Microbial Communities in Ferromanganese Sediments from the Northern Basin of Lake Baikal (Russia)
Abstract
:1. Introduction
2. Materials and Methods
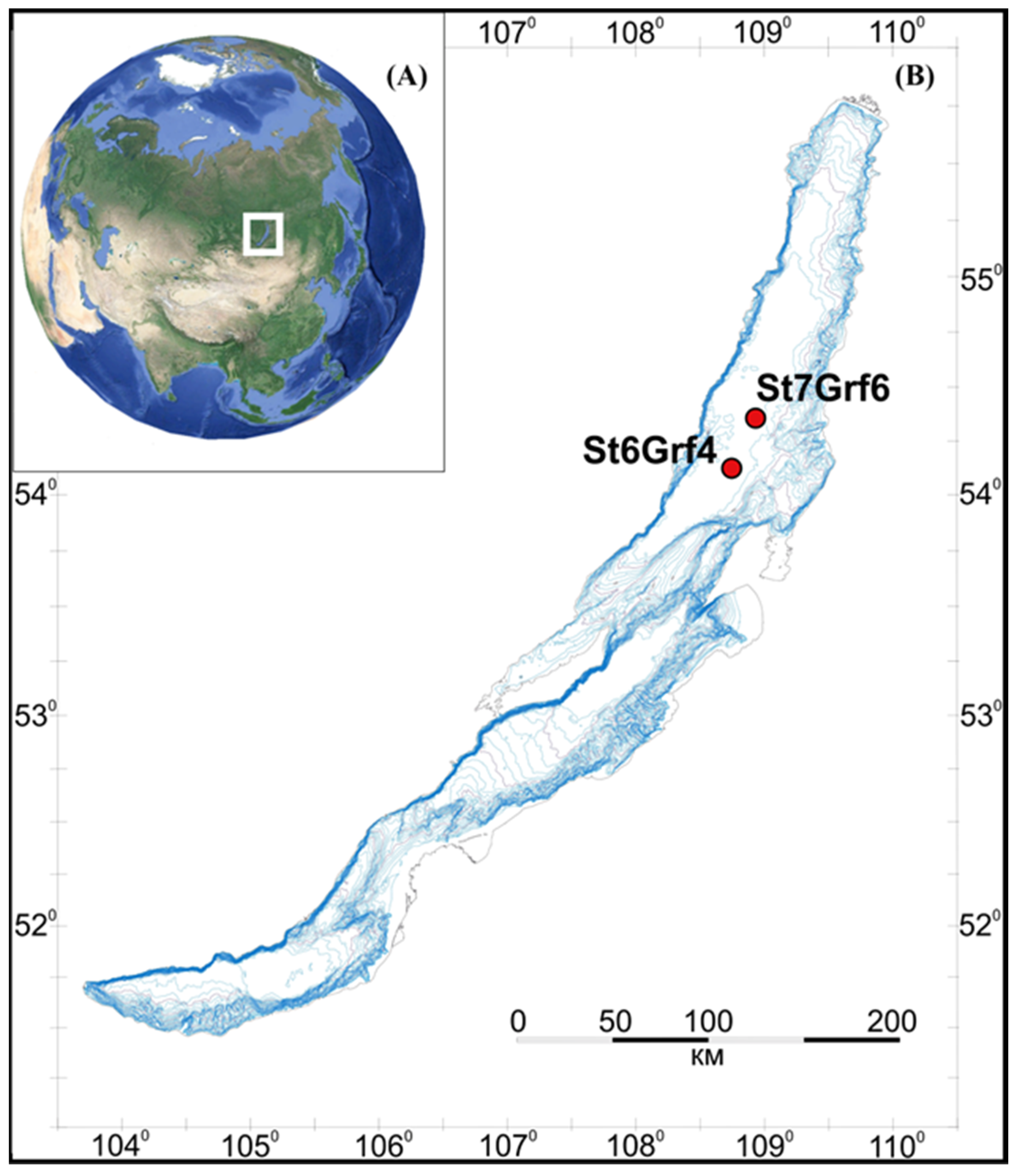
2.1. Description of Sampling Sites, Sample Collection, and Pretreatment
2.2. Chemical Analysis
2.3. Methane Concentration
2.4. DNA Isolation, PCR, and Sequencing of 16S rRNA Genes
2.5. 16S rRNA Sequence Processing and Statistical Analysis
2.6. Preparation of Enrichment Culture from the Sedimentary Layer Enriched in Fe and Mn for Genomic Analysis
2.7. Shotgun Sequencing, Assembly, Binning, and Annotation
3. Results
3.1. Lithology
3.2. Chemical Composition of Pore Water
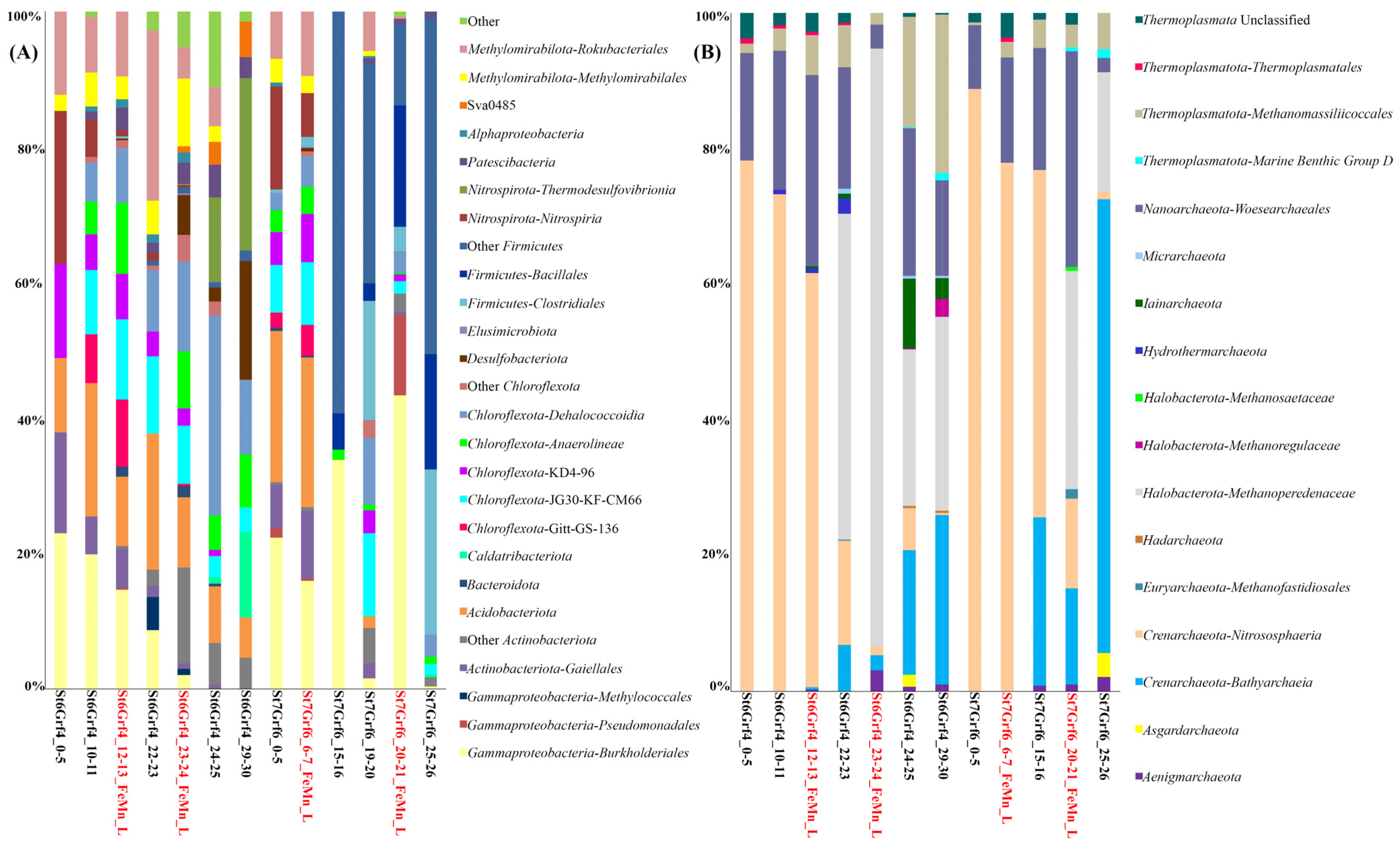
3.3. Alfa Diversity
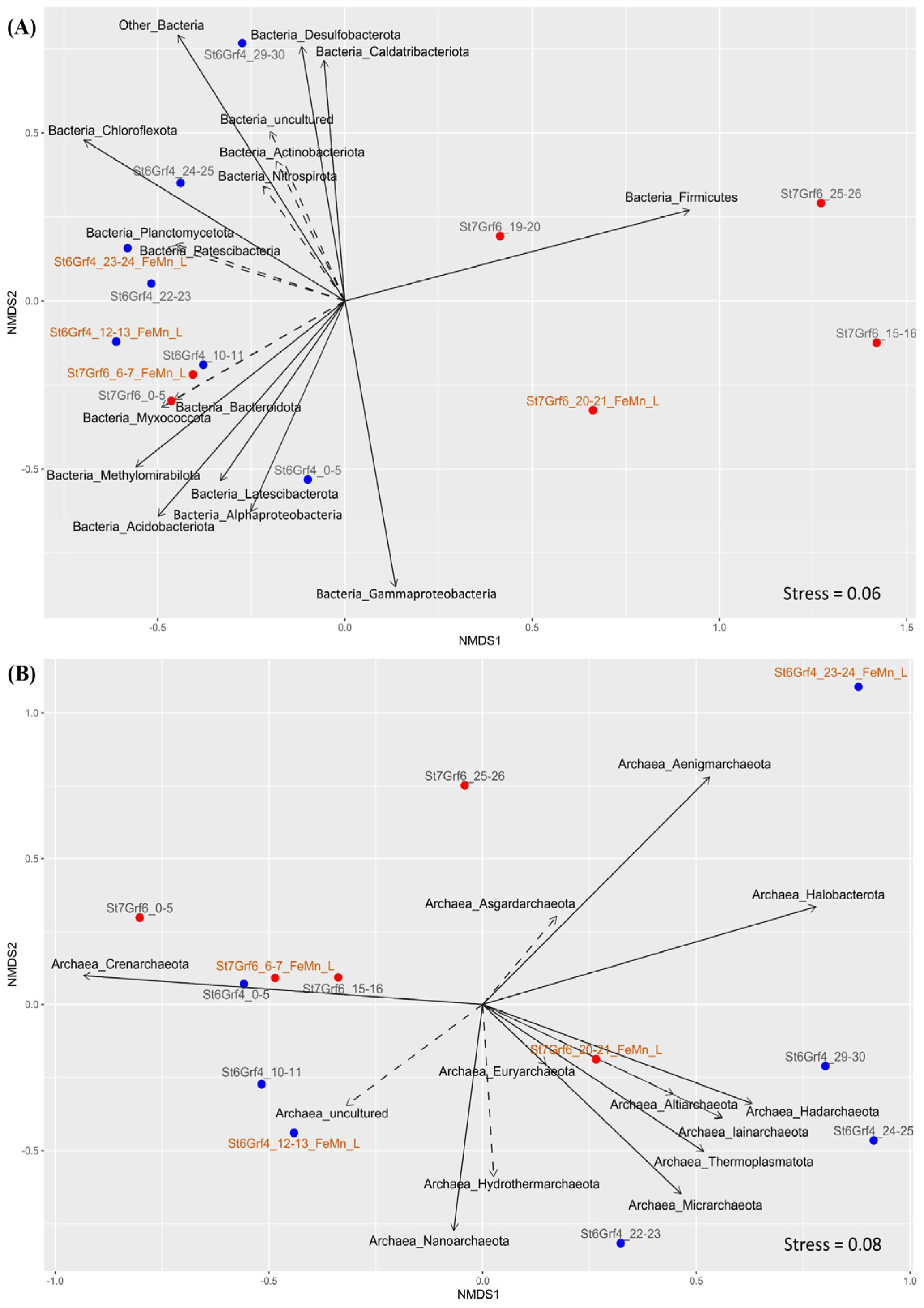
3.4. Beta Diversity
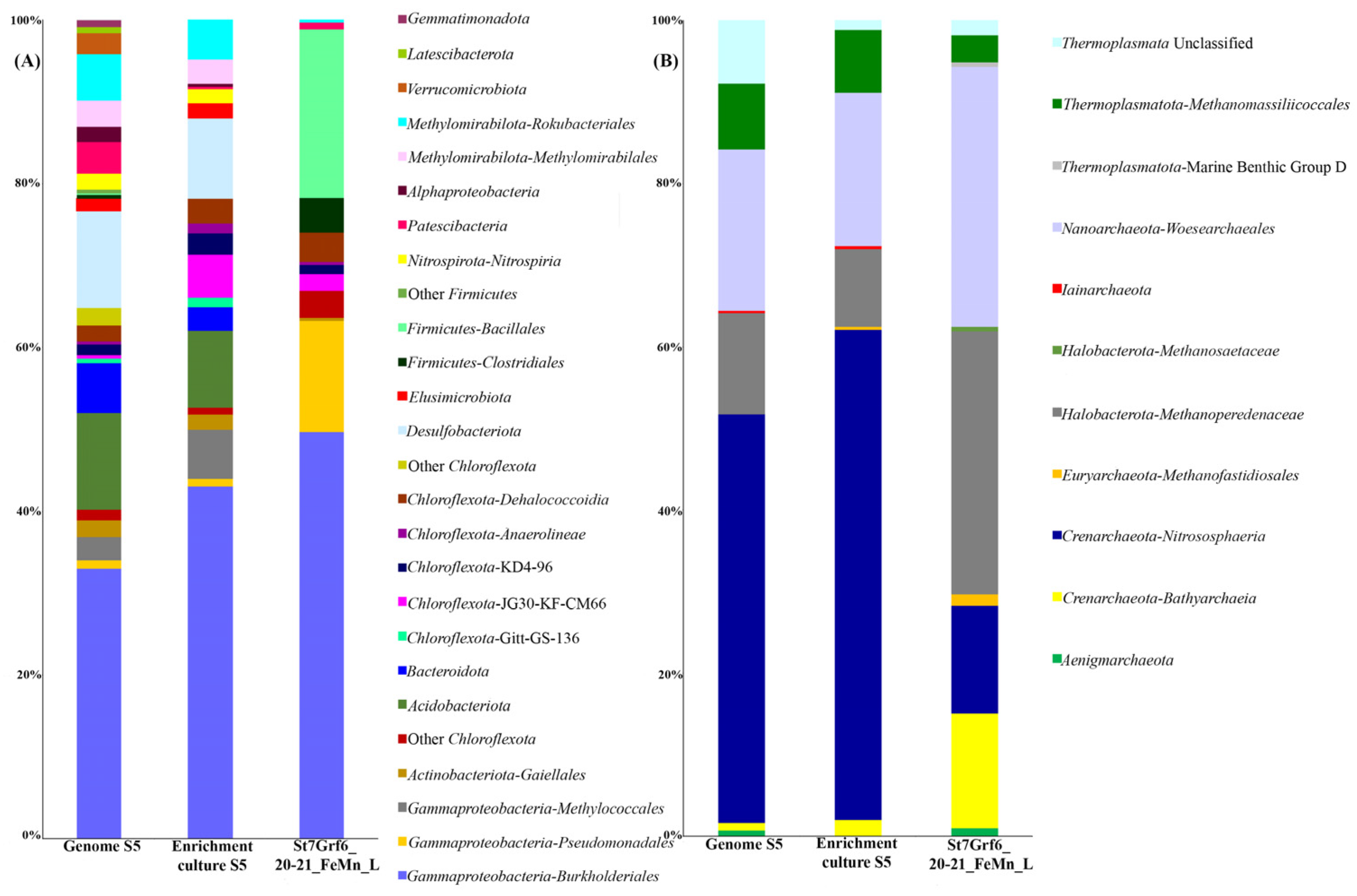
3.5. Genomic Research
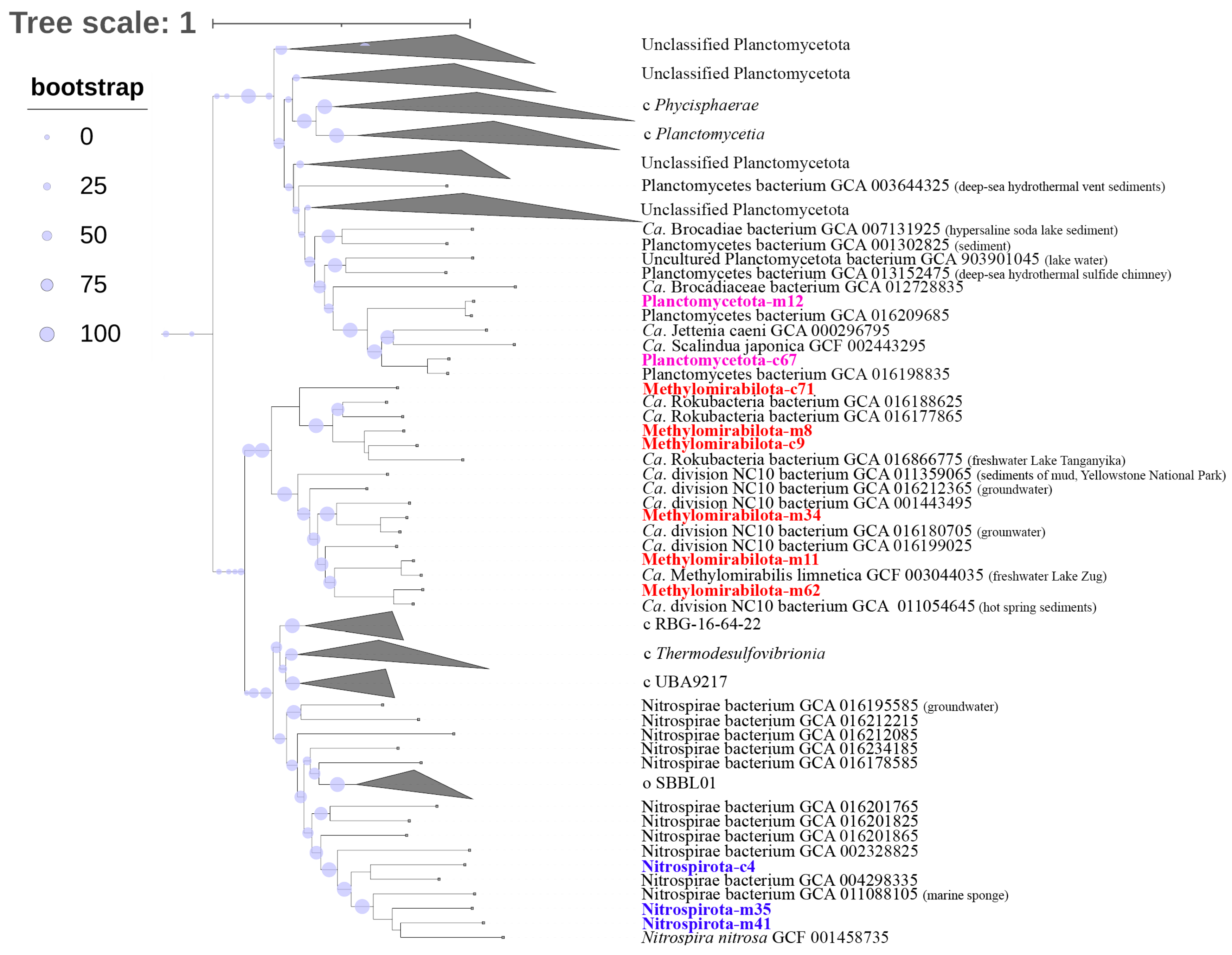
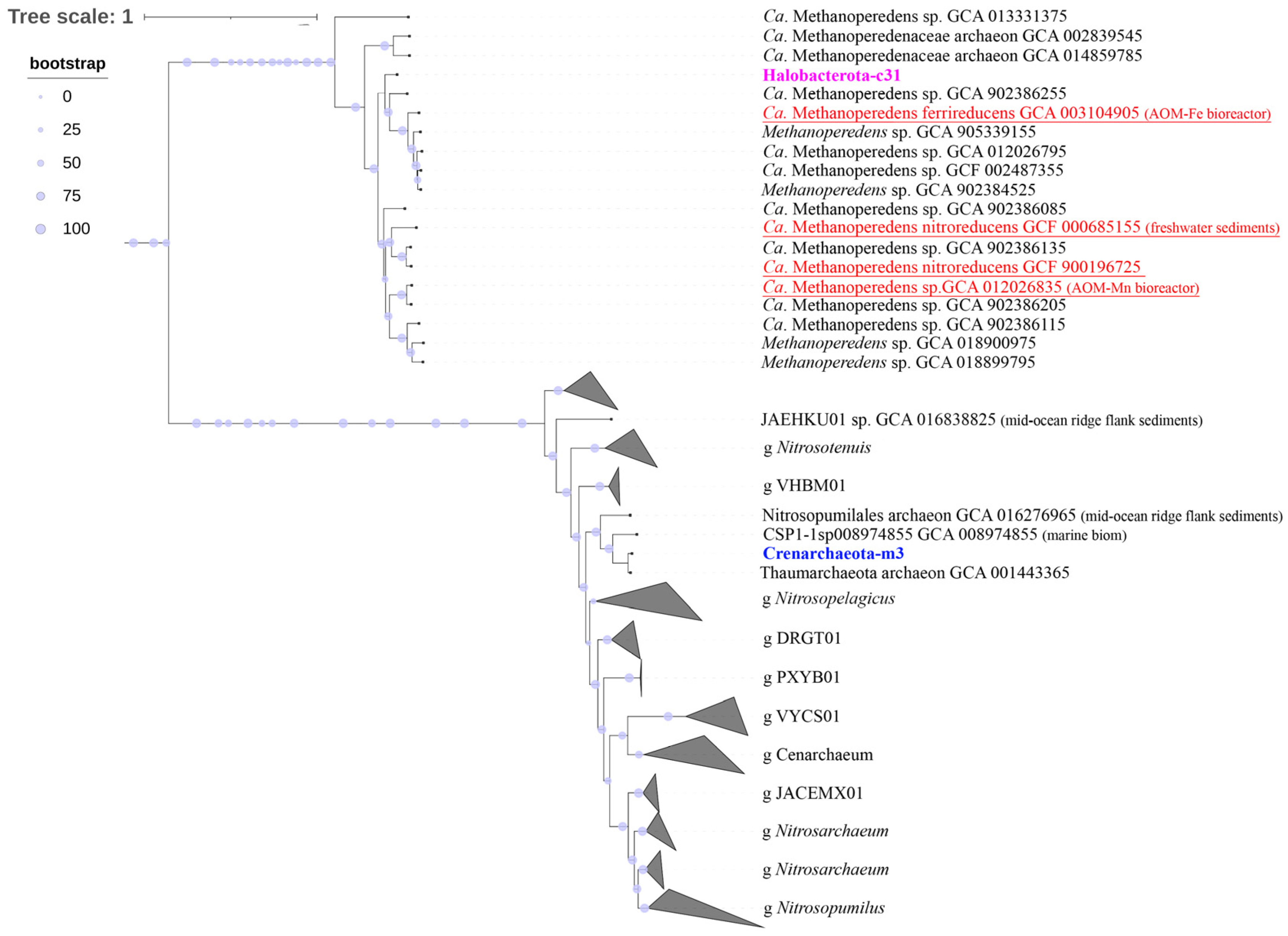
3.6. Phylogenomic Analysis
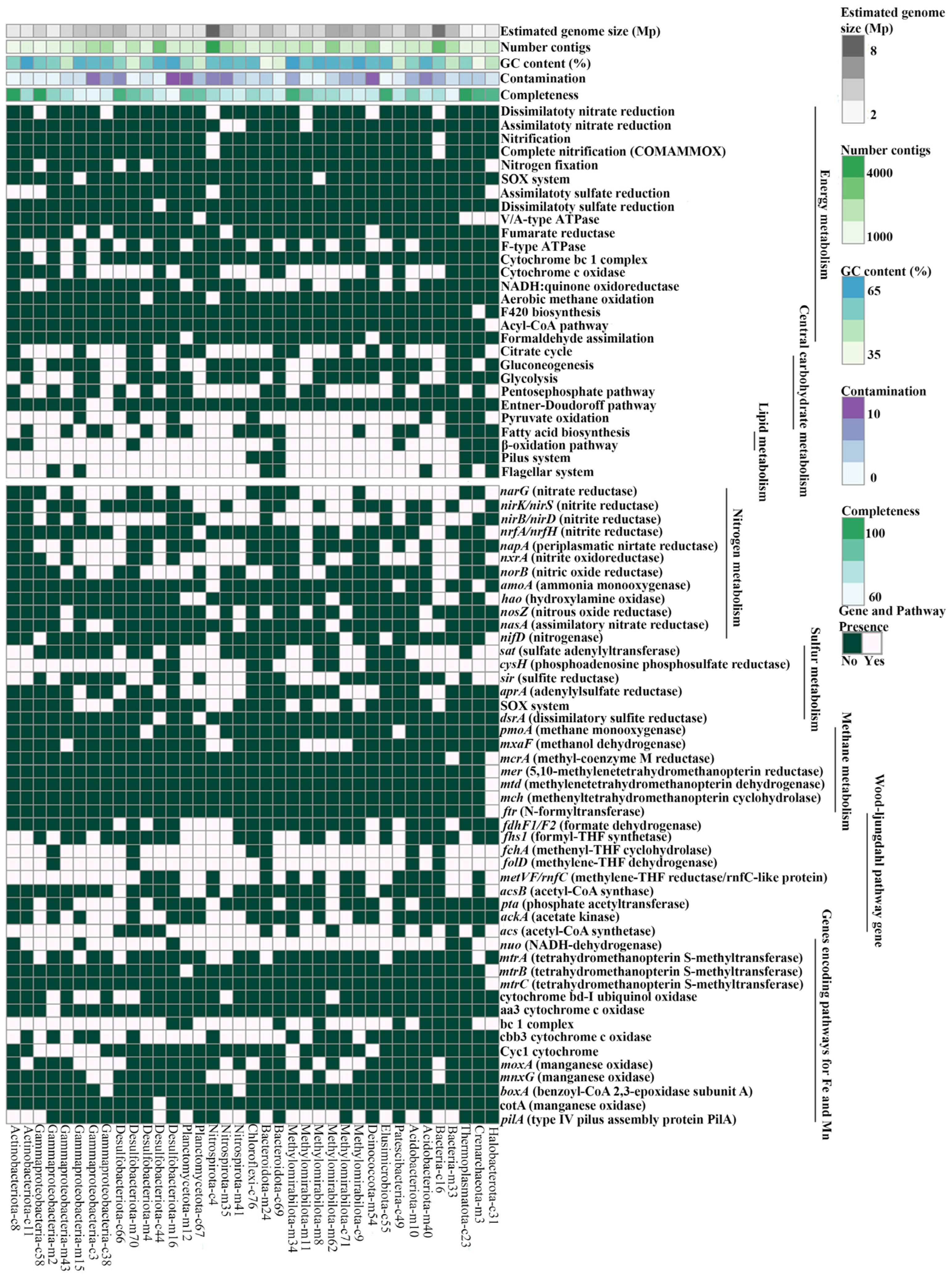
3.7. Key Metabolic Pathways of MAGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hawks, R.C.; Troup, A.G. Chemistry and origin of freshwater ferromanganese concretions. Limnol. Oceanogr. 1970, 15, 702–712. [Google Scholar] [CrossRef]
- Sommers, M.; Dollhopf, M.; Douglas, S. Freshwater ferromanganese stromatolites from Lake Vermilion, Minnesota: Microbialculturing and environmental scanning electron microscopy investigations. Geomicrobiol. J. 2002, 19, 407–427. [Google Scholar] [CrossRef]
- Kepkay, P.E. Kinetics of microbial manganese oxidation and trace metal binding in sediments: Results from an in situ dialysis technique. Limnol. Oceanogr. 1985, 30, 713–726. [Google Scholar] [CrossRef]
- Dittrich, M.; Moreau, L.; Gordon, J.; Quazi, S.; Palermo, C.; Fulthorpe, R.; Katsev, S.; Bollmann, J.; Chesnyuk, A. Geomicrobiology of iron layers in the sediment of Lake Superior. Aquat. Geochem. 2015, 21, 123–140. [Google Scholar] [CrossRef]
- Edgington, D.N.; Callender, E. Minor element geochemistry of Lake Michigan ferromanganese nodules. Earth Planet. Sci. Lett. 1970, 8, 97–100. [Google Scholar] [CrossRef] [Green Version]
- Hauck, S.; Benz, M.; Brune, A.; Schink, B. Ferrous iron oxidation by denitrifying bacteria in profundal sediments of a deep lake (Lake Constance). FEMS Microbiol. Ecol. 2001, 37, 127–134. [Google Scholar] [CrossRef]
- Baturin, G.; Yushina, I.; Zolotykh, E. Variations in the elemental composition of ferromanganese structures from Lake Baikal. Oceanology 2009, 49, 505–514. (In Russian) [Google Scholar] [CrossRef]
- Granina, L.Z. Early Diagenesis in Bottom Sediments of Lake Baikal; Academic Publishing House Geo: Novosibirsk, Russia, 2008; 159p. [Google Scholar]
- Och, L.M.; Müller, B.; Voegelin, A.; Ulrich, A.; Göttlicher, J.; Steiniger, R.; Mangold, S.; Vologina, E.G.; Sturm, M. New insights into the formation and burial of Fe/Mn accumulations in Lake Baikal sediments. Chem. Geol. 2012, 330–331, 244–259. [Google Scholar] [CrossRef]
- Strakhovenko, V.; Subetto, D.; Ovdina, E.; Belkina, N.; Efremenko, N. Distribution of elements in iron-manganese formations in bottom sediments of Lake Onego (NW Russia) and small lakes (Shotozero and Surgubskoe) of Adjacent Territories. Minerals 2020, 10, 440. [Google Scholar] [CrossRef]
- Torres, N.T.; Och, L.M.; Hauser, P.C.; Furrer, G.; Brandl, H.; Vologina, E.; Sturm, M.; Burgmann, H.; Beat Muller, B. Early diagenetic processes generate iron and manganese oxide layers in the sediments of Lake Baikal, Siberia. Environ. Sci. 2014, 16, 879–889. [Google Scholar] [CrossRef] [Green Version]
- Hein, J.R.; Mizell, K.; Koschinsky, A.; Conrad, T.A. Deep-ocean mineral deposits as a source of critical metals for high- and green-technology applications: Comparison with land-based resources. Ore Geol. Rev. 2013, 51, 1–14. [Google Scholar] [CrossRef]
- Baturin, G.N.; Dubinchuk, V.T.; Novigatsky, A.N. Phase distribution of elements in ferromanganese nodules of the Kara Sea. Dokl. Earth Sci. 2016, 471, 1199–1203. (In Russian) [Google Scholar] [CrossRef]
- Zhong, Y.; Chen, Z.; González, F.J.; Hein, J.R.; Zheng, X.; Li, G.; Luo, Y.; Mo, A.; Tian, Y.; Wang, S. Composition and genesis of ferromanganese deposits from the northern South China Sea. J. Asian Earth Sci. 2017, 138, 110–128. [Google Scholar] [CrossRef] [Green Version]
- Bukharov, A.A.; Fialkov, V.A. The Geologic Structure of the Baikal Bottom; Nauka: Moscow, Russia, 1996. [Google Scholar]
- Granina, L.Z.; Mats, V.D.; Phedorin, M.A. Iron-manganese formations in the Baikal region. Russ. Geol. Geophys. 2010, 51, 650–660. [Google Scholar] [CrossRef]
- Deike, R.G.; Granina, L.; Callender, E.; McGee, J.J. Formation of iron crusts in Quaternary sediments of Lake Baikal, Russia, and its implications for paleoclimate. Mar. Geol. 1997, 139, 21–46. [Google Scholar] [CrossRef]
- Och, L.M.; Müller, B.; März, C.; Wichser, A.; Vologina, E.G.; Sturm, M. Elevated uranium concentrations in Lake Baikal sediments: Burial and early diagenesis. Chem. Geol. 2016, 441, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Zhmodik, S.M.; Verkhovtseva, N.V.; Soloboeva, E.V.; Mironov, A.G.; Nemirovskaya, N.A.; Ilić, R.; Khlystov, O.M.; Titov, A.T. The study of distribution and forms of uranium occurrences in Lake Baikal sediments by the SSNTD method. Radiat. Meas. 2005, 40, 532–538. [Google Scholar] [CrossRef]
- Manceau, A.; Kersten, M.; Marcus, M.A.; Geoffroy, N.; Granina, L. Ba and Ni speciation in a nodule of binary Mn oxide phase composition from Lake Baikal. Geoch. Cosmochim. Acta 2007, 71, 1967–1981. [Google Scholar] [CrossRef]
- Granina, L.; Karabanov, E.; Callender, E. Relics of oxidised ferromanganese formations in the bottom sediments of Lake Baikal. IPPCCE Newsl. 1993, 7, 32–39. [Google Scholar]
- Zemskaya, T.; Konstantinova, N.; Shubenkova, O.; Pogodaeva, T.; Ivanov, V.; Bukin, S.; Khabuev, A.; Khlystov, O.; Vilkin, G.; Lomakina, A. Microbial communities of ferromanganese sedimentary layers and nodules of Lake Baikal (Bolshoy Ushkany Island). Diversity 2022, 14, 868. [Google Scholar] [CrossRef]
- Jianga, X.-D.; Sun, X.-M.; Yao, G. Biogenic mineralization in the ferromanganese nodules and crusts from the South China Sea. J. Asian Earth Sci. 2019, 171, 46–59. [Google Scholar] [CrossRef]
- Wang, X.H.; Schlossmacher, U.; Natalio, F.; Schröder, H.C.; Wolf, S.E.; Tremel, W.; Müller, W.E. Evidence for biogenic processes during formation of ferromanganese crusts from the Pacific Ocean: Implications of biologically induced mineralization. Micron 2009, 40, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Okumura, T.; Uematsu, K.; Hirai, M.; Iijima, K.; Usui, A.; Suzuki, K. Heterogeneity of microbial communities on deep-sea ferromanganese crusts in the Takuyo-Daigo seamount. Microbes Environ. 2018, 33, 366–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, S.; Hirai, M.; Ohkuma, M.; Suzuki, K. Microbial metabolisms in an abyssal ferromanganese crust from the Takuyo-Daigo Seamount as revealed by metagenomics. PLoS ONE 2019, 14, e0224888. [Google Scholar] [CrossRef]
- Bergo, N.M.; Bendia, A.G.; Neiva Ferreira, J.C.; Murton, B.J.; Brandini, F.P.; Pellizari, V.H. Microbial diversity of deep-sea ferromanganese crust field in the Rio Grande Rise, Southwestern Atlantic Ocean. Microb. Ecol. 2021, 82, 344–355. [Google Scholar] [CrossRef]
- Lowenstam, H.A. Minerals formed by organisms. Science 1987, 211, 1126–1131. [Google Scholar] [CrossRef] [Green Version]
- Tebo, B.M.; Bargar, J.R.; Clement, B.G.; Dick, G.J.; Murray, K.J.; Parker, D.L.; Verity, R.; Webb, S.M. Biogenic manganese oxides: Properties and mechanisms of formation. Annu. Rev. Earth Planet. Sci. 2004, 32, 287–328. [Google Scholar] [CrossRef] [Green Version]
- Chapnick, S.D.; Moore, W.S.; Nealson, K.H. Microbially mediated manganese oxidation in a freshwater lake. Limnol. Oceanogr. 1982, 27, 1004–1014. [Google Scholar] [CrossRef]
- Schink, B. Microbially driven redox reactions in anoxic environments: Pathways, energetics, and biochemical consequences. Eng. Life Sci. 2006, 6, 228–233. [Google Scholar] [CrossRef] [Green Version]
- Thamdrup, B.; Fossing, H.; Jørgensen, B.B. Manganese, iron and sulfur cycling in a coastal marine sediment, Aarhus Bay, Denmark. Geochim. Cosmochim. Acta 1994, 58, 5115–5129. [Google Scholar] [CrossRef]
- Berg, J.S.; Jeґzeґque, D.; Duverger, A.; Lamy, D.; Laberty-Robert, C.; Miot, J. Microbial diversity involved in iron and crypticsulfur cycling in the ferruginous, low-sulfate waters of Lake Pavin. PLoS ONE 2019, 14, e0212787. [Google Scholar] [CrossRef]
- Nealson, K.H.; Saffarini, D. Iron and manganese in anaerobic respiration: Environmental significance, physiology and regulation. Annu. Rev. Microbiol. 1994, 48, 311–343. [Google Scholar] [CrossRef]
- Finneran, K.T.; Johnsen, C.V.; Lovley, D.R. Rhodoferax ferrireducens sp. nov., a psychrotolerant, facultatively anaerobic bacterium that oxidizes acetate with the reduction of Fe(III). Int. J. Syst. Evol. Microbiol. 2003, 53, 669–673. [Google Scholar] [CrossRef] [Green Version]
- Strous, M.; Pelletier, E.; Mangenot, S.; Rattei, T.; Lehner, A.; Taylor, M.W.; Horn, M.; Daims, H.; Bartol-Mavel, D.; Wincker, P.; et al. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 2006, 440, 790–794. [Google Scholar] [CrossRef]
- Shi, L.; Dong, H.; Reguera, G.; Beyenal, H.; Lu, A.; Liu, J.; Yu, H.-Q.; Fredrickson, J.K. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Microbiol. 2016, 14, 651–662. [Google Scholar] [CrossRef]
- Jiang, Y.; Shi, M.; Shi, L. Molecular underpinnings for microbial extracellular electron transfer during biogeochemical cycling of earth elements. Sci. China Life Sci. 2019, 62, 1275–1286. [Google Scholar] [CrossRef]
- Beal, E.J.; House, C.H.; Orphan, V.J. Manganese- and iron-dependent marine methane oxidation. Science 2009, 325, 184. [Google Scholar] [CrossRef] [Green Version]
- Cai, C.; Leu, A.O.; Xie, G.-J.; Guo, J.; Feng, Y.; Zhao, J.-X.; Tyson, G.W.; Yuan, Z.; Hu, S. A methanotrophic archaeon couples anaerobic oxidation of methane to Fe (III) reduction. ISME J. 2018, 2, 1929–1939. [Google Scholar] [CrossRef]
- Ettwig, K.F.; Zhu, B.; Speth, D.; Keltjens, J.T.; Jetten, M.S.; Kartal, B. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc. Natl. Acad. Sci. USA 2016, 113, 12792–12796. [Google Scholar] [CrossRef]
- Leu, A.O.; Cai, C.; McIlroy, S.J.; Southam, G.; Orphan, V.J.; Yuan, Z.; Hu, S.; Tyson, G.W. Anaerobic methane oxidation coupled to manganese reduction by members of the Methanoperedenaceae. ISME J. 2020, 14, 1030–1041. [Google Scholar] [CrossRef] [Green Version]
- Slobodkina, G.B.; Kolganova, T.V.; Querellou, J.; Bonch-Osmolovskaya, E.A.; Slobodkin, A.I. Geoglobus acetivorans sp. nov., an iron(III)-reducing archaeon from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2009, 59, 2880–2883. [Google Scholar] [CrossRef] [PubMed]
- Dubinina, G.A. Study of the ecology of iron bacteria in fresh water basins. Izv. Acad. Sci. USSR Ser. Biol. 1976, 4, 575–592. (In Russian) [Google Scholar]
- Zakharova, Y.R.; Parfenova, V.V.; Granina, L.Z.; Kravchenko, O.S.; Zemskaya, T.I. Distribution of iron and manganese-oxidizing bacteria in the bottom sediments of Lake Baikal. Inland Water Biol. 2010, 3, 313–321. [Google Scholar] [CrossRef]
- Wetzel, R.G.; Likens, G.E. Limnological Analyses; Springer: New York, NY, USA, 1991; 391p. [Google Scholar] [CrossRef]
- Baam, G.I.; Vereshchagin, A.L.; Golobokova, L.P. Microcolumn high performance liquid chromatography with UV detect in for the determination of anion in environmental materials. J. Anal. Chem. 1999, 54, 854–857. [Google Scholar]
- Bol’shakov, A.M.; Egorov, A.V. On the application of phase equilibrium degassing for gasometric research in water areas. Okeanologiya 1987, 27, 861–862. (In Russian) [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1987. [Google Scholar]
- Sahm, K.; John, P.; Nacke, H.; Wemheuer, B.; Grote, R.; Daniel, R.; Antranikian, G. High abundance of heterotrophic prokaryotes in hydrothermal springs of the Azores as revealed by a network of 16S rRNA gene-based methods. Extremophiles 2013, 17, 649–662. [Google Scholar] [CrossRef]
- Yu, Y.; Lee, C.; Kim, J.; Hwang, S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 2005, 89, 670–679. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2020. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 11 April 2022).
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor workflow for microbiome data analysis: From raw reads to community analyses. F1000 Res. 2016, 5, 1492. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, M.H.H.; Henry, M. The vegan package. Commun. Ecol. Packag. 2007, 10, 719. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; p. 260. [Google Scholar]
- Stein, L.Y.; La Duc, M.T.; Grundl, T.J.; Nealson, K.H. Bacterial and archaeal populations associated with freshwater ferromanganous micronodules and sediments. Environ. Microbiol. 2001, 3, 10–18. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [Green Version]
- Tamames, J.; Puente-Sánchez, F. SqueezeMeta, a highly portable, fully automatic metagenomic analysis pipeline. Front. Microbiol. 2019, 9, 3349. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Barrnap 0.9: Rapid Ribosomal RNA Prediction. GPLv3. 2013. Available online: https://github.com/tseemann/barrnap (accessed on 11 April 2023).
- Hyatt, D.; Chen, G.L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009, 1, 205–211. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alneberg, J.; Bjarnason, B.; de Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning metagenomic contigs by coverage and composition. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef]
- Wu, Y.W.; Simmons, B.A.; Singer, S.W. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 2016, 32, 605–607. [Google Scholar] [CrossRef]
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. Peer J. 2019, 7, e7359. [Google Scholar] [CrossRef]
- Sieber, C.M.K.; Probst, A.J.; Sharrar, A.; Thomas, B.C.; Hess, M.; Tringe, S.G.; Banfield, J.F. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 2018, 3, 836–843. [Google Scholar] [CrossRef] [Green Version]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 2019, 36, 1925–1927. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Chaumeil, P.A.; Rinke, C.; Mussing, A.J.; Hugenholtz, P. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 2020, 38, 1079–1086. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [Green Version]
- Nawrocki, E.P. Structural RNA Homology Search and Alignment Using Covariance Models. Ph.D. Thesis, Washington University School of Medicine, St. Louis, MO, USA, 2009. [Google Scholar]
- Garber, A.I.; Nealson, K.H.; Okamoto, A.; McAllister, S.M.; Chan, C.S.; Barco, R.A.; Merino, N. FeGenie: A Comprehensive tool for the identification of iron genes and iron gene neighborhoods in genome and metagenome assemblies. Front. Microbiol. 2020, 11, 37. [Google Scholar] [CrossRef] [Green Version]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Wurzbacher, C.; Nilsson, R.; Rautio, M.; Peura, S. Poorly known microbial taxa dominate the microbiome of permafrost thaw ponds. ISME J. 2017, 11, 1938–1941. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Schubert, C.J.; Fiskal, A.; Dubois, N.; Lever, M.A. Eutrophication as a driver of microbial community structure in lake sediments. Environ. Microbiol. 2020, 22, 3446–3462. [Google Scholar] [CrossRef]
- Zemskaya, T.I.; Lomakina, A.V.; Mamaeva, E.V.; Zakharenko, A.S.; Likhoshvai, A.V.; Galachyants, Y.P.; Miller, B. Composition of microbial communities in sediments from southern Baikal containing Fe/Mn concretions. Microbiology 2018, 87, 382–392. (In Russian) [Google Scholar] [CrossRef]
- Walker, C.B.; Torre, J.R.; Klotz, M.G.; Urakawa, H.; Pinel, N.; Arp, D. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. USA 2010, 107, 8818–8823. [Google Scholar] [CrossRef]
- Molari, M.; Janssen, F.; Vonnahme, T.; Wenzhöfer, F.; Boetius, A. The contribution of microbial communities in polymetallic nodules to the diversity of the deep-sea microbiome of the Peru Basin (4130–4198 m depth). Biogeosciences 2020, 17, 3203–3222. [Google Scholar] [CrossRef]
- Nitahara, S.; Kato, S.; Urabe, T.; Usui, A.; Yamagishi, A. Molecular characterization of the microbial community in hydrogenetic ferromanganese crusts of the Takuyo-Daigo Seamount, northwest Pacific. FEMS Microbiol. Lett. 2011, 321, 121–129. [Google Scholar] [CrossRef]
- Aylward, F.O.; Santoro, A.E. Heterotrophic Thaumarchaea with small genomes are widespread in the Dark Ocean. mSystems 2020, 5, e00415-20. [Google Scholar] [CrossRef]
- Li, S.; Diao, M.; Wang, S.; Zhu, X.; Dong, X.; Strous, M.; Ji, G. Distinct oxygen isotope fractionations driven by different electron donors during microbial nitrate reduction in lake sediments. Environ. Microbiol. Rep. 2022, 14, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Lomakina, A.; Pogodaeva, T.; Kalmychkov, G.; Chernitsyna, S.; Zemskaya, T. Diversity of NC10 bacteria and ANME-2d archaea in sediments of fault zones at Lake Baikal. Diversity 2020, 12, 10. [Google Scholar] [CrossRef] [Green Version]
- Fincker, M.; Huber, J.A.; Orphan, V.J.; Rappé, M.S.; Teske, A.; Spormann, A.M. Metabolic strategies of marine subseafloor Chloroflexi inferred from genome reconstructions. Environ. Microbiol. 2020, 22, 3188–3204. [Google Scholar] [CrossRef] [PubMed]
- Mehrshad, M.; Salcher, M.M.; Okazaki, Y.; Nakano, S.; Šimek, K.; Andrei, A.-S.; Ghai, R. Hidden in plain sight—Highly abundant and diverse planktonic freshwater Chloroflexi. Microbiome 2018, 6, 176. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, Y.; Cápiro, N.L.; Yan, J. Genomic characteristics distinguish geographically distributed Dehalococcoidia. Front. Microbiol. 2020, 11, 546063. [Google Scholar] [CrossRef]
- Vuillemin, A.; Kerrigan, Z.; D’Hondt, S.; Orsi, W. Exploring the abundance, metabolic potential and gene expression of subseafloor Chloroflexi in million-year-old oxic and anoxic abyssal clay. FEMS Microbiol. Ecol. 2020, 96, fiaa223. [Google Scholar] [CrossRef]
- Karaevskaya, E.S.; Demidov, N.; Kazantsev, V.S.; Elizarov, I.M.; Khaloshin, A.G.; Petrov, A.L.; Karlov, D.S.; Schirrmeister, L.; Belov, A.A.; Wetterich, S. Bacterial communities of frozen quaternary sediments of marine origin on the coast of Western Spitsbergen. Izv. Atmos. Ocean. Phys. 2021, 20, 895–917. [Google Scholar] [CrossRef]
- Kanevskiy, M.; Fortier, D.; Shur, Y.; Bray, M.; Jorgenson, T. Detailed cryostratigraphic studies of syngenetic permafrost in the winze of the CRREL. Permafrost Tunnel, Fox, Alaska. In Proceedings of the Ninth International Conference on Permafrost, Fairbanks, Alaska, 29 June–3 July 2008; pp. 889–894. [Google Scholar]
- Mackelprang, R.; Burkert, A.; Haw, M.; Mahendrarajah, T.; Conaway, C.H.; Douglas, T.A.; Waldrop, M.P. Microbial survival strategies in ancient permafrost: Insights from metagenomics. ISME J. 2017, 11, 2305–2318. [Google Scholar] [CrossRef] [Green Version]
- Cherbunina, M.Y.; Karaevskaya, E.S.; Vasil’chuk, Y.K.; Tananaev, N.I.; Shmelev, D.G.; Budantseva, N.A.; Merkel, A.Y.; Rakitin, A.L.; Mardanov, A.V.; Brouchkov, A.V.; et al. Microbial and geochemical evidence of permafrost formation at Mamontova Gora and Syrdakh, Central Yakutia. Front. Earth Sci. 2021, 9, 739365. [Google Scholar] [CrossRef]
- Vologina, E.G.; Sturm, M.; Vorobyeva, S.S. Holocene/Pleistocene: Sediments of basins, ridges and near -coast areas of Lake Baikal, Russia. Geol. Res. Abstr. 2011, 2, 2263. [Google Scholar]
- Evangelinos, D.; Nelson, C.H.; Escutia, C.; De Batist, M.; Khlystov, O. Late quaternary climatic control of Lake Baikal (Russia) turbidite systems: Implications for turbidite systems worldwide. Geology 2017, 45, 179–182. [Google Scholar] [CrossRef]
- Karabanov, E.B.; Prokopenko, A.A.; Williams, D.F.; Khursevich, G.K. A new record of Holocene climate change from the bottom sediments of Lake Baikal. J. Paleogeogr. Paleoclimatol. Paleoecol. 2000, 56, 211–224. [Google Scholar] [CrossRef]
- Riedinger, N.; Formolo, M.J.; Lyons, T.W.; Henkel, S.; Beck, A.; Kasten, S. An inorganic geochemical argument for coupled an aerobic oxidation of methane and iron reduction in marine sediments. Geobiology 2014, 12, 172–181. [Google Scholar] [CrossRef]
- Winkel, M.; Sepulveda-Jauregui, A.; Martinez-Cruz, K.; Heslop, J.K.; Rijkers, R.; Horn, F.; Liebner, S.; Walter Anthony, K.M. First evidence for cold-adapted anaerobic oxidation of methane in deep sediments of thermokarst lakes. Environ. Res. Commun. 2019, 1, 021002. [Google Scholar] [CrossRef]
- Segarra, K.E.; Comerford, C.; Slaughter, J.; Joye, S.B. Impact of electron acceptor availability on the anaerobic oxidation of methane in coastal freshwater and brackish wetland sediments. Geochim. Cosmochim. Acta 2013, 115, 15–30. [Google Scholar] [CrossRef]
- Sivan, O.; Adler, M.; Pearson, A.; Gelman, F.; Bar-Or, I.; John, S.G.; Ecker, W. Geochemical evidence for iron-mediated anaerobic oxidation of methane. Limnol. Oceanogr. 2011, 56, 1536–1544. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Liu, J.; Yang, J.; Wu, G.; Hua, Z.; Dong, H.; Hedlund, B.P.; Baker, B.J.; Jiang, H. Compositional and metabolic responses of autotrophic microbial community to salinity in lacustrine environments. mSystems 2022, 30, e0033522. [Google Scholar] [CrossRef]
- Kanaparthi, D.; Pommerenke, B.; Casper, P.; Dumont, M.G. Chemolithotrophic nitrate-dependent Fe(II)-oxidizing nature of actinobacterial subdivision lineage TM3. ISME J. 2013, 7, 1582–1594. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, K.; Izallalen, M.; Mouser, P.; Richter, H.; Risso, C.; Mahadevan, R.; Lovley, D.R. Genome-scale dynamic modeling of the competition between Rhodoferax and Geobacter in anoxic subsurface environments. ISME J. 2011, 5, 305–316. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef]
- Arshad, A.; Speth, D.R.; de Graaf, R.M.; Op den Camp, H.J.; Jetten, M.S.; Welte, C.U. A metagenomics-based metabolic model of nitrate-dependent anaerobic oxidation of methane by Methanoperedens-like archaea. Front. Microbiol. 2015, 6, 1423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Destan, E.; Yuksel, B.; Tolar, B.B.; Ayan, E.; Deutsch, S.; Yoshikuni, Y.; Wakatsuki, S.; Francis, C.A.; DeMirci, H. Structural insights into bifunctional thaumarchaeal crotonyl-CoA hydratase and 3-hydroxypropionyl-CoA dehydratase from Nitrosopumilus maritimus. Sci. Rep. 2021, 11, 22849. [Google Scholar] [CrossRef] [PubMed]
- Cabello-Yeves, P.J.; Zemskaya, T.I.; Zakharenko, A.S.; Sakirko, M.V.; Ivanov, V.G.; Ghai, R.; Rodriguez-Valera, F. Microbiome of the deep Lake Baikal, a unique oxic bathypelagic habitat. Limnol. Oceanogr. 2020, 65, 1471–1488. [Google Scholar] [CrossRef] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lomakina, A.; Bukin, S.; Shubenkova, O.; Pogodaeva, T.; Ivanov, V.; Bukin, Y.; Zemskaya, T. Microbial Communities in Ferromanganese Sediments from the Northern Basin of Lake Baikal (Russia). Microorganisms 2023, 11, 1865. https://doi.org/10.3390/microorganisms11071865
Lomakina A, Bukin S, Shubenkova O, Pogodaeva T, Ivanov V, Bukin Y, Zemskaya T. Microbial Communities in Ferromanganese Sediments from the Northern Basin of Lake Baikal (Russia). Microorganisms. 2023; 11(7):1865. https://doi.org/10.3390/microorganisms11071865
Chicago/Turabian StyleLomakina, Anna, Sergei Bukin, Olga Shubenkova, Tatyana Pogodaeva, Vyacheslav Ivanov, Yuri Bukin, and Tamara Zemskaya. 2023. "Microbial Communities in Ferromanganese Sediments from the Northern Basin of Lake Baikal (Russia)" Microorganisms 11, no. 7: 1865. https://doi.org/10.3390/microorganisms11071865
APA StyleLomakina, A., Bukin, S., Shubenkova, O., Pogodaeva, T., Ivanov, V., Bukin, Y., & Zemskaya, T. (2023). Microbial Communities in Ferromanganese Sediments from the Northern Basin of Lake Baikal (Russia). Microorganisms, 11(7), 1865. https://doi.org/10.3390/microorganisms11071865






