Kolde, R.; Franzosa, E.; Vlamakis, H.; Xavier, R.; Huttenhower, C. Population structure discovery in meta-analyzed microbial communities and inflammatory bowel disease using MMUPHin. Genome Biol. 2022, 23, 208. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Sternes, P.R.; Wang, M.; Song, J.; Morrison, M.; Li, T.; Zhou, L.; Wu, X.; He, F.; Zhu, J.; et al. Shotgun metagenomics reveals an enrichment of potentially cross-reactive bacterial epitopes in ankylosing spondylitis patients, as well as the effects of TNFi therapy upon microbiome composition. Ann. Rheum. Dis. 2019, 79, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Min, S.; Lee, B.; Yoon, S. Deep learning in bioinformatics. Brief. Bioinform. 2016, 18, 851–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Ferrero, E.; Agapow, P.-M.; Zietz, M.; Hoffman, M.M.; et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 2018, 15, 20170387. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| CD (n = 127) | UC (n = 175) | HC (n = 100) | |
|---|---|---|---|
| Age (year), mean ± SD | 37.6 ± 11.6 | 39.4 ± 15.6 | 37.4 ± 14.5 |
| Male, n (%) | 97 (76.4) | 124 (70.9) | 50 (50) |
| BMI (kg/m2), mean ± SD | 20.8 ± 4.1 | 23 ± 3.1 | |
| Smoking status, n (%) | |||
| Current | 20 (15.7) | 23 (13.1) | |
| Former | 5 (3.9) | 37 (21.1) | |
| Never | 101 (79.5) | 112 (64) | |
| Unknown | 1 (0.8) | 3 (1.7) | |
| Disease location, n (%) | Ileum, 28 (22) | Proctitis, 69 (39.4) | |
| Colon, 25 (19.7) | Distal, 57 (32.6) | ||
| Ileocolon, 71 (55.9) | Extensive, 45 (25.7) | ||
| Ileocolon + upper GI, 1 (0.8) | |||
| Unknown, 2 (1.6) | Unknown, 4 (2.3) |
| Accuray | CD Sens. | CD Prec. | UC Sens. | UC Prec. | HC Sens. | HC Prec. | AUC | |
|---|---|---|---|---|---|---|---|---|
| Min. | 0.525 | 0.405 | 0.442 | 0.377 | 0.574 | 0.633 | 0.476 | 0.650 |
| 1st Qu. | 0.617 | 0.595 | 0.565 | 0.491 | 0.665 | 0.8 | 0.619 | 0.759 |
| Median | 0.658 | 0.676 | 0.624 | 0.528 | 0.705 | 0.867 | 0.659 | 0.801 |
| Mean | 0.653 | 0.666 | 0.615 | 0.532 | 0.7 | 0.85 | 0.652 | 0.819 |
| 3rd Qu. | 0.683 | 0.73 | 0.659 | 0.566 | 0.738 | 0.9 | 0.693 | 0.898 |
| Max. | 0.758 | 0.919 | 0.808 | 0.755 | 0.852 | 1 | 0.839 | 0.968 |
| Accuracy | Sensitivity | Specificity | Precision | AUC | |
|---|---|---|---|---|---|
| Min. | 0.808 | 0.8 | 0.633 | 0.887 | 0.916 |
| 1st Qu. | 0.892 | 0.9 | 0.833 | 0.943 | 0.954 |
| Median | 0.908 | 0.917 | 0.867 | 0.955 | 0.967 |
| Mean | 0.908 | 0.919 | 0.974 | 0.957 | 0.966 |
| 3rd Qu. | 0.925 | 0.944 | 0.933 | 0.977 | 0.979 |
| Max. | 0.967 | 1 | 1 | 1 | 0.995 |
| Accuracy | Sensitivity | Specificity | Precision | AUC | |
|---|---|---|---|---|---|
| Min. | 0.744 | 0.595 | 0.679 | 0.646 | 0.829 |
| 1st Qu. | 0.822 | 0.784 | 0.83 | 0.767 | 0.899 |
| Median | 0.844 | 0.824 | 0.87 | 0.816 | 0.928 |
| Mean | 0.846 | 0.82 | 0.864 | 0.812 | 0.923 |
| 3rd Qu. | 0.878 | 0.872 | 0.906 | 0.857 | 0.949 |
| Max. | 0.922 | 0.973 | 0.962 | 0.936 | 0.978 |
| Accuray | CD Sens. | CD Prec. | UC Sens. | UC Prec. | HC Sens. | HC Prec. | |
|---|---|---|---|---|---|---|---|
| Min. | 0.658 | 0.595 | 0.6 | 0.547 | 0.682 | 0.633 | 0.581 |
| 1st Qu. | 0.783 | 0.73 | 0.738 | 0.717 | 0.799 | 0.833 | 0.741 |
| Median | 0.8 | 0.797 | 0.795 | 0.764 | 0.837 | 0.867 | 0.784 |
| Mean | 0.803 | 0.791 | 0.792 | 0.77 | 0.833 | 0.875 | 0.791 |
| 3rd Qu. | 0.825 | 0.865 | 0.844 | 0.83 | 0.867 | 0.933 | 0.839 |
| Max. | 0.892 | 0.919 | 0.968 | 0.925 | 0.932 | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, S.-B.; Kim, H.; Kim, S.; Kim, J.; Park, S.-K.; Lee, C.-W.; Kim, K.O.; Seo, G.-S.; Kim, M.S.; Cha, J.M.; et al. Potential Oral Microbial Markers for Differential Diagnosis of Crohn’s Disease and Ulcerative Colitis Using Machine Learning Models. Microorganisms 2023, 11, 1665. https://doi.org/10.3390/microorganisms11071665
Kang S-B, Kim H, Kim S, Kim J, Park S-K, Lee C-W, Kim KO, Seo G-S, Kim MS, Cha JM, et al. Potential Oral Microbial Markers for Differential Diagnosis of Crohn’s Disease and Ulcerative Colitis Using Machine Learning Models. Microorganisms. 2023; 11(7):1665. https://doi.org/10.3390/microorganisms11071665
Chicago/Turabian StyleKang, Sang-Bum, Hyeonwoo Kim, Sangsoo Kim, Jiwon Kim, Soo-Kyung Park, Chil-Woo Lee, Kyeong Ok Kim, Geom-Seog Seo, Min Suk Kim, Jae Myung Cha, and et al. 2023. "Potential Oral Microbial Markers for Differential Diagnosis of Crohn’s Disease and Ulcerative Colitis Using Machine Learning Models" Microorganisms 11, no. 7: 1665. https://doi.org/10.3390/microorganisms11071665
APA StyleKang, S.-B., Kim, H., Kim, S., Kim, J., Park, S.-K., Lee, C.-W., Kim, K. O., Seo, G.-S., Kim, M. S., Cha, J. M., Koo, J. S., & Park, D.-I. (2023). Potential Oral Microbial Markers for Differential Diagnosis of Crohn’s Disease and Ulcerative Colitis Using Machine Learning Models. Microorganisms, 11(7), 1665. https://doi.org/10.3390/microorganisms11071665






