First Detection of Influenza D Virus Infection in Cattle and Pigs in the Republic of Korea
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Strategy
2.2. Reference Viruses
2.3. Hemagglutination Inhibition Assay
2.4. Real-Time RT-PCR
2.5. Genomic Sequencing and Phylogenetic Analysis
2.6. Data Analysis
3. Results
3.1. Seropositivity of IDV in Cattle and Pigs
3.2. Molecular Detection of IDV RNA in Cattle and Pig Samples
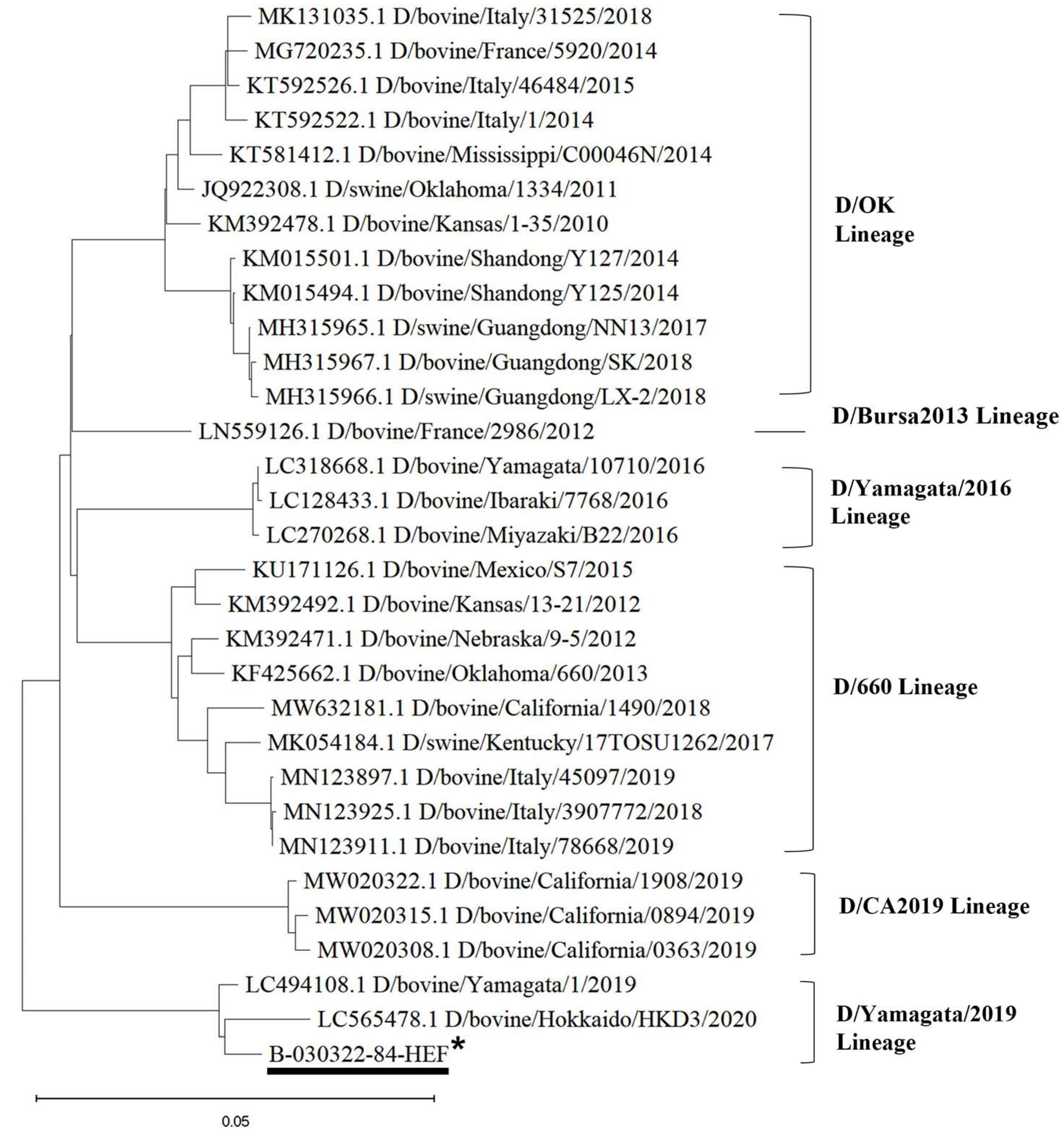
3.3. Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guan, M.; Blackmon, S.; Olivier, A.K.; Zhang, X.; Liu, L.; Woolums, A.; Crenshaw, M.A.; Liao, S.F.; Webby, R.; Epperson, W.; et al. Time-Dependent Proinflammatory Responses Shape Virus Interference during Coinfections of Influenza A Virus and Influenza D Virus. Viruses 2022, 14, 224. [Google Scholar] [CrossRef]
- Gaudino, M.; Chiapponi, C.; Moreno, A.; Zohari, S.; O’Donovan, T.; Quinless, E.; Sausy, A.; Oliva, J.; Salem, E.; Fusade-Boyer, M.; et al. Evolutionary and Temporal Dynamics of Emerging Influenza D Virus in Europe (2009–22). Virus Evol. 2022, 8, veac081. [Google Scholar] [CrossRef]
- Leibler, J.H.; Abdelgadir, A.; Seidel, J.; White, R.F.; Johnson, W.E.; Reynolds, S.J.; Gray, G.C.; Schaeffer, J.W. Influenza D Virus Exposure among US Cattle Workers: A Call for Surveillance. Zoonoses Public Health 2023, 70, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Osterhaus, A.D.; Rimmelzwaan, G.F.; Martina, B.E.; Bestebroer, T.M.; Fouchier, R.A. Influenza B Virus in Seals. Science 2000, 288, 1051–1053. [Google Scholar] [CrossRef]
- Collin, E.A.; Sheng, Z.; Lang, Y.; Ma, W.; Hause, B.M.; Li, F. Cocirculation of Two Distinct Genetic and Antigenic Lineages of Proposed Influenza D Virus in Cattle. J. Virol. 2015, 89, 1036–1042. [Google Scholar] [CrossRef]
- Yesilbag, K.; Toker, E.B.; Ates, O. Recent Strains of Influenza D Virus Create a New Genetic Cluster for European Strains. Microb. Pathog. 2022, 172, 105769. [Google Scholar] [CrossRef] [PubMed]
- Hause, B.M.; Ducatez, M.; Collin, E.A.; Ran, Z.; Liu, R.; Sheng, Z.; Armien, A.; Kaplan, B.; Chakravarty, S.; Hoppe, A.D.; et al. Isolation of a Novel Swine Influenza Virus from Oklahoma in 2011 Which Is Distantly Related to Human Influenza C Viruses. PLoS Pathog. 2013, 9, e1003176. [Google Scholar] [CrossRef] [PubMed]
- Sanogo, I.N.; Kouakou, C.; Batawui, K.; Djegui, F.; Byarugaba, D.K.; Adjin, R.; Adjabli, K.; Wabwire-Mangen, F.; Erima, B.; Atim, G.; et al. Serological Surveillance of Influenza D Virus in Ruminants and Swine in West and East Africa, 2017–2020. Viruses 2021, 13, 1749. [Google Scholar] [CrossRef]
- Nemanichvili, N.; Berends, A.J.; Tomris, I.; Barnard, K.N.; Parrish, C.R.; Gröne, A.; Rijks, J.M.; Verheije, M.H.; De Vries, R.P. Influenza D Binding Properties Vary amongst the Two Major Virus Clades and Wildlife Species. Vet. Microbiol. 2022, 264, 109298. [Google Scholar] [CrossRef]
- Guan, M.; Jacobson, O.; Sarafianos, G.; Baroch, J.; Deliberto, T.J.; Wan, X.-F. Exposure of White-Tailed Deer in North America to Influenza D Virus. Virology 2022, 573, 111–117. [Google Scholar] [CrossRef]
- Jiang, W.-M.; Wang, S.-C.; Peng, C.; Yu, J.-M.; Zhuang, Q.-Y.; Hou, G.-Y.; Liu, S.; Li, J.-P.; Chen, J.-M. Identification of a Potential Novel Type of Influenza Virus in Bovine in China. Virus Genes 2014, 49, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Endoh, M.; Kobayashi, T.; Takenaka-Uema, A.; Chambers, J.K.; Uchida, K.; Nishihara, M.; Hause, B.; Horimoto, T. Influenza D Virus Infection in Herd of Cattle, Japan. Emerg. Infect. Dis. 2016, 22, 1517–1519. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, A.; Umar, S.; Turan, N.; Aydin, O.; Tali, H.E.; Oguzoglu, T.C.; Yilmaz, H.; Richt, J.A.; Ducatez, M.F. First Report of Influenza D Virus Infection in Turkish Cattle with Respiratory Disease. Res. Vet. Sci. 2020, 130, 98–102. [Google Scholar] [CrossRef]
- Chen, Z.; Zeng, Y.; Wei, Y.; Wang, Q.; Liu, M.; Zhang, B.; Liu, J.; Zhu, Q.; Xu, S. Influenza D Virus Matrix Protein 1 Restricts the Type I Interferon Response by Degrading TRAF6. Virology 2022, 568, 1–11. [Google Scholar] [CrossRef]
- Saegerman, C.; Gaudino, M.; Savard, C.; Broes, A.; Ariel, O.; Meyer, G.; Ducatez, M.F. Influenza D Virus in Respiratory Disease in Canadian, Province of Québec, Cattle: Relative Importance and Evidence of New Reassortment between Different Clades. Transbound. Emerg. Dis. 2022, 69, 1227–1245. [Google Scholar] [CrossRef]
- Zhai, S.-L.; Zhang, H.; Chen, S.-N.; Zhou, X.; Lin, T.; Liu, R.; Lv, D.-H.; Wen, X.-H.; Wei, W.-K.; Wang, D.; et al. Influenza D Virus in Animal Species in Guangdong Province, Southern China. Emerg. Infect. Dis. 2017, 23, 1392–1396. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Li, F.; Wang, D. The First Decade of Research Advances in Influenza D Virus. J. Gen. Virol. 2020, 102, jgv001529. [Google Scholar] [CrossRef] [PubMed]
- Korean Statistical Information Service. Available online: https://kostat.go.kr/board.es?mid=a10301010000&bid=225&tag=&act=view&list_no=423107&ref_bid= (accessed on 7 June 2023).
- Kim, T.-Y.; Kim, Y.-S.; Kim, J.-K.; Shon, H.-J.; Lee, Y.-H.; Kang, C.-B.; Park, J.-S.; Kang, K.-S.; Lee, Y.-S. Risk Analysis of Bovine Spongiform Encephalopathy in Korea. J. Vet. Med. Sci. 2005, 67, 743–752. [Google Scholar] [CrossRef]
- Ducatez, M.F.; Pelletier, C.; Meyer, G. Influenza D Virus in Cattle, France, 2011–2014. Emerg. Infect. Dis. 2015, 21, 368–371. [Google Scholar] [CrossRef]
- Sreenivasan, C.; Thomas, M.; Sheng, Z.; Hause, B.M.; Collin, E.A.; Knudsen, D.E.B.; Pillatzki, A.; Nelson, E.; Wang, D.; Kaushik, R.S.; et al. Replication and Transmission of the Novel Bovine Influenza D Virus in a Guinea Pig Model. J. Virol. 2015, 89, 11990–12001. [Google Scholar] [CrossRef]
- Moreno, A.; Lelli, D.; Lavazza, A.; Sozzi, E.; Zanni, I.; Chiapponi, C.; Foni, E.; Capucci, L.; Brocchi, E. MAb-Based Competitive ELISA for the Detection of Antibodies against Influenza D Virus. Transbound. Emerg. Dis. 2019, 66, 268–276. [Google Scholar] [CrossRef] [PubMed]
- WOAH. Manual for Swine Influenza A Virus. Available online: https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/3.09.07_INT_A_SWINE.pdf (accessed on 26 June 2023).
- Snoeck, C.J.; Oliva, J.; Pauly, M.; Losch, S.; Wildschutz, F.; Muller, C.P.; Hübschen, J.M.; Ducatez, M.F. Influenza D Virus Circulation in Cattle and Swine, Luxembourg, 2012–2016. Emerg. Infect. Dis. 2018, 24, 1388–1389. [Google Scholar] [CrossRef]
- Foni, E.; Chiapponi, C.; Baioni, L.; Zanni, I.; Merenda, M.; Rosignoli, C.; Kyriakis, C.S.; Luini, M.V.; Mandola, M.L.; Bolzoni, L.; et al. Influenza D in Italy: Towards a Better Understanding of an Emerging Viral Infection in Swine. Sci. Rep. 2017, 7, 11660. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Stech, J.; Guan, Y.; Webster, R.G.; Perez, D.R. Universal Primer Set for the Full-Length Amplification of All Influenza A Viruses. Arch. Virol. 2001, 146, 2275–2289. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.-M.; Spirason, N.; Iannello, P.; Jelley, L.; Lau, H.; Barr, I.G. A Simplified Sanger Sequencing Method for Full Genome Sequencing of Multiple Subtypes of Human Influenza A Viruses. J. Clin. Virol. 2015, 68, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Trombetta, C.M.; Marchi, S.; Manini, I.; Kistner, O.; Li, F.; Piu, P.; Manenti, A.; Biuso, F.; Sreenivasan, C.; Druce, J.; et al. Influenza D Virus: Serological Evidence in the Italian Population from 2005 to 2017. Viruses 2019, 12, 30. [Google Scholar] [CrossRef]
- White, S.K.; Ma, W.; McDaniel, C.J.; Gray, G.C.; Lednicky, J.A. Serologic Evidence of Exposure to Influenza D Virus among Persons with Occupational Contact with Cattle. J. Clin. Virol. 2016, 81, 31–33. [Google Scholar] [CrossRef]
- Dane, H.; Duffy, C.; Guelbenzu, M.; Hause, B.; Fee, S.; Forster, F.; McMenamy, M.J.; Lemon, K. Detection of Influenza D Virus in Bovine Respiratory Disease Samples, UK. Transbound. Emerg. Dis. 2019, 66, 2184–2187. [Google Scholar] [CrossRef]
- Chiapponi, C.; Faccini, S.; De Mattia, A.; Baioni, L.; Barbieri, I.; Rosignoli, C.; Nigrelli, A.; Foni, E. Detection of Influenza D Virus among Swine and Cattle, Italy. Emerg. Infect. Dis. 2016, 22, 352–354. [Google Scholar] [CrossRef]
- Salem, E.; Cook, E.A.J.; Lbacha, H.A.; Oliva, J.; Awoume, F.; Aplogan, G.L.; Hymann, E.C.; Muloi, D.; Deem, S.L.; Alali, S.; et al. Serologic Evidence for Influenza C and D Virus among Ruminants and Camelids, Africa, 1991–2015. Emerg. Infect. Dis. 2017, 23, 1556–1559. [Google Scholar] [CrossRef]
- Silveira, S.; Falkenberg, S.M.; Kaplan, B.S.; Crossley, B.; Ridpath, J.F.; Bauermann, F.B.; Fossler, C.P.; Dargatz, D.A.; Dassanayake, R.P.; Vincent, A.L.; et al. Serosurvey for Influenza D Virus Exposure in Cattle, United States, 2014–2015. Emerg. Infect. Dis. 2019, 25, 2074–2080. [Google Scholar] [CrossRef] [PubMed]
- Lauterbach, S.E.; Nelson, S.W.; Robinson, M.E.; Lorbach, J.N.; Nolting, J.M.; Bowman, A.S. Assessing Exhibition Swine as Potential Disseminators of Infectious Disease through the Detection of Five Respiratory Pathogens at Agricultural Exhibitions. Vet. Res. 2019, 50, 63. [Google Scholar] [CrossRef]
- Hause, B.M.; Collin, E.A.; Liu, R.; Huang, B.; Sheng, Z.; Lu, W.; Wang, D.; Nelson, E.A.; Li, F. Characterization of a Novel Influenza Virus in Cattle and Swine: Proposal for a New Genus in the Orthomyxoviridae Family. mBio 2014, 5, e00031-14. [Google Scholar] [CrossRef]
- Oliva, J.; Eichenbaum, A.; Belin, J.; Gaudino, M.; Guillotin, J.; Alzieu, J.-P.; Nicollet, P.; Brugidou, R.; Gueneau, E.; Michel, E.; et al. Serological Evidence of Influenza D Virus Circulation Among Cattle and Small Ruminants in France. Viruses 2019, 11, 516. [Google Scholar] [CrossRef]
- Gaudino, M.; Moreno, A.; Snoeck, C.J.; Zohari, S.; Saegerman, C.; O’Donovan, T.; Ryan, E.; Zanni, I.; Foni, E.; Sausy, A.; et al. Emerging Influenza D Virus Infection in European Livestock as Determined in Serology Studies: Are We Underestimating Its Spread over the Continent? Transbound. Emerg. Dis. 2021, 68, 1125–1135. [Google Scholar] [CrossRef]
- Mekata, H.; Yamamoto, M.; Hamabe, S.; Tanaka, H.; Omatsu, T.; Mizutani, T.; Hause, B.M.; Okabayashi, T. Molecular Epidemiological Survey and Phylogenetic Analysis of Bovine Influenza D Virus in Japan. Transbound. Emerg. Dis. 2018, 65, e355–e360. [Google Scholar] [CrossRef]
- Da Silva, M.S.; Mosena, A.C.S.; Baumbach, L.; Demoliner, M.; Gularte, J.S.; Pavarini, S.P.; Driemeier, D.; Weber, M.N.; Spilki, F.R.; Canal, C.W. Cattle Influenza D Virus in Brazil Is Divergent from Established Lineages. Arch. Virol. 2022, 167, 1181–1184. [Google Scholar] [CrossRef] [PubMed]
- Trombetta, C.M.; Montomoli, E.; Di Bartolo, I.; Ostanello, F.; Chiapponi, C.; Marchi, S. Detection of Antibodies against Influenza D Virus in Swine Veterinarians in Italy in 2004. J. Med. Virol. 2022, 94, 2855–2859. [Google Scholar] [CrossRef]
- Ferguson, L.; Luo, K.; Olivier, A.K.; Cunningham, F.L.; Blackmon, S.; Hanson-Dorr, K.; Sun, H.; Baroch, J.; Lutman, M.W.; Quade, B.; et al. Influenza D Virus Infection in Feral Swine Populations, United States. Emerg. Infect. Dis. 2018, 24, 1020–1028. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Li, T.; Wen, Z.; Wu, S.; Wang, Z.; Zheng, J.; Chen, M.; Chen, F.; Wei, W.-K.; Zhai, S.-L.; et al. Identification of D/Yama2019 Lineage-Like Influenza D Virus in Chinese Cattle. Front. Vet. Sci. 2022, 9, 939456. [Google Scholar] [CrossRef] [PubMed]
- Holwerda, M.; Kelly, J.; Laloli, L.; Stürmer, I.; Portmann, J.; Stalder, H.; Dijkman, R. Determining the Replication Kinetics and Cellular Tropism of Influenza D Virus on Primary Well-Differentiated Human Airway Epithelial Cells. Viruses 2019, 11, 377. [Google Scholar] [CrossRef] [PubMed]
- Borkenhagen, L.K.; Mallinson, K.A.; Tsao, R.W.; Ha, S.-J.; Lim, W.-H.; Toh, T.-H.; Anderson, B.D.; Fieldhouse, J.K.; Philo, S.E.; Chong, K.-S.; et al. Surveillance for Respiratory and Diarrheal Pathogens at the Human-Pig Interface in Sarawak, Malaysia. PLoS ONE 2018, 13, e0201295. [Google Scholar] [CrossRef] [PubMed]
- Bailey, E.S.; Choi, J.Y.; Zemke, J.; Yondon, M.; Gray, G.C. Molecular Surveillance of Respiratory Viruses with Bioaerosol Sampling in an Airport. Trop. Dis. Travel. Med. Vaccines 2018, 4, 11. [Google Scholar] [CrossRef] [PubMed]
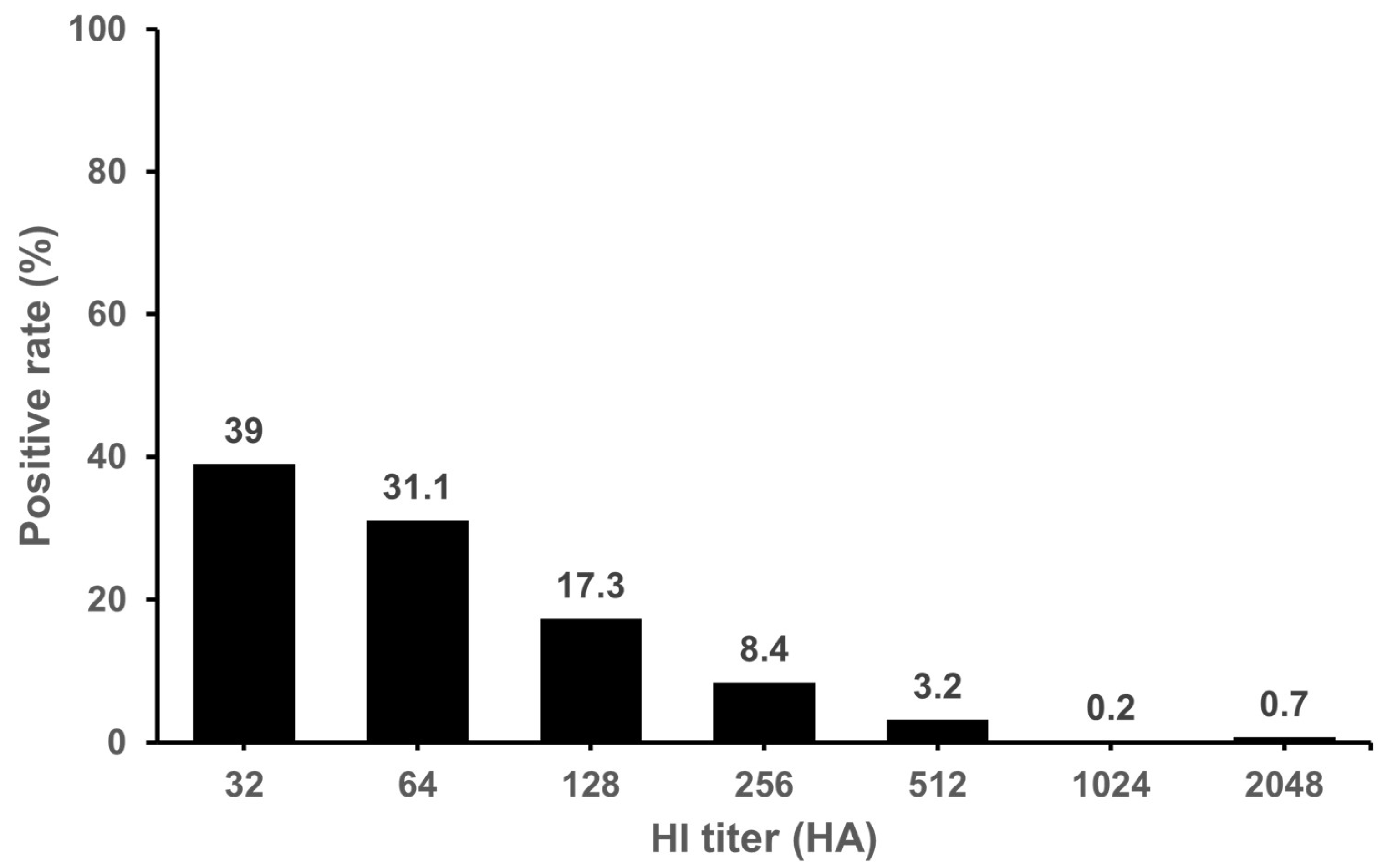
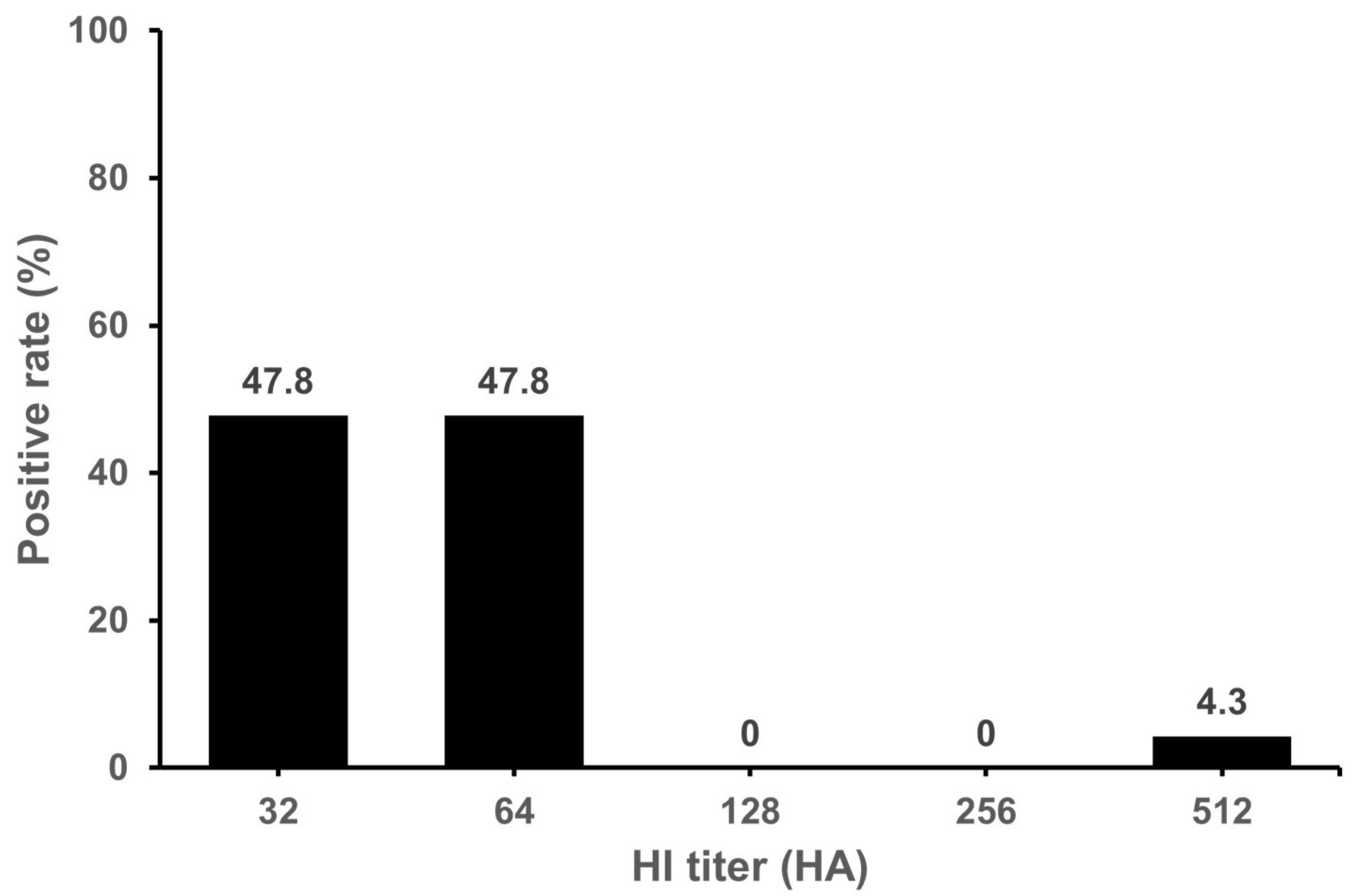
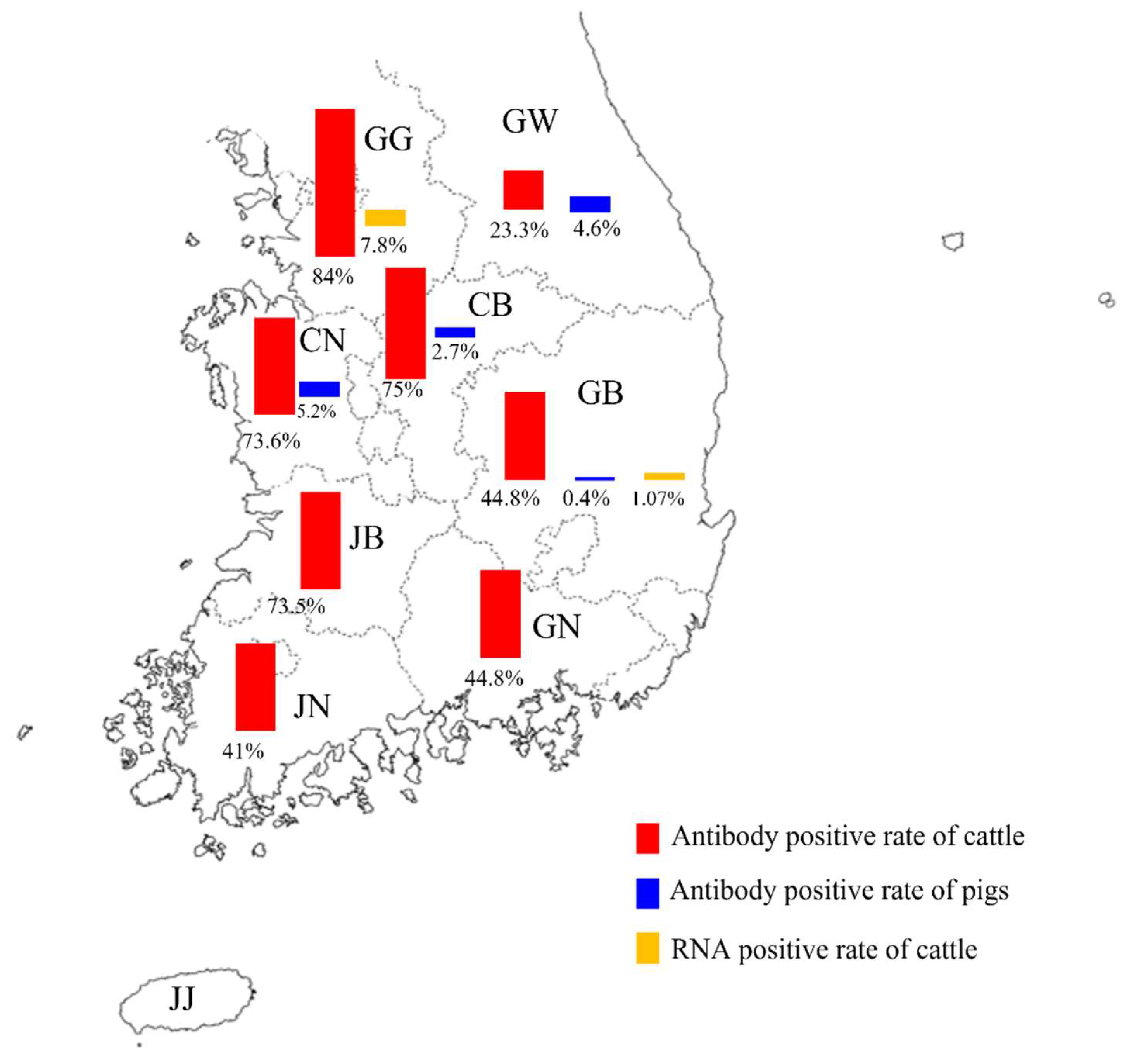
| Province * | Number of Tested Samples | Number of Positive Samples | Positive Rate (%) | GMT |
|---|---|---|---|---|
| Gyeonggi (GG) | 119 | 100 | 84.0 | 70.0 |
| Chungbuk (CB) | 56 | 42 | 75.0 | 69.3 |
| Chungnam (CG) | 72 | 53 | 73.6 | 50.5 |
| Jeonbuk (JB) | 34 | 25 | 73.5 | 73.0 |
| Gyeongbuk (GB) | 261 | 117 | 44.8 | 55.0 |
| Gyeongnam (GN) | 58 | 26 | 44.8 | 82.1 |
| Jeonnam (JN) | 78 | 32 | 41.0 | 113.8 |
| Gangwon (GW) | 43 | 10 | 23.3 | 111.4 |
| Jeju Island (JJ) ** | 21 | 0 | 0.0 | 0.0 |
| Total | 742 | 405 | 54.6 | 68.3 |
| Province * | Number of Tested Samples | Number of Positive Samples | Positive Rate (%) | GMT |
|---|---|---|---|---|
| Chungbuk (CB) | 80 | 5 | 6.3 | 84.4 |
| Gangwon (GW) | 80 | 4 | 5.0 | 45.2 |
| Chungnam (CG) | 259 | 12 | 4.6 | 45.3 |
| Gyeongbuk (GB) | 526 | 2 | 0.4 | 32.0 |
| Gyeonggi (GG) | 339 | 0 | 0 | - |
| Gyeongnam (GN) | 40 | 0 | 0 | - |
| Jeonbuk (JB) | 80 | 0 | 0 | - |
| Jeonnam (JN) | 41 | 0 | 0 | - |
| Jeju Island (JJ) ** | 182 | 0 | 0 | - |
| Total | 1627 | 23 | 1.4 | 48.5 |
| Samples | Cattle | Pigs | ||
|---|---|---|---|---|
| Number of Tested Samples | Number of Positive Samples(Positive Rate %) | Number of Tested Samples | Number of Positive Samples | |
| Lung tissues | 45 | 0 | 159 | 0 |
| Nasal swabs | 954 | 14 (1.5%) | 2232 | 0 |
| Total | 999 | 14 (1.4%) | 2391 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, E.H.; Lim, S.-I.; Kim, M.J.; Kwon, M.; Kim, M.-J.; Lee, K.-B.; Choe, S.; An, D.-J.; Hyun, B.-H.; Park, J.-Y.; et al. First Detection of Influenza D Virus Infection in Cattle and Pigs in the Republic of Korea. Microorganisms 2023, 11, 1751. https://doi.org/10.3390/microorganisms11071751
Lim EH, Lim S-I, Kim MJ, Kwon M, Kim M-J, Lee K-B, Choe S, An D-J, Hyun B-H, Park J-Y, et al. First Detection of Influenza D Virus Infection in Cattle and Pigs in the Republic of Korea. Microorganisms. 2023; 11(7):1751. https://doi.org/10.3390/microorganisms11071751
Chicago/Turabian StyleLim, Eui Hyeon, Seong-In Lim, Min Ji Kim, MiJung Kwon, Min-Ji Kim, Kwan-Bok Lee, SeEun Choe, Dong-Jun An, Bang-Hun Hyun, Jee-Yong Park, and et al. 2023. "First Detection of Influenza D Virus Infection in Cattle and Pigs in the Republic of Korea" Microorganisms 11, no. 7: 1751. https://doi.org/10.3390/microorganisms11071751
APA StyleLim, E. H., Lim, S.-I., Kim, M. J., Kwon, M., Kim, M.-J., Lee, K.-B., Choe, S., An, D.-J., Hyun, B.-H., Park, J.-Y., Bae, Y.-C., Jeoung, H.-Y., Lee, K.-K., & Lee, Y.-H. (2023). First Detection of Influenza D Virus Infection in Cattle and Pigs in the Republic of Korea. Microorganisms, 11(7), 1751. https://doi.org/10.3390/microorganisms11071751






