Exploring the Synergistic Potential of Radiomics and Laboratory Biomarkers for Enhanced Identification of Vulnerable COVID-19 Patients
Abstract
1. Introduction
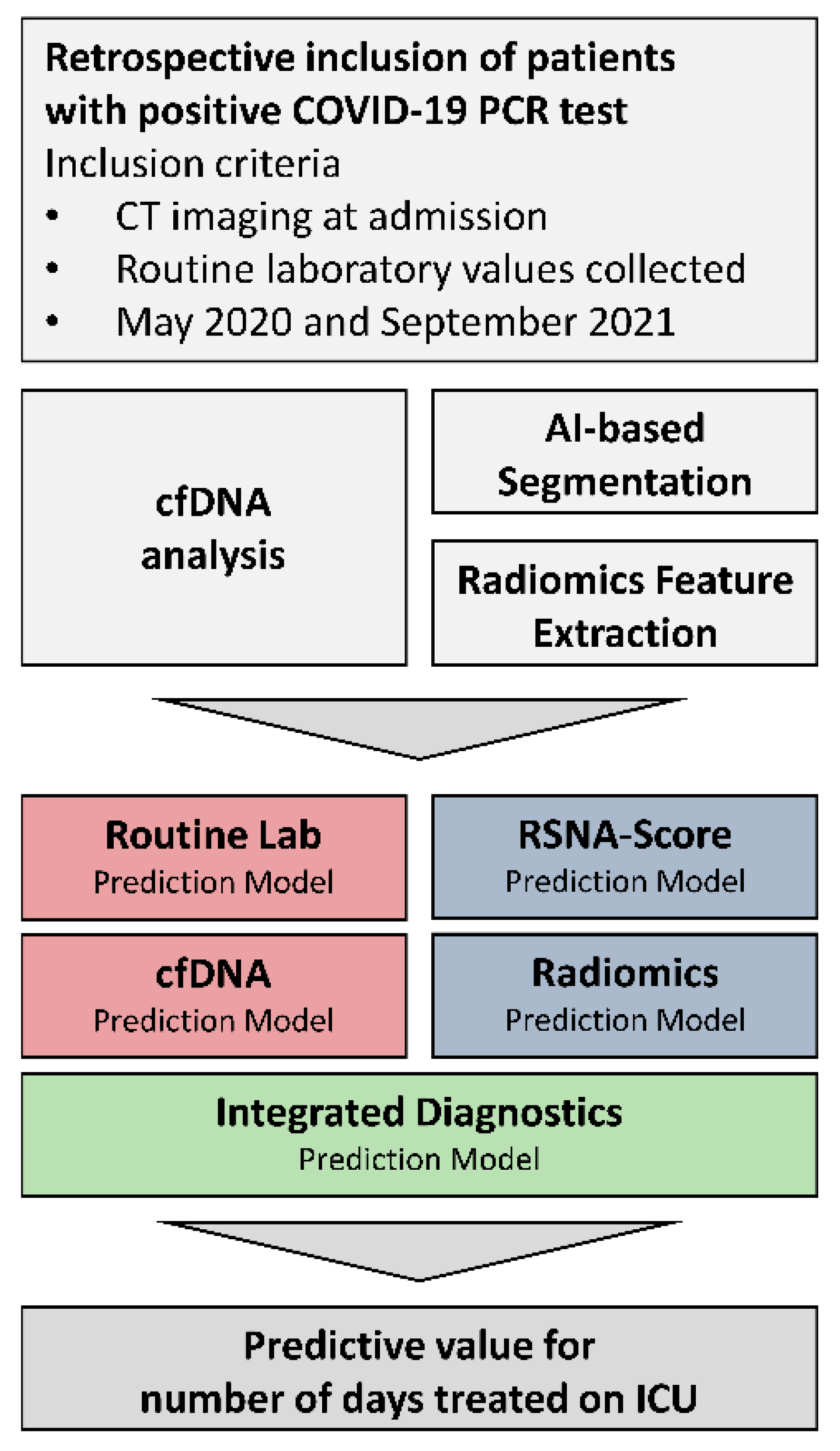
2. Materials and Methods
2.1. Participant Recruitment
2.2. Routine Laboratory Analysis
2.3. Sample Collection and cfDNA Analysis
2.4. Chest CT Imaging
2.5. Chest CT Imaging Analysis
2.6. Performance of Feature Selection and Statistical Analysis
3. Results
3.1. Demographics and Clinical Aspects
3.2. Prognostic Value of Laboratory Parameters
Creation of the Laboratory Prediction Model
3.3. Prognostic Value of Radiological Parameters
Creation of the Radiological Prediction Model
3.4. Prognostic Value of Integrated Diagnostics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALAT | Alanine aminotransferase |
| aPTT | activated partial thromboplastin time |
| ARDS | Acute respiratory distress syndrome |
| ASAT | Aspartate aminotransferase |
| BGA | Blood gas analysis |
| cfDNA | Circulation free Deoxyribonucleic acid |
| CAHA | COVID-19-associated hemostatic abnormalities |
| COVID-19 | Coronavirus disease 2019 |
| CRP | C-reactive protein |
| CT | Computed tomography |
| DIC | Disseminated intravascular coagulopathy |
| EDTA | Ethylene diamine tetraacetic acid |
| GFR | Glomerular filtration rate |
| GGT | Gamma-glutamyltransferase |
| ICU | Intensive Care Unit |
| IL | Interleukin |
| ISTH | International Society of Thrombosis and Hemostatic |
| LC | Lymphocyte count |
| LDH | Lactate dehydrogenase |
| MCH | Mean corpuscular hemoglobin |
| MCHC | Mean corpuscular hemoglobin concentration, |
| MCV | Mean corpuscular volume |
| MOF | Multiple organ failure |
| NC | Neutrophil count |
| NLR | Neutrophil: lymphocyte ratio |
| PCT | Procalcitonin |
| PE | Pulmonary embolism |
| qPCR | quantitative polymerase chain reaction |
| RSNA | Radiological Society of North America |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus type 2 |
| WBC | White blood cells |
References
- Mouffak, S.; Shubbar, Q.; Saleh, E.; El-Awady, R. Recent advances in management of COVID-19: A review. Biomed. Pharmacother. 2021, 143, 112107. [Google Scholar] [CrossRef]
- Wang, Q.; Cheng, J.; Shang, J.; Wang, Y.; Wan, J.; Yan, Y.Q.; Liu, W.B.; Zhang, H.P.; Wang, J.P.; Wang, X.Y.; et al. Clinical value of laboratory indicators for predicting disease progression and death in patients with COVID-19: A retrospective cohort study. BMJ Open 2021, 11, e043790. [Google Scholar] [CrossRef]
- Bivona, G.; Agnello, L.; Ciaccio, M. Biomarkers for Prognosis and Treatment Response in COVID-19 Patients. Ann. Lab. Med. 2021, 41, 540–548. [Google Scholar] [CrossRef]
- Guan, X.; Zhang, B.; Fu, M.; Li, M.; Yuan, X.; Zhu, Y.; Peng, J.; Guo, H.; Lu, Y. Clinical and inflammatory features based machine learning model for fatal risk prediction of hospitalized COVID-19 patients: Results from a retrospective cohort study. Ann. Med. 2021, 53, 257–266. [Google Scholar] [CrossRef]
- Cavalier, E.; Guiot, J.; Lechner, K.; Dutsch, A.; Eccleston, M.; Herzog, M.; Bygott, T.; Schomburg, A.; Kelly, T.; Holdenrieder, S. Circulating Nucleosomes as Potential Markers to Monitor COVID-19 Disease Progression. Front. Mol. Biosci. 2021, 8, 600881. [Google Scholar] [CrossRef]
- Hammad, R.; Eldosoky, M.A.E.R.; Fouad, S.H.; Elgendy, A.; Tawfeik, A.M.; Alboraie, M.; Abdelmaksoud, M.F. Circulating cell-free DNA, peripheral lymphocyte subsets alterations and neutrophil lymphocyte ratio in assessment of COVID-19 severity. Innate Immun. 2021, 27, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Rubin, G.D.; Ryerson, C.J.; Haramati, L.B.; Sverzellati, N.; Kanne, J.P.; Raoof, S.; Schluger, N.W.; Volpi, A.; Yim, J.J.; Martin, I.B.K.; et al. The Role of Chest Imaging in Patient Management during the COVID-19 Pandemic: A Multinational Consensus Statement from the Fleischner Society. Radiology 2020, 296, 172–180. [Google Scholar] [CrossRef]
- Ooi, G.C.; Khong, P.L.; Müller, N.L.; Yiu, W.C.; Zhou, L.J.; Ho, J.C.; Lam, B.; Nicolaou, S.; Tsang, K.W. Severe Acute Respiratory Syndrome: Temporal Lung Changes at Thin-Section CT in 30 Patients. Radiology 2004, 230, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Simpson, S.; Kay, F.U.; Abbara, S.; Bhalla, S.; Chung, J.H.; Chung, M.; Henry, T.S.; Kanne, J.P.; Kligerman, S.; Ko, J.P.; et al. Radiological Society of North America Expert Consensus Document on Reporting Chest CT Findings Related to COVID-19: Endorsed by the Society of Thoracic Radiology, the American College of Radiology, and RSNA. Radiol. Cardiothorac. Imaging 2020, 2, e200152. [Google Scholar] [CrossRef]
- Gouda, W.; Yasin, R. COVID-19 disease: CT Pneumonia Analysis prototype by using artificial intelligence, predicting the disease severity. Egypt. J. Radiol. Nucl. Med. 2020, 51, 196. [Google Scholar] [CrossRef]
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging—“how-to” guide and critical reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef]
- Lippi, G.; Luca Salvagno, G.; Montagnana, M.; Brocco, G.; Guidi, G.C. Influence of hemolysis on routine clinical chemistry testing. Clin. Chem. Lab. Med. CCLM 2006, 44, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Simundic, A.-M.; Baird, G.; Cadamuro, J.; Costelloe, S.J.; Lippi, G. Managing hemolyzed samples in clinical laboratories. Crit. Rev. Clin. Lab. Sci. 2020, 57, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, F.; Seifert, S.; Szymczak, S. Evaluation of variable selection methods for random forests and omics data sets. Brief. Bioinform. 2019, 20, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Acharjee, A.; Larkman, J.; Xu, Y.; Cardoso, V.R.; Gkoutos, G.V. A random forest based biomarker discovery and power analysis framework for diagnostics research. BMC Med. Genom. 2020, 13, 178. [Google Scholar] [CrossRef]
- R: A Language and Environment for Statistical Computing. Statistical Software. Available online: https://posit.co/downloads/ (accessed on 25 June 2023).
- Liu, H.; Ren, H.; Wu, Z.; Xu, H.; Zhang, S.; Li, J.; Hou, L.; Chi, R.; Zheng, H.; Chen, Y.; et al. CT radiomics facilitates more accurate diagnosis of COVID-19 pneumonia: Compared with CO-RADS. J. Transl. Med. 2021, 19, 29. [Google Scholar] [CrossRef]
- Zhang, M.; Zeng, X.; Huang, C.; Liu, J.; Liu, X.; Xie, X.; Wang, R. An AI-based radiomics nomogram for disease prognosis in patients with COVID-19 pneumonia using initial CT images and clinical indicators. Int. J. Med. Inf. 2021, 154, 104545. [Google Scholar] [CrossRef]
- Lassau, N.; Ammari, S.; Chouzenoux, E.; Gortais, H.; Herent, P.; Devilder, M.; Soliman, S.; Meyrignac, O.; Talabard, M.P.; Lamarque, J.P.; et al. Integrating deep learning CT-scan model, biological and clinical variables to predict severity of COVID-19 patients. Nat. Commun. 2021, 12, 634. [Google Scholar] [CrossRef]
- Hectors, S.J.; Riyahi, S.; Dev, H.; Krishnan, K.; Margolis, D.J.A.; Prince, M.R. Multivariate analysis of CT imaging, laboratory, and demographical features for prediction of acute kidney injury in COVID-19 patients: A Bi-centric analysis. Abdom. Radiol. 2021, 46, 1651–1658. [Google Scholar] [CrossRef]
- Weikert, T.; Rapaka, S.; Grbic, S.; Re, T.; Chaganti, S.; Winkel, D.J.; Anastasopoulos, C.; Niemann, T.; Wiggli, B.J.; Bremerich, J.; et al. Prediction of Patient Management in COVID-19 Using Deep Learning-Based Fully Automated Extraction of Cardiothoracic CT Metrics and Laboratory Findings. Korean J. Radiol. 2021, 22, 994. [Google Scholar] [CrossRef]
- Do, T.D.; Skornitzke, S.; Merle, U.; Kittel, M.; Hofbaur, S.; Melzig, C.; Kauczor, H.U.; Wielpütz, M.O.; Weinheimer, O. COVID-19 pneumonia: Prediction of patient outcome by CT-based quantitative lung parenchyma analysis combined with laboratory parameters. PLoS ONE 2022, 17, e0271787. [Google Scholar] [CrossRef]
- Naymagon, L.; Zubizarreta, N.; Feld, J.; van Gerwen, M.; Alsen, M.; Thibaud, S.; Kessler, A.; Venugopal, S.; Makki, I.; Qin, Q.; et al. Admission D-dimer levels, D-dimer trends, and outcomes in COVID-19. Thromb. Res. 2020, 196, 99–105. [Google Scholar] [CrossRef]
- Zhang, L.; Yan, X.; Fan, Q.; Liu, H.; Liu, X.; Liu, Z.; Zhang, Z. D-dimer levels on admission to predict in-hospital mortality in patients with COVID-19. J. Thromb. Haemost. 2020, 18, 1324–1329. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Botran, R.; Furmanek, S.; Ambadapoodi, R.S.; Expósito González, E.; Cahill, M.; Carrico, R.; Akca, O.; Ramírez, J.A. Association and predictive value of biomarkers with severe outcomes in hospitalized patients with SARS-CoV-2 infection. Cytokine 2022, 149, 155755. [Google Scholar] [CrossRef]
- Hoteit, L.; Deeb, A.-P.; Andraska, E.A.; Kaltenmeier, C.; Yazdani, H.O.; Tohme, S.; Neal, M.D.; Mota, R.I. The Pathobiological Basis for Thrombotic Complications in COVID-19: A Review of the Literature. Curr. Pathobiol. Rep. 2021, 9, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Gatto, M.C.; Oliva, A.; Palazzolo, C.; Picariello, C.; Garascia, A.; Nicastri, E.; Girardi, E.; Antinori, A. Efficacy and Safety of Anticoagulant Therapy in COVID-19-Related Pulmonary Embolism with Different Extension. Biomedicines 2023, 11, 1282. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Halalau, A.; Dalal, B.; Abbas, A.E.; Ivascu, F.; Amin, M.; Nair, G.B. Machine learning methods to predict mechanical ventilation and mortality in patients with COVID-19. PLoS ONE 2021, 16, e0249285. [Google Scholar] [CrossRef]
- Chieregato, M.; Frangiamore, F.; Morassi, M.; Baresi, C.; Nici, S.; Bassetti, C.; Bnà, C.; Galelli, M. A hybrid machine learning/deep learning COVID-19 severity predictive model from CT images and clinical data. Sci. Rep. 2022, 12, 4329. [Google Scholar] [CrossRef]
- Giraudo, C.; Frattin, G.; Fichera, G.; Motta, R.; Stramare, R. A practical integrated radiomics model predicting intensive care hospitalization in COVID-19. Crit. Care 2021, 25, 145. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. Comparison of methods for the quantification of cell-free DNA isolated from cell culture supernatant. Tumor Biol. 2019, 41, 101042831986636. [Google Scholar] [CrossRef]
- Simonnet, A.; Chetboun, M.; Poissy, J.; Raverdy, V.; Noulette, J.; Duhamel, A.; Labreuche, J.; Mathieu, D.; Pattou, F.; Jourdain, M. High Prevalence of Obesity in Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) Requiring Invasive Mechanical Ventilation. Obesity 2020, 28, 1195–1199. [Google Scholar] [CrossRef]
- Nishimoto, S.; Fukuda, D.; Higashikuni, Y.; Tanaka, K.; Hirata, Y.; Murata, C.; Kim-Kaneyama, J.R.; Sato, F.; Bando, M.; Yagi, S.; et al. Obesity-induced DNA released from adipocytes stimulates chronic adipose tissue inflammation and insulin resistance. Sci. Adv. 2016, 2, e1501332. [Google Scholar] [CrossRef] [PubMed]
- Menni, C.; Valdes, A.M.; Freidin, M.B.; Sudre, C.H.; Nguyen, L.H.; Drew, D.A.; Ganesh, S.; Varsavsky, T.; Cardoso, M.J.; El-Sayed Moustafa, J.S.; et al. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat. Med. 2020, 26, 1037–1040. [Google Scholar] [CrossRef] [PubMed]
- Gerhards, C.; Thiaucourt, M.; Kittel, M.; Becker, C.; Ast, V.; Hetjens, M.; Neumaier, M.; Haselmann, V. Longitudinal assessment of anti-SARS-CoV-2 antibody dynamics and clinical features following convalescence from a COVID-19 infection. Int. J. Infect. Dis. 2021, 107, 221–227. [Google Scholar] [CrossRef] [PubMed]





| All Patients | Non-ICU Cohort | ICU Cohort | p Value | |
|---|---|---|---|---|
| n = 52 | n = 16 | n = 36 | ||
| Age (mean (SD)) | 68.46 (13.56) | 73.38 (14.94) | 66.28 (12.50) | 0.081 |
| Gender F/M (%) | 22/31 (42.3/57.7) | 10/6 (62.5/37.5) | 12/25 (33.3/66.7) | 0.070 |
| Symptoms | ||||
| Fever (%) | 14 (30.4) | 3 (20.0) | 11 (35.5) | 0.331 |
| Subfebrile (%) | 1 (2.2) | 1 (6.7) | 0 (0.0) | 0.326 |
| Night sweat (%) | 1 (2.2) | 0 (0.0) | 1 (3.2) | 1.000 |
| Reduced condition (%) | 4 (8.7) | 2 (13.3) | 2 (6.5) | 0.587 |
| Diarrhoea (%) | 5 (10.9) | 2 (13.3) | 3 (9.7) | 1.000 |
| Cough (%) | 16 (34.8) | 5 (33.3) | 11 (35.5) | 1.000 |
| Sore throat (%) | 2 (4.3) | 0 (0.0) | 2 (6.5) | 1.000 |
| Dyspnea (%) | 16 (34.8) | 6 (40.0) | 10 (32.3) | 0.744 |
| Fatigue (%) | 7 (15.2) | 2 (13.3) | 5 (16.1) | 1.000 |
| Nausea (%) | 2 (4.3) | 0 (0.0) | 2 (6.5) | 1.000 |
| Anosmia (%) | 3 (6.5) | 1 (6.7) | 2 (6.5) | 1.000 |
| Ageusia (%) | 4 (8.7) | 1 (6.7) | 3 (9.7) | 1.000 |
| Severity-Score (mean (SD)) | 1.65 (1.17) | 1.60 (1.02) | 1.68 (1.26) | 0.837 |
| Treatment | ||||
| ICU days (mean (SD)) | 9.52 [0.00, 22.66] | 0.00 [0.00, 0.00] | 16.17 [7.92, 27.06] | <0.001 |
| Deceased (%) | 3 (5.9) | 0 (0.0) | 3 (8.6) | 0.543 |
| Ventilation (%) | 24 (46.2) | 0 (0.0) | 24 (66.7) | <0.001 |
| CVC (%) | 28 (53.8) | 0 (0.0) | 28 (77.8) | <0.001 |
| Reanimation (%) | 5 (9.6) | 0 (0.0) | 5 (13.9) | 0.308 |
| ICU complex (%) | 28 (53.8) | 0 (0.0) | 28 (77.8) | <0.001 |
| Transfusion | ||||
| erythrocytes/platelets (%) | 12 (23.1) | 0 (0.0) | 12 (33.3) | 0.010 |
| plasma (%) | 1 (1.9) | 0 (0.0) | 1 (2.8) | 1.000 |
| ECMO (%) | 6 (11.5) | 0 (0.0) | 6 (16.7) | 0.160 |
| Hemodiafiltration (%) | 9 (17.3) | 1 (6.2) | 8 (22.2) | 0.245 |
| Tracheostomy (%) | 11 (21.2) | 0 (0.0) | 11 (30.6) | 0.012 |
| Operation (%) | 9 (17.3) | 1 (6.2) | 8 (22.2) | 0.245 |
| Laboratory parameters | ||||
| cfDNA (median [IQR]), ng/mL | 118.85 [70.58, 292.87] | 68.54 [25.73, 93.33] | 220.18 [102.19, 25.54] | <0.001 |
| Quick (mean (SD)), % | 87.75 (17.47) | 90.69 (12.32) | 86.44 (19.34) | 0.424 |
| PTT (median [IQR]), sec. | 25.70 [22.03, 34.92] | 23.60 [22.17, 26.30] | 27.15 [21.65, 38.85] | 0.115 |
| D-dimer (median [IQR]), mg/L | 1.63 [0.76, 3.90] | 1.49 [0.96, 1.73] | 1.93 [0.72, 4.06] | 0.619 |
| Fibrinogen (mean (SD)), g/L | 6.33 (1.91) | 5.08 (NA) | 6.37 (1.94) | NA |
| Platelets (mean (SD)), 109/L | 270.06 (121.64) | 254.25 (130.85) | 277.08 (118.57) | 0.537 |
| RBC (mean (SD)), 1012/L | 3.49 (0.71) | 3.75 (0.61) | 3.38 (0.73) | 0.084 |
| Hemoglobin (mean (SD)), g/dL | 10.31 (2.18) | 10.71 (2.13) | 10.14 (2.21) | 0.389 |
| MCV (mean (SD)), fl | 88.44 (7.36) | 84.06 (7.14) | 90.38 (6.66) | 0.003 |
| MCH (median [IQR]), pg | 30.10 [28.62, 31.02] | 29.05 [27.88, 30.22] | 30.65 [29.23, 31.27] | 0.026 |
| MCHC (mean (SD)), g/dL | 33.49 (1.34) | 33.92 (1.36) | 33.29 (1.31) | 0.118 |
| WBC (median [IQR]), 109/L | 8.71 [6.26, 11.65] | 5.86 [3.92, 8.32] | 9.66 [7.94, 15.05] | <0.001 |
| CRP (median [IQR]), mg/L | 83.50 [41.75, 149.75] | 38.50 [27.00, 76.75] | 95.55 [64.00, 173.00] | 0.001 |
| GFR (mean (SD)), mL/min/1.73 m2 | 62.69 (32.35) | 62.94 (34.60) | 62.58 (31.81) | 0.971 |
| Creatinine (median [IQR]), mg/dL | 1.03 [0.73, 1.62] | 0.92 [0.68, 1.25] | 1.03 [0.75, 1.85] | 0.258 |
| Urea (median [IQR]), mg/dL | 49.10 [37.55, 90.22] | 35.60 [30.92, 58.75] | 52.45 [41.45, 96.95] | 0.012 |
| AST (median [IQR]), U/L | 38.00 [27.00, 61.00] | 29.00 [26.00, 38.00] | 48.00 [32.50, 77.75] | 0.016 |
| ALT (median [IQR]), U/L | 32.00 [23.00, 60.00] | 26.00 [17.00, 40.00] | 39.50 [24.75, 61.50] | 0.094 |
| GGT (median [IQR]), U/L | 96.00 [37.00, 161.00] | 45.00 [28.00, 107.00] | 131.00 [38.75, 181.75] | 0.014 |
| Cholinesterase (mean (SD)), U/L | 5722.41 (2140.03) | 6459.33 (1974.75) | 5588.42 (2169.98) | 0.366 |
| Albumin (mean (SD)), g/L | 24.13 (5.48) | 29.62 (4.02) | 21.84 (4.27) | <0.001 |
| Bilirubin (median [IQR]), mg/dL | 0.41 [0.30, 0.67] | 0.44 [0.28, 0.53] | 0.40 [0.30, 0.82] | 0.464 |
| LDH (median [IQR]), U/L | 382.00 [293.50, 447.00] | 331.00 [228.00, 389.50] | 399.50 [323.50, 469.25] | 0.016 |
| Internal Cross-Validation I | ||||
|---|---|---|---|---|
| Laboratory Values | Dataset 1 Training n = 23 Validation n = 9 | Dataset 2 Training n = 22 Validation n = 10 | Dataset 3 Training n = 22 Validation n = 10 | Ranking frequencies |
| partial thromboplastin time | 0.364 | 0.156 | 0.003 | 3 |
| Albumin | 0.198 | 0.656 | 0.679 | 3 |
| C-reactive protein | 0.903 | 0.712 | 0.462 | 3 |
| gamma-glutamyltransferase | 0.224 | 0.517 | 0.158 | 3 |
| alanine aminotransferase | 0.286 | 0.163 | 2 | |
| Platelets | 0.850 | 0.806 | 2 | |
| white blood cells | 0.375 | 0.467 | 2 | |
| Urea | 0.344 | 1 | ||
| glomerular filtration rate | 0.065 | 1 | ||
| creatinine | 0.512 | 1 | ||
| red blood cells | 0.736 | 1 | ||
| mean corpuscular hemoglobin concentration | 0.482 | 1 | ||
| lactate dehydrogenase | 0.495 | 1 | ||
| Internal cross-validation II | ||||
| Radiomics | dataset 1 training n = 20 validation n = 10 | dataset 2 training n = 20 validation n = 10 | dataset 3 training n = 20 validation n = 10 | Ranking frequencies |
| original_firstorder_10Percentile | 0.921 | 0.643 | 0.508 | 3 |
| original_gldm_LargeDependenceLowGrayLevelEmphasis | 0.236 | 0.565 | 0.484 | 3 |
| original_shape_Maximum2DdiameterSlice | 0.355 | 0.871 | 2 | |
| original_firstorder_Energy | 0.707 | 0.724 | 2 | |
| original_firstorder_TotalEnergy | 0.050 | 0.673 | 2 | |
| original_glcm_ClusterShade | 0.299 | 0.923 | 2 | |
| original_glcm_DifferenceVariance | 0.325 | 0.972 | 2 | |
| original_glrlm_RunEntropy | 0.590 | 0.967 | 2 | |
| original_glrlm_RunLengthNonUniformity | 0.891 | 0.925 | 2 | |
| original_ngtdm_Busyness | 0.117 | 0.493 | 2 | |
| original_ngtdm_Contrast | 0.454 | 0.050 | 2 | |
| original_shape_Elongation | 0.277 | 0.654 | 2 | |
| original_shape_Flatness | 0.214 | 0.387 | 2 | |
| original_shape_LeastAxisLength | 0.094 | 0.462 | 2 | |
| original_shape_MajorAxisLength | 0.461 | 0.961 | 2 | |
| original_shape_Maximum3Ddiameter | 0.519 | 0.823 | 2 | |
| original_firstorder_90Percentile | 0.576 | 1 | ||
| original_glcm_DifferenceEntropy | 0.942 | 1 | ||
| original_glcm_MaximumProbability | 0.849 | 1 | ||
| original_gldm_DependenceNonUniformity | 0.905 | 1 | ||
| original_gldm_SmallDependenceHighGrayLevelEmphasis | 0.235 | 1 | ||
| original_glszm_GrayLevelNonUniformity | 0.731 | 1 | ||
| original_glszm_ZoneEntropy | 0.524 | 1 | ||
| original_shape_SphericalDisproportion | 0.328 | 1 | ||
| original_shape_VoxelVolume | 0.251 | 1 | ||
| original_glcm_ClusterTendency | 0.941 | 1 | ||
| original_glcm_Imc1 | 0.254 | 1 | ||
| original_glcm_MCC | 0.022 | 1 | ||
| original_gldm_GrayLevelNonUniformity | 0.946 | 1 | ||
| original_gldm_SmallDependenceEmphasis | 0.577 | 1 | ||
| original_glszm_GrayLevelVariance | 0.382 | 1 | ||
| original_ngtdm_Complexity | 0.229 | 1 | ||
| original_shape_MinorAxisLength | 0.318 | 1 | ||
| original_shape_SurfaceVolumeRatio | 0.037 | 1 | ||
| original_firstorder_Skewness | 0.564 | 1 | ||
| original_glrlm_GrayLevelNonUniformity | 0.726 | 1 | ||
| original_glszm_GrayLevelNonUniformityNormalized | 0.922 | 1 | ||
| original_glszm_LowGrayLevelZoneEmphasis | 0.666 | 1 | ||
| original_glszm_SizeZoneNonUniformity | 0.492 | 1 | ||
| original_shape_Compactness1 | 0.347 | 1 | ||
| original_shape_Maximum2DdiameterRow | 0.270 | 1 | ||
| Prediction of ICU Demand | |
|---|---|
| Model | Accuracy |
| cfDNA | 0.47 |
| Radiomics | 0.54 |
| Routine lab | 0.74 |
| Integrated diagnostics | 0.87 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gerhards, C.; Haselmann, V.; Schaible, S.F.; Ast, V.; Kittel, M.; Thiel, M.; Hertel, A.; Schoenberg, S.O.; Neumaier, M.; Froelich, M.F. Exploring the Synergistic Potential of Radiomics and Laboratory Biomarkers for Enhanced Identification of Vulnerable COVID-19 Patients. Microorganisms 2023, 11, 1740. https://doi.org/10.3390/microorganisms11071740
Gerhards C, Haselmann V, Schaible SF, Ast V, Kittel M, Thiel M, Hertel A, Schoenberg SO, Neumaier M, Froelich MF. Exploring the Synergistic Potential of Radiomics and Laboratory Biomarkers for Enhanced Identification of Vulnerable COVID-19 Patients. Microorganisms. 2023; 11(7):1740. https://doi.org/10.3390/microorganisms11071740
Chicago/Turabian StyleGerhards, Catharina, Verena Haselmann, Samuel F. Schaible, Volker Ast, Maximilian Kittel, Manfred Thiel, Alexander Hertel, Stefan O. Schoenberg, Michael Neumaier, and Matthias F. Froelich. 2023. "Exploring the Synergistic Potential of Radiomics and Laboratory Biomarkers for Enhanced Identification of Vulnerable COVID-19 Patients" Microorganisms 11, no. 7: 1740. https://doi.org/10.3390/microorganisms11071740
APA StyleGerhards, C., Haselmann, V., Schaible, S. F., Ast, V., Kittel, M., Thiel, M., Hertel, A., Schoenberg, S. O., Neumaier, M., & Froelich, M. F. (2023). Exploring the Synergistic Potential of Radiomics and Laboratory Biomarkers for Enhanced Identification of Vulnerable COVID-19 Patients. Microorganisms, 11(7), 1740. https://doi.org/10.3390/microorganisms11071740





