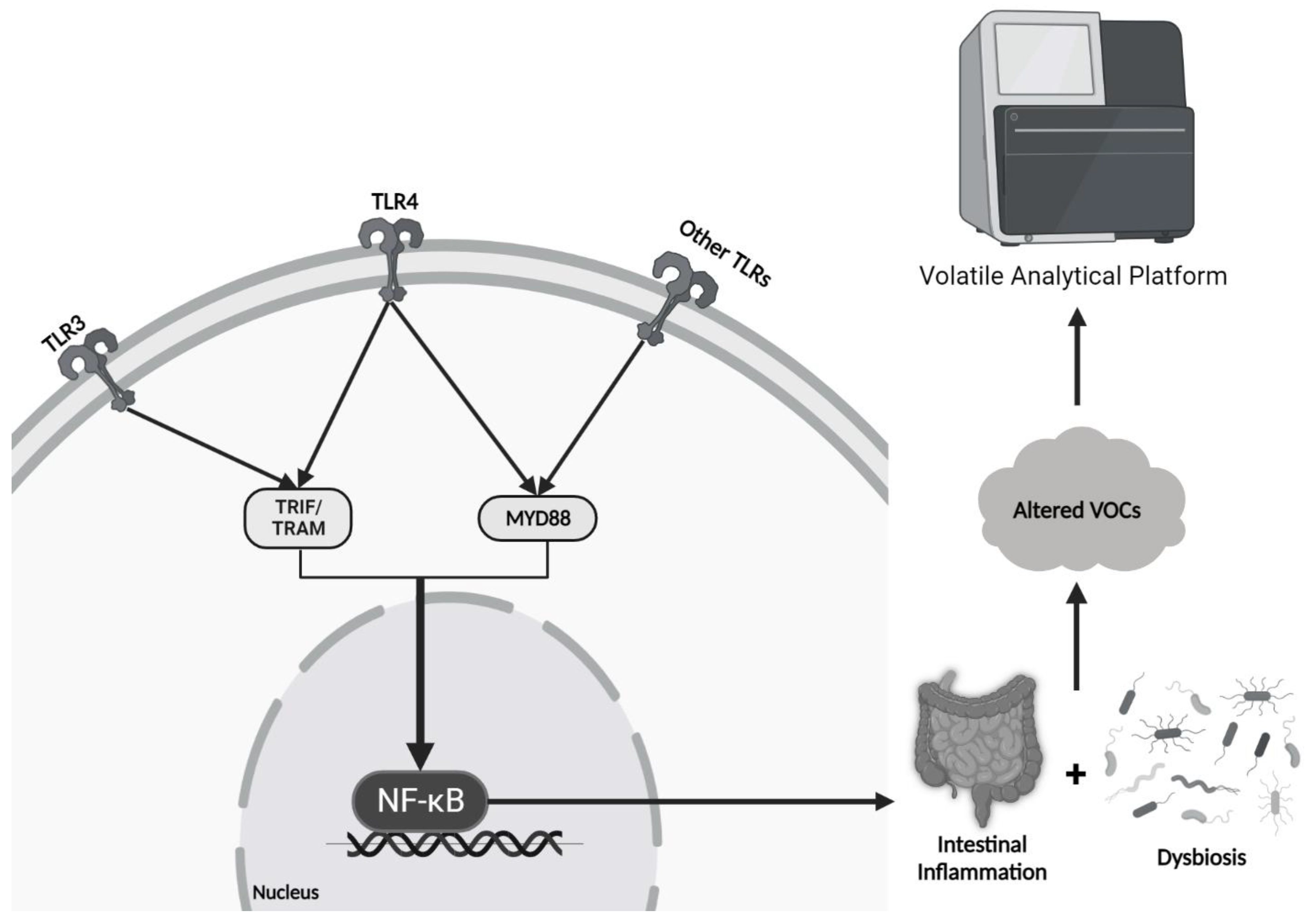
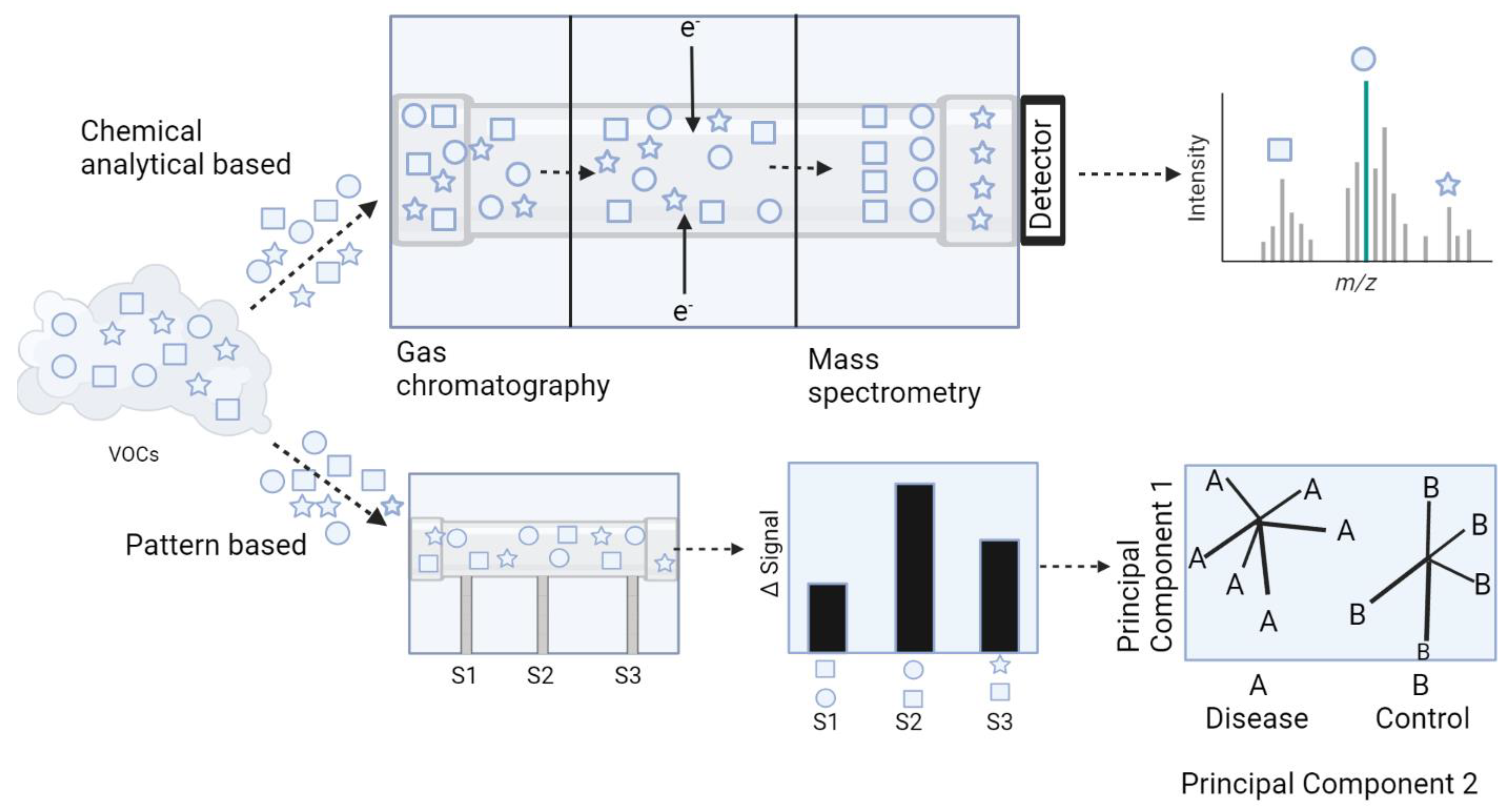
3.1. Inflammatory Bowel Disease and Irritable Bowel Syndrome
Inflammatory bowel disease (IBD) is characterized by chronic inflammation of the GI tract with patterns of flare-ups and remissions. IBD can be split into two main subtypes: ulcerative colitis (UC) and Crohn’s disease (CD). While the pathophysiology of IBD is not completely understood, an association with dysbiosis is presumed [
59]. Alterations in short-chain fatty acid (SCFA)-producing bacteria such as
Clostridium and
Faecalibacterium prausnitzzi, an increase in mucolytic bacteria such as
Ruminococci, and increasing sulfate-reducing bacteria such as
Desulfovibrio have been associated with the symptoms of IBD [
60]. A study conducted by Walton et al. [
61] found increased concentrations of indoles, alcohols, and esters in patients with active CD. Once these patients were appropriately treated, the concentrations decreased to a point that more closely resembled healthy individuals. Additionally, specific VOC biomarkers have been identified that demonstrate promising results for diagnosing and monitoring patients with IBD, namely propan-1-ol and 1-methyl-4-propan-2-ylcyclohexa-1,4-diene [
62]. The dysfunctions associated with IBD include decreased energy supply for enterocytes and degradation of the protective mucosal barrier. These aberrancies lead to enterocyte inflammation and/or death and increased bacterial invasion [
60]. The most common volatile analytical platforms used in detecting IBD via VOC analysis include GC-MS, FAIMS, SIFT-MS, and E-nose technology (
Table 3).
De Meij and colleagues [
63] performed VOC analysis on pediatric patients to differentiate between IBD and healthy controls during active and latent disease states using the Cyranose 320
®, which is a conducting polymer sensor-based E-nose. To accomplish this, a total of 55 children newly diagnosed with IBD (26 with UC and 29 with CD) each provided two stool samples, one at baseline and one during remission. These stool samples were compared to 28 controls. Active disease states were assessed using three parameters: elevated serum C-reactive protein, global physician’s assessment, and fecal calprotectin (FCP) analysis. Results demonstrated that Cyranose 320
® was able to successfully differentiate IBD active disease states from healthy, non-diseased patients. These results also held true when differentiating IBD remission cases from healthy individuals. In a smaller study by De Meij [
64], 19 pediatric patients diagnosed with IBD (10 with UC and 9 with CD) were assessed by E-nose. Stool samples from these patients were compared to pediatric controls that did not have any known GI disease. The results showed that Cyranose 320
® was more aptly able to differentiate the CD cases from the controls compared to the UC cases.
Shepherd et al. [
65] performed fecal headspace VOC profile analysis on healthy patients and those with IBD or irritable bowel syndrome (IBS) using GC coupled to a metal oxide sensor, a type of sensor commonly used in E-nose technology. This hybrid approach was able to differentiate IBD from IBS patients with a sensitivity of 76% and a specificity of 78%. Furthermore, differentiation of IBD patients from healthy individuals yielded a sensitivity of 79%. Similarly, Cauchi et al. [
66] used GC-MS to analyze VOC profiles from patients with CD, UC, IBS, and controls. VOCs were obtained via urine, stool, serum, and breath samples, and they found that stool samples provided the best indicator for disease activity. CD was the most distinguishable condition compared to the others using GC-MS.
Other studies have utilized the Fox 4000 E-nose (MOX-based E-nose) along with FAIMS technology to assess urine VOCs from patients with IBD and healthy controls [
68] Data was assessed via PCA and DFA using the E-nose, and solely by DFA while using FAIMS. PCA and DFA accuracies for the E-nose were 79.4% and 90.4%, respectively. Both modalities yielded an accuracy of over 75%, highlighting a relatively similar capability in differentiating IBD and healthy patients.
VOC analysis has had mixed reviews in its ability to discriminate between active IBD flare and a state of remission. Fecal headspace VOC analysis was performed using GC coupled to IMS (GC-IMS). Researchers classified patients diagnosed with IBD as either in an active disease state or in remission based on fecal calprotectin (FCP) levels [
70]. Patients with FCP levels ≥250 mg/g were considered active, whereas patients with levels <100 mg/g were considered in remission. Results demonstrated high accuracy for differentiating both CD and UC from controls in both active and remission states, smaller differences when attempting to differentiate CD from UC, and no differences between active and remission states within each disease. Despite these poor differences, follow up studies suggested that in the active disease state group, VOC profiles were significantly different between those that were predicted to remain active and those that were predicted to enter remission (AUC 0.86) [
69]. In the remission group, there was also a significant difference in the VOC profile for those that would remain in remission and those that would re-enter an active state (AUC 0.75). This study suggested that analysis of VOC profiles using GC-IMS could potentially predict disease course in patients with IBD.
Other VOC assessment modalities including SIFT-MS and FAIMS have been reported to detect differences in IBD patients. Hicks et al. [
71] conducted a study to identify, quantify, and analyze VOCs emitted from patients with IBD and controls utilizing SIFT-MS. They found that certain VOCs (e.g., dimethyl sulfide, hydrogen sulfide, hydrogen cyanide, ammonia, butanal, and nonanal) were found in significant quantities in IBD-positive patients. Van Gaal et al. [
72] performed a study on pediatric patients with IBD and analyzed fecal headspace VOCs using FAIMS. Results demonstrated that FAIMS was able to successfully differentiate headspace VOCs between CD profiles and controls and UC profiles and controls, but not between CD and UC cases.
Although sometimes confused with inflammatory bowel disease (Crohn’s and ulcerative colitis), irritable bowel syndrome (IBS) is a chronic, functional GI disorder characterized by abdominal pain, bloating, diarrhea (IBS-D), and/or constipation (IBS-C). While the exact pathogenesis has not been established, it is speculated that dysbiosis plays a role in the development of IBS [
73]. While there are conflicting data regarding which exact bacterial species are in abundance or deficient in the setting of IBS, a well-established relationship is the proportionality of
Actinobacteria and
Bifidobacteria [
73]. For example, multiple studies demonstrated that in a high percentage of IBS patients, there is either a significant reduction in
Bifidobacteria or relative abundance in
Actinobacteria [
74,
75,
76,
77,
78]. One study found significantly elevated levels of serum (but not fecal) propionate and butyrate in patients with IBS-D, suggesting that these SCFAs may play a role in the pathogenesis of IBS-D [
79].
Given that IBS is a functional GI disorder, researchers are struggling to significantly differentiate VOC profiles from patients with IBS and controls (
Table 4). Ahmed et al. [
80] analyzed fecal VOC profiles from IBS patients with predominately diarrheal symptoms (IBS-D) using GC-MS. They found the profiles from IBS patients were significantly different from those with IBD and healthy individuals, with AUCs and sensitivities all greater than 90% for each group comparison. Specifically, they found an elevated concentration in organic acids and esters from the IBS-D samples, while alternatively there was an increase in the number of aldehydes in the samples from patients with IBD.
Other studies have used multi-capillary column ion mobility spectrometry (MCC-IMS) to investigate both breath and fecal VOC samples from patients with IBS [
81]. MCC-IMS is a relatively cheaper, more user-friendly version of GC-MS; however, it only allows for pseudo identification of VOCs, and therefore requires combination with other volatile analytical platforms for precise detection of compounds. MCC-IMS was able to successfully differentiate IBS from healthy individuals using both breath and fecal samples.
Given that IBS is a diagnosis of exclusion, it often requires patients to undergo invasive testing to rule out other GI diseases (e.g., IBD). Bosch et al. [
82] performed a study in the pediatric population in an attempt to identify potential fecal VOC biomarkers using FAIMS. Specifically, fifteen patients with IBS, thirty patients with IBD, and thirty healthy controls were recruited, and results demonstrated an AUC of 0.94 when comparing IBS and IBD samples; however, IBS samples could not be reliably differentiated from healthy controls (AUC 0.59). Therefore, FAIMS may be a potential non-invasive method to differentiate IBS from IBD.
3.2. Colorectal Cancer (CRC)
Colorectal cancer is the third most common cancer among men and women and has the second highest mortality rate in cancer-related deaths worldwide. CRC is expensive to diagnose and treat; the total annual cost of CRC in the US was USD 14.1 billion, as measured in 2010 dollars [
83]. In the US, the current most effective screening method for CRC is routine colonoscopy, although many individuals are reluctant to get screened. In 2015, only 6 in 10 people in the US who were eligible received a screening test [
83]. This reluctance has been determined to be multifactorial, and includes, but is not limited to, the invasive nature of colonoscopy, fear of embarrassment, fear of pain and/or catching a disease during the procedure, lack of insurance, and older age [
83,
84]. Therefore, there is a need for a non-invasive screening test for CRC that has high sensitivity and specificity. Indeed, studies investigating potential VOC biomarkers emitted from murine colorectal adenocarcinoma cell lines using SPME and GC-MS have found significant elevations in the concentrations of 1-methoxy-hexane, 2,4-dimethyl-heptane, acetone, butylated hydroxytoluene, and many others [
85,
86]. Bond et al. [
87] performed GC-MS fecal headspace VOC analysis from patients with CRC, adenomatous polyps, and healthy individuals, and found that a person was six times more likely to have CRC if propan-2-ol, hexan-2-one, and ethyl 3-methyl-butanoate were all present in the fecal headspace. The major findings of CRC VOC analysis are summarized in
Table 5.
Arasaradnam et al. [
88] assessed FAIMS ability to differentiate patients with CRC from healthy controls via urine VOC analysis, and results demonstrated a sensitivity of 88% and a specificity of 60%. The relatively higher sensitivity value was promising for screening purposes, even surpassing the values seen in fecal immunochemistry testing (FITs), which is the primary screening method in Europe [
98]. Future clinical applications will likely utilize both FITs and VOC analysis for cancer screening [
99]. For example, Widlak et al. [
100] found a sensitivity and specificity of 80% and 93%, respectively, for detecting CRC using FITs. However, when paired with urinary VOC analysis, a new sensitivity and specificity of 97% and 72% was achieved. This study provides evidence supporting the combined use of VOC analysis with FITs to increase screening accuracy.
Batty et al. [
89] analyzed fecal headspace VOCs using SIFT-MS in patients with a positive fecal occult blood test (FOBT) to determine its efficacy in classifying the stool as high-risk or low-risk for CRC. Diagnoses were confirmed with colonoscopy. They found that SIFT-MS was able to correctly classify 75% of the cases. In a study with a similar design, Mozdiak et al. [
101] compared the classification capability of FAIMS and GC-IMS in patients following a FOBT and found that both FAIMS and GC-IMS were able to classify CRC patients from controls (AUCs 0.98 and 0.82, respectively) and adenomas (AUC range 0.83–0.92); however, classification of adenomas from controls was relatively weak (AUC range 0.54–0.61). However, a conflicting study found that GC-IMS was able to accurately differentiate patients with adenomas from healthy controls but not CRC [
96]. In a separate study comparing GC-MS, FAIMS, and SIFT-MS in detecting CRC and polyps from healthy individuals via urine VOC analysis, GC-MS yielded the highest clinical utility, with a sensitivity and specificity of 88% and an AUC of 0.90 [
93]. These studies provide evidence that following a positive FOBT, SIFT-MS, FAIMS, and/or GC-IMS could potentially be used for risk stratification of colorectal cancer.
Alustiza et al. used magnetic headspace adsorptive extraction (MAG-HSAE) followed by thermal desorption-gas chromatography-mass spectrometry (TD-GC-MS) to investigate potential VOC biomarkers from the stool samples from patients with CRC, adenomatous polyps, and healthy controls [
92]. MAG-HSAE is a subtype of solid-phase extraction technique in which one end of a small neodymium magnet contains graphene oxide decorated with iron oxide magnetic nanoparticles that function as a sorbent, in which VOCs from the headspace can be directly collected and analyzed with a coupled modality (e.g., TD-GC-MS here) [
102]. Using this technique, they found significantly elevated levels of p-Cresol and 3(4H)-dibenzofuranone,4a,9b-dihydro-8,9b-dimethyl- (3(4H)-DBZ) in the CRC group compared to the polyp and control groups. Additionally, they found that p-Cresol is a potential biomarker for premalignant lesions, yielding an AUC 0.69, sensitivity 83%, and specificity 63%.
E-noses and hybrid approaches using E-noses and traditional VOC analytical platforms have also been used to detect CRC. A study by Tyagi et al. used a hybrid approach of the portable E-nose 3 (PEN3), which is a QMB sensor-based E-nose, and GC coupled to time of flight-MS (GC-TOF-MS) in attempt to differentiate urinary VOCs from CRC-positive patients from controls. [
94] TOF-MS has the same purpose as any MS, where particles are separated based on their m/z ratio; however, in TOF-MS, this is achieved by accelerating all particles at the same electric potential and subsequently measuring how long it takes them to reach the detector. Results were promising, with GC-TOF-MS correctly differentiating CRC patients from controls an AUC of 0.81.
De Meij et al. [
97] performed Cyranose 320
® VOC analysis in CRC-positive patients, patients with advanced adenomas, and control patients to differentiate between their disease states. A total of 157 stool samples were collected from individuals who were undergoing an elective colonoscopy, with 60 being from CRC-positive patients, 40 from patients with AAs, and 57 from HCs (determined by no abnormalities detected during colonoscopy). Results of the study demonstrated that fecal VOC profiles differed significantly between CRC-positive patients and HCs and between AAs and HCs.
Given that symptoms of CRC can often mimic IBD, there is a need to develop non-invasive testing to distinguish these [
90]. An optical and electrochemical sensor-based E-nose was used to analyze urine VOCs to differentiate patients with CRC from IBS, and results showed sensitivity and specificity values approaching 80%, highlighting E-nose technology as a potential non-invasive modality for diagnosing CRC. Van Keulen et al. [
91] also performed an E-nose study (Aeonose, MOX) to detect CRC and patients with advanced adenomas (AAs) via exhaled VOC profile analysis after confirmation with colonoscopy. Results demonstrated that the Aeonose yielded a higher AUC, sensitivity, and specificity in the CRC group compared to the AA group, likely indicating exhaled VOC profiles change more as disease progresses.
3.3. Infectious Diarrhea
While many microorganisms are associated with the development of diarrhea, the ones that have been studied with VOC profile analysis include
Clostridioides difficile (previously
Clostridium difficile),
Vibrio cholera,
Campylobacter jejuni, and
Rotavirus, with
C. difficile being the most studied [
57]. While sulfur-containing VOCs are strongly associated with
C. difficile-infected stool, which gives it its characteristic odor, other molecules have also been identified, including straight- and branched-chain carboxylic acids, isocaproic acid, furan species, p-Cresol, and fluorine-containing molecules (e.g., 2-fluoro-4-methylphenol) [
103,
104,
105,
106]. Probert et al. [
104] used GC-MS and found that
C. jejuni-infected stool was shown to be associated with increased concentrations of phenols, organic acids, and indoles, while
Rotavirus-infected stool was associated with ethyl dodecanoate, although the latter relationship was ubiquitous (
Table 6) [
57,
104].
Garner et al. [
67] used GC-MS to identify VOCs in patients with UC,
C. difficile,
C. jejuni infections, and healthy patients, and found hundreds of different volatile biomarkers. Because of the large quantity of VOCs identified, cluster samples for each disease state were made from selected biomarkers via discriminant analysis, resulting in a sensitivity of 96%. Additionally, the total number of VOCs decreased in UC (N = 145),
C. difficile (N = 149), and
C. jejuni (N = 183) compared to healthy individuals (N = 297), and it is hypothesized that this finding is likely due to the faster flow of stool through the GI tract in diseased states, and therefore there is less time for VOC production from the microbiome.
As opposed to using clusters of VOCs per disease state, Tait et al. [
106] used SPME coupled to GC-MS to isolate a single volatile biomarker in patients with C. difficile infection: 2-fluoro-4-methylphenol. Although they were able to do so with a high sensitivity and specificity, the process took approximately 18 h in duration, greatly lowering its clinical utility as there are currently faster, more practical modalities for diagnosing
C. difficile infections (e.g., E-noses and PCR testing).
Given the difficulty associated with traditional analytical systems, E-noses have potential for diagnosis of infectious colitis. McGuire et al. [
107] used GC, an MOX sensor-based E-nose, and an artificial neural network (ANN) software for volatile profiling in patients with
C. difficile infection and HCs. This method yielded a sensitivity and specificity of 85% and 80%, respectively. In a similar study by Chan et al. [
108] using only Enose technology (Aeonose, specifically) coupled to an ANN, results were similar, yielding a sensitivity and specificity of 80% and 85%, respectively. These studies provide evidence in favor of using pattern recognition of VOC clusters as opposed to the individual biomarker identification seen in GC-MS.
3.4. Celiac Disease
Celiac disease (CelD) occurs when a susceptible individual consumes gluten and a T-cell-mediated immune reaction develops in response, resulting in small intestinal inflammation and potentially chronic malabsorption [
109]. Diagnosis typically depends on serology to look for disease-specific antibodies (e.g., anti-tissue transglutaminase IgM antibodies) and duodenal biopsy to look for intraepithelial lymphocytosis, villous atrophy, and crypt hyperplasia [
110]. The use of headspace VOCs as a potential diagnostic modality is an area of increasing research, as this would eliminate the need for the current invasive methods. A pilot study conducted by Di Cagno et al. [
111] showed that gluten-free diets in children with diagnosed CelD resulted in modulation of the microbiota and subsequent VOC profiles, which was analyzed with bacterial 16S-DNA sequencing and GC-MS-SPME, respectively. This study demonstrated that dietary intake may not only impact the microbiota, but also the headspace VOC profiles. Indeed, McFarlane et al. [
112] used both FAIMS and GC-TOF-MS to analyze urinary VOC profiles from patients with CelD on a gluten-free diet, patients with CelD consuming 3 g of gluten per day, and HCs, and found that minimal gluten exposure to patients with CelD resulted in significant VOC profile changes. Specifically, six VOCs were persistently altered for 2 weeks post-exposure, with one VOC (N-methyltaurine) remaining altered after the 2-week mark.
Table 7 summarizes the studies using VOC profiles to diagnose CelD.
Rouvroye et al. [
113] performed GC-IMS fecal headspace VOC analysis in patients with CelD on a gluten-free diet, refractory CelD (RCD), and HCs. Results showed a significant VOC profile difference between the CelD and RCD groups as well as the CelD and HC groups. However, there were no significant differences seen between the RCD and HC groups (
p = 0.310). This study is limited by the relatively low number of participants (30 total), but provides evidence in favor of using VOC profiles as a potential non-invasive diagnostic modality for RCD.
CelD can often be mistaken for IBS, given that symptoms often overlap and can be difficult to differentiate without invasive testing. To identify a potential non-invasive method, Arasaradnam et al. [
114] performed FAIMS urinary VOC analysis in patients with CelD and IBS-D and found that it was able to differentiate them with high accuracy, sensitivity, and specificity. Additionally, GC-MS was performed on fecal samples to identify potential VOC biomarkers, and they found that 1,3,5,7-cyclooctatetraene was only found associated with CelD and not IBS-D. This study provides evidence in favor of using FAIMS to differentiate CelD from IBS-D and that the presence of 1,3,5,7-cyclooctatetraene in CelD specimens requires further validation.
3.5. Late-Onset Sepsis and Necrotizing Enterocolitis
While it is important to detect VOC profile differences in already-diseased individuals from controls, a far more clinically useful application would be predicting disease onset. Two diseases that show promise thus far are neonatal late-onset sepsis and necrotizing enterocolitis. Neonatal late-onset sepsis (LOS) is defined as sepsis that occurs at least 72 h after birth. In developed and developing countries, coagulase-negative
Staphylococci (CONS) spp. are the leading cause of LOS, accounting for 53.7–77.7% and 35.5–47.4% of cases, respectively [
115]. Following CONS, in the United States, the most frequently reported LOS-causing pathogens include
S. aureus,
Candida spp.,
E. coli, Klebsiella spp., and
Enterobacter spp., with Gram-positive organisms being more common, but Gram-negative organisms having a higher mortality rate [
115]. LOS primarily affects preterm infants, having incidence rates that vary anywhere from 20 to 38% in the first 4 months of life and a mortality rate between 13 and 19% [
116]. This makes LOS one of the leading causes of death in neonatal intensive care units (NICUs). Infants that do survive LOS are at an increased risk for long-term effects, including neurodevelopmental delay (i.e., cerebral palsy, vision impairment, poor psychomotor skills, impaired head growth), growth abnormalities, intestinal and respiratory infections, and others [
116,
117]. While there are screening options for LOS, they often involve invasive procedures, which ironically increase the risk of developing LOS independently [
118]. Central venous catheters (CVCs) have been attributed to being a major source of LOS, although studies have demonstrated that the organisms isolated from blood cultures do not perfectly match genetically with the bacteria cultured from the catheters [
119,
120]. Instead, there is a higher genetic similarity between the cultured LOS pathogen and isolates from the GI tract, indicating that the gut is a potential origin of these pathogens [
121]. Using the notion that dysbiosis precedes LOS, studies have shown that isolated pathogens from blood cultures can be detected in the GI tract several days before the onset of clinical symptoms (
Table 8) [
122,
123,
124,
125].
Berkhout et al. [
126] performed a prospective, multicenter cohort study to analyze fecal headspace VOCs from preterm infants born at a gestational age (GA) ≤ 30 weeks. Fecal samples from preterm infants were collected daily from the NICU up until a postnatal age of 28 days old. A total of 843 preterm infants were included, with 127 cases of LOS and 127 matched controls. The most frequent causative pathogens included
E. coli,
Staph aureus, and
Staph epidermidis. FAIMS was able to significantly differentiate fecal VOCs 3 days (t
-3), 2 days (t
-2), and 1 day (t
-1) preceding the onset of clinical symptoms for all three pathogens.
As discussed earlier, there is evidence suggesting a higher genetic similarity between pathogens that cause LOS and isolates from the GI tract rather than isolates from blood cultures associated with CVCs. In a similar methodology as the previous study described, Berkhout et al. [
127] used FAIMS to analyze fecal headspace VOCs from preterm infants with LOS without a CVC and HCs. FAIMS was able to significantly differentiate VOC profiles at t
-3 and t
-1, but not at t
-2 (
p = 0.061). This study provides further evidence supporting aberrations in the gut microbiota may play a role in the pathogenesis of LOS.
Frerichs et al. [
128] Previous studies using GC-IMS to predict preclinical onset of neonatal LOS found that fecal VOC profiles were most profoundly different at two days prior to clinical onset (t
-2) and one day prior to clinical onset (t
-1) for
E. coli LOS and at three days prior to clinical onset (t
-3) and t
-2 for all Gram-negative LOS. Additionally, GC-TOF-MS was used to identify unique metabolites. For proper discrimination of Gram-negative LOS from HCs, ethyl acetate, ethyl 2-(methylamino)acetate, ethyl 2-hydroxypropanoate, prop-1-ene, butane-2,3-dione, and 2,2,4,4-tetramethylpentane were found to be most important, while heptanal was found to be the most important for the discrimination of CONS-LOS. Fecal VOC profiles of 36 infants with LOS were compared to 40 matched controls using Cyranose 320
® [
121]. Results showed that the Cyranose 320
® was able to discriminate between LOS-positive infants from the controls 3 days, 2 days, and 1 day before the clinical onset of LOS. However, the E-nose was unable to differentiate between LOS-positive infants from controls beyond 3 days prior to LOS onset.
Necrotizing Enterocolitis (NEC) is a gastrointestinal emergency primarily affecting preterm, formula-fed infants, and is characterized by intestinal inflammation that can result in necrosis and perforation of the bowel, sepsis, and death. The total annual cost in the US is estimated to be between USD 500 million and USD 1 billion, largely due to the current diagnostic methods used, prolonged hospital admission required for treatment, and the possible need for surgery [
129]. While the pathophysiology of NEC is not completely understood, there are established risk factors, including preterm delivery, low birth weight, formula-feeding as opposed to breastfeeding, and dysbiosis [
130]. In 2009, Garner et al. [
131] performed a pilot study investigating potential VOC biomarkers associated with NEC via SPME coupled to GC-MS. Results of the study demonstrated a decreased total number of VOCs along with an absence of 2-ethylhexyl acetic ester, decanoic acid ethyl ester, dodecanoic acid ethyl ester, and hexadecanoic acid ethyl ester in patients with NEC compared to controls, suggesting VOC profiles could potentially be used in screening for NEC. Indeed, Probert et al. [
132] used SPME coupled to GC-MS to identify potential fecal VOC biomarkers associated with the onset of NEC, and found a strong prediction accuracy (AUC 0.75–0.76) associated with (Z)-hept-2-enal, pent-1-en-3-one, 2-ethylfuran, pentanal, and 2-pentylfuran up to 3–4 days prior to clinical onset. Furthermore, Hosfield et al. [
133] performed 16S rRNA microbial sequencing and fecal VOC analysis of mouse pups with with NEC and breastfed controls using Cyranose 320
® and found a significant abundance of
Lactobacillus and decreased concentration of
E. coli in the HCs compared to the pups with NEC (
p < 0.05).
Diagnosis of NEC relies primarily on abdominal X-rays to detect the presence of air in the intestines, portal vein of the liver, and/or peritoneum as well as other diagnostic and clinical markers [
134]. Not only are X-rays expensive and expose the infant to radiation, but they are also not ordered until clinical symptoms of NEC are exhibited, often when the disease has already significantly progressed. Thus, VOC analysis provides a potential solution for earlier detection of NEC (
Table 9).
De Meij et al. [
135] assessed VOCs from frozen stool samples of 128 preterm neonates who had NEC, sepsis, or were considered healthy controls [
84]. VOC profile analysis of these stool samples was performed with Cyranose 320
®. Fecal samples were clustered into three time windows: 5 and 4 days before diagnosis (t
-5,
-4), 3 and 2 days before diagnosis (t
-3,
-2), and 1 day before and the day of diagnosis (t
-1,
0). Results demonstrated that the E-nose was able to accurately discriminate the VOC profiles between infants with NEC from the controls at both t
-3,
-2 and at t
-1,
0. The accuracy of the VOC profile analysis increased as time approached NEC clinical onset. Additionally, NEC VOC profiles were significantly differentiated from the VOC profiles of the infants with sepsis at t
-3,
-2 but not at t
-1,
0. This study supports the use of E-noses as a screening test for NEC from 2–3 days before the onset of symptoms.