Transcriptome Analysis of Glycerin Regulating Reuterin Production of Lactobacillus reuteri
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation, Morphological Observation, and Antibacterial Activity Analysis of L. reuteri
2.2. Heterologous Expression of GDHt
2.3. Extraction and Purification of GDHt and Evaluation of Effect of Glycerol Concentration on GDHt Activity
2.4. Transcriptome Sequencing and Sequence Data Analysis
2.5. Data Analysis
3. Results
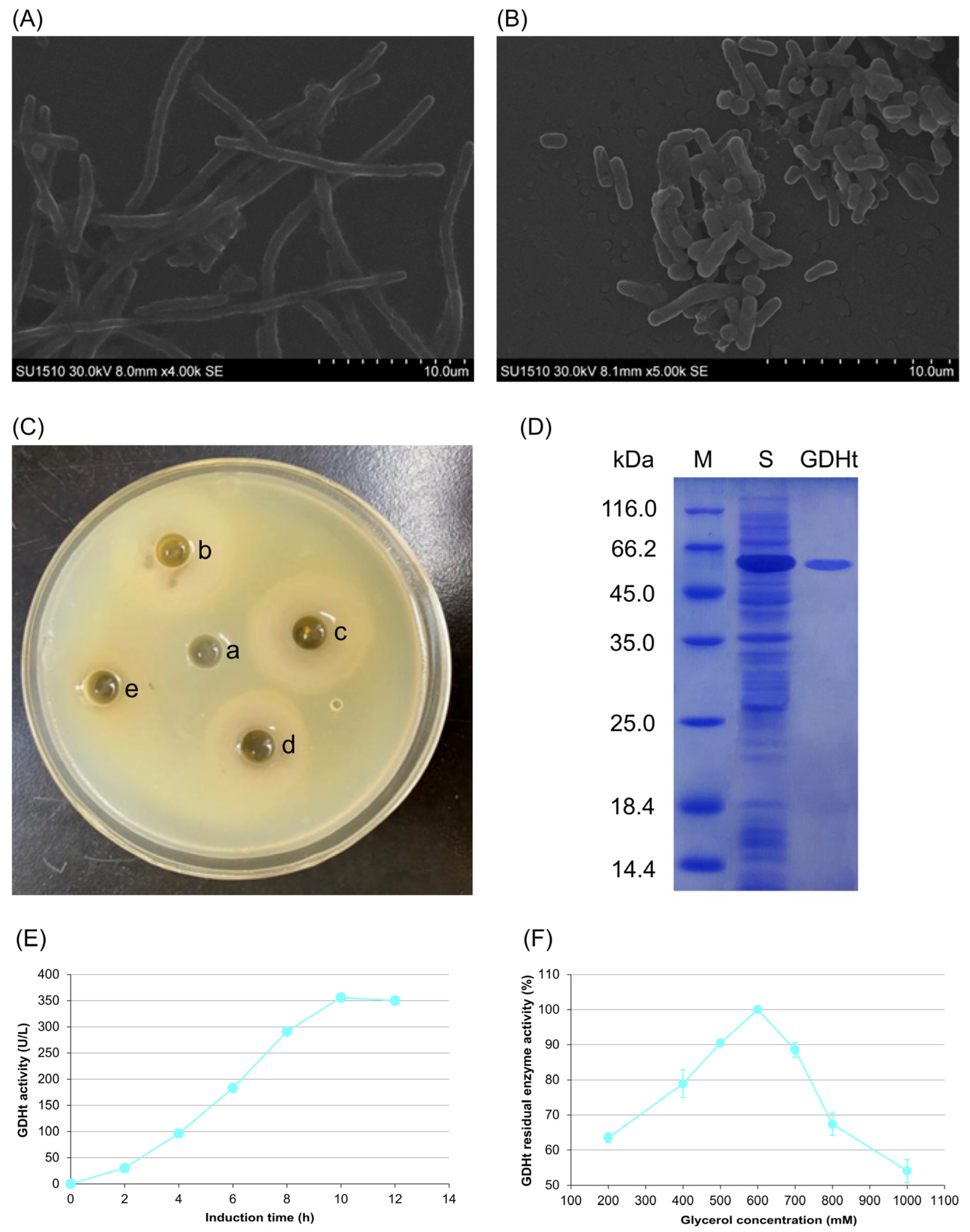
3.1. Effects of Glycerol on Morphology, Antimicrobial Properties, and GDHt Activity of L. reuteri LR301
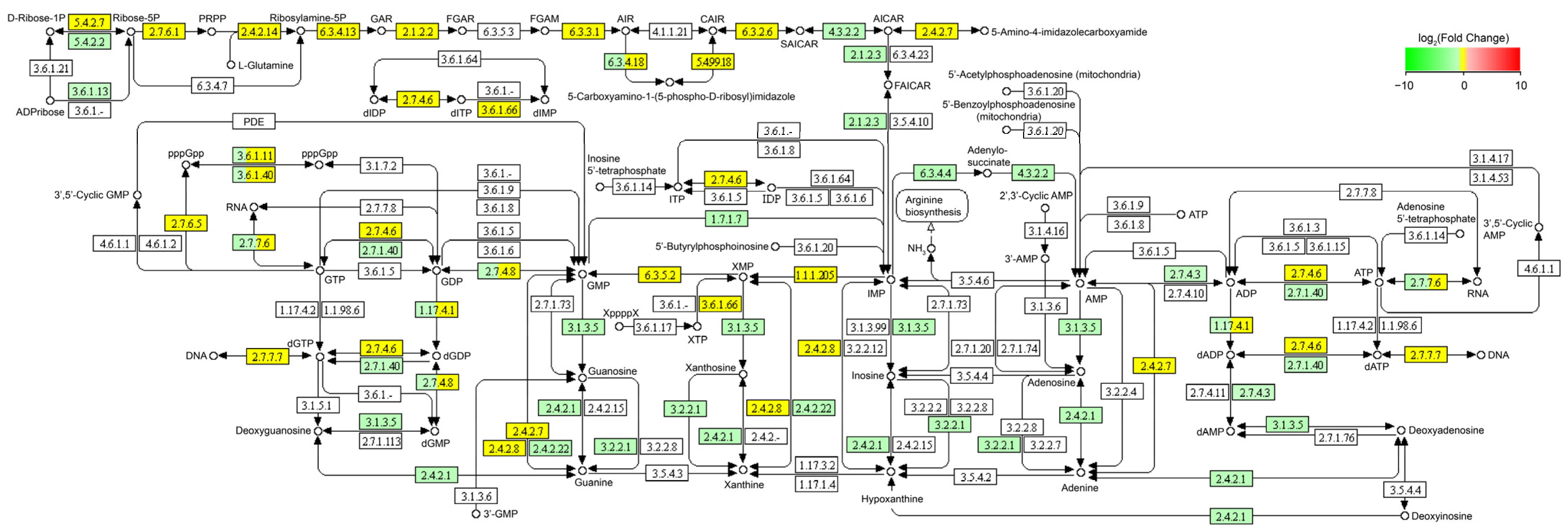
3.2. Effect of Glycerol on L. reuteri Transcriptome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ortiz-Rivera, Y.; Sánchez-Vega, R.; Gutiérrez-Méndez, N.; León-Félix, J.; Acosta-Muñiz, C.; Sepulveda, D.R. Production of reuterin in a fermented milk product by Lactobacillus reuteri: Inhibition of pathogens, spoilage microorganisms, and lactic acid bacteria. J. Dairy Sci. 2017, 100, 4258–4268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vimont, A.; Fernandez, B.; Ahmed, G.; Fortin, H.P.; Fliss, I. Quantitative antifungal activity of reuterin against food isolates of yeasts and moulds and its potential application in yogurt. Int. J. Food Microbiol. 2019, 289, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Sung, V.; D’Amico, F.; Cabana, M.D.; Chau, K.; Koren, G.; Savino, F.; Szajewska, H.; Deshpande, G.; Dupont, C.; Indrio, F.; et al. Lactobacillus reuteri to treat infant Colic: A meta-analysis. Pediatrics 2018, 141, e20171811. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Rivera, Y.; Sánchez-Vega, R.; Acosta-Muñiz, C.H.; Gutiérrez-Méndez, N.; León-Félix, J.; Sepulveda, D.R. Influence of environmental and genetic factors on 3-hydoxypropionaldehyde production by Lactobacillus reuteri. J. Basic Microbiol. 2018, 58, 1053–1060. [Google Scholar] [CrossRef]
- Lindlbauer, K.A.; Marx, H.; Sauer, M. 3-Hydroxypropionaldehyde production from crude glycerol by Lactobacillus diolivorans with enhanced glycerol uptake. Biotechnol. Biofuels 2017, 10, 295. [Google Scholar] [CrossRef] [Green Version]
- Nasir, A.; Ashok, S.; Shim, J.Y.; Park, S.; Yoo, T.H. Recent progress in the understanding and engineering of coenzyme B12-dependent glycerol dehydratase. Front. Bioeng. Biotechnol. 2020, 8, 500867. [Google Scholar] [CrossRef] [PubMed]
- Sardari, R.R.R.; Dishisha, T.; Pyo, S.H.; Hatti-Kaul, R. Improved production of 3-hydroxypropionaldehyde by complex formation with bisulfite during biotransformation of glycerol. Biotechnol. Bioeng. 2013, 110, 1243–1248. [Google Scholar] [CrossRef] [PubMed]
- Dishisha, T.; Pereyra, L.P.; Pyo, S.H.; Britton, R.A.; Hatti-Kaul, R. Flux analysis of the Lactobacillus reuteri propanediol-utilization pathway for production of 3-hydroxypropionaldehyde, 3-hydroxypropionic acid and 1,3-propanediol from glycerol. Microb. Cell Fact. 2014, 13, 76. [Google Scholar] [CrossRef] [Green Version]
- Seo, M.Y.; Seo, J.W.; Heo, S.Y.; Baek, J.O.; Rairakhwada, D.; Oh, B.R.; Seo, P.S.; Choi, M.H.; Kim, C.H. Elimination of by-product formation during production of 1,3-propanediol in Klebsiella pneumoniae by inactivation of glycerol oxidative pathway. Appl. Microbiol. Biotechnol. 2009, 84, 527–534. [Google Scholar] [CrossRef]
- Ramakrishnan, G.G.; Nehru, G.; Suppuram, P.; Balasubramaniyam, S.; Gulab, B.R.; Subramanian, R. Bio-transformation of glycerol to 3-hydroxypropionia acid using resting cells of Lactobacillus reuteri. Curr. Microbiol. 2015, 71, 517–523. [Google Scholar] [CrossRef]
- Mao, L.T.; Huang, Z.C.; Lu, Z.Z.; Lin, G.G.; Chen, Z.F.; Yang, Z. Multi-species Bacillus inoculant and its effect on potato growth and control of potato (Solanum tuberosum L.) blackleg disease. Appl. Ecol. Environ. Res. 2022, 20, 1571–1583. [Google Scholar] [CrossRef]
- Lin, X.B.; Lohans, C.T.; Duar, R.; Zheng, J.; Vederas, J.C.; Walter, J.; Gänzle, M. Genetic determinants of reutericyclin biosynthesis in Lactobacillus reuteri. Appl. Environ. Microbiol. 2015, 81, 2032–2041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okonechnikov, K.; Conesa, A.; Garcia-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.-W.; Chen, C.-N.; Liang, H.-F.; Hong, M.-H. A natural compound (reuterin) produced by Lactobacillus reuteri for biological-tissue fixation. Biomaterials 2003, 24, 1335–1347. [Google Scholar] [CrossRef]
- Greppi, A.; Asare, P.T.; Schwab, C.; Zemp, N.; Stephan, R.; Lacroix, C. Isolation and comparative genomic analysis of reuterin-producing Lactobacillus reuteri from the chicken gastrointestinal tract. Front. Microbiol. 2020, 11, 1166. [Google Scholar] [CrossRef]
- Sun, M.-C.; Hu, Z.-Y.; Li, D.-D.; Chen, Y.-X.; Xi, J.-H.; Zhao, C.-H. Application of the reuterin system as food preservative or health-promoting agent: A critical review. Foods 2022, 11, 4000. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Gutierrez-Maddox, N.; Mutukumira, A.N.; Maddox, I.S.; Shu, Q. Influence of operating conditions on reuterin production using resting cells of Limosilactobacillus reuteri DPC16. Fermentation 2022, 8, 227. [Google Scholar] [CrossRef]
- Cleusix, V.; Lacroix, C.; Vollenweider, S.; Duboux, M.; Le Blay, G. Inhibitory activity spectrum of reuterin produced by Lactobacillus reuteri against intestinal bacteria. BMC Microbiol. 2007, 7, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevens, M.; Vollenweider, S.; Lacroix, C. The potential of reuterin produced by Lactobacillus reuteri as a broad spectrum preservative in food. In Protective Cultures, Antimicrobial Metabolites and Bacteriophages for Food and Beverage Biopreservation; Woodhead Publishing Series in Food Science, Technology and Nutrition; Elsevier: Amsterdam, The Netherlands, 2011; pp. 129–160. [Google Scholar] [CrossRef]
- Toraya, T.; Kuno, S.; Fukui, S. Distribution of coenzyme B12-dependent diol dehydratase and glycerol dehydratase in selected genera of Enterobacteriaceae and Propionibacteriaceae. J. Bacteriol. 1980, 141, 1439–1442. [Google Scholar] [CrossRef]
- Talarico, T.L.; Casas, I.A.; Chung, T.C.; Dobrogosz, W.J. Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 1988, 32, 1854–1858. [Google Scholar] [CrossRef] [Green Version]
- Kovačević, B.; Barić, D.; Babić, D.; Bilić, L.; Hanževački, M.; Sandala, G.M.; Radom, L.; Smith, D.M. Computational tale of two enzymes: Glycerol dehydration with or without B12. J. Am. Chem. Soc. 2018, 140, 8487–8496. [Google Scholar] [CrossRef] [Green Version]
- Lüthi-Peng, Q.; Dileme, F.; Puhan, Z. Effect of glucose on glycerol bioconversion by Lactobacillus reuteri. Appl. Microbiol. Biotechnol. 2002, 59, 289–296. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Yin, Q.; Bai, H.; Wang, W.; Chen, Y.; Zhou, M.; Zhang, R.; Ding, G.; Xu, Z.; Zhang, Y. Transcriptome Analysis of Glycerin Regulating Reuterin Production of Lactobacillus reuteri. Microorganisms 2023, 11, 2007. https://doi.org/10.3390/microorganisms11082007
Wang J, Yin Q, Bai H, Wang W, Chen Y, Zhou M, Zhang R, Ding G, Xu Z, Zhang Y. Transcriptome Analysis of Glycerin Regulating Reuterin Production of Lactobacillus reuteri. Microorganisms. 2023; 11(8):2007. https://doi.org/10.3390/microorganisms11082007
Chicago/Turabian StyleWang, Jingjing, Qiang Yin, Han Bai, Wei Wang, Yajun Chen, Minghui Zhou, Ran Zhang, Guoao Ding, Zhongdong Xu, and Yan Zhang. 2023. "Transcriptome Analysis of Glycerin Regulating Reuterin Production of Lactobacillus reuteri" Microorganisms 11, no. 8: 2007. https://doi.org/10.3390/microorganisms11082007




