Transient SARS-CoV-2 RNA-Dependent RNA Polymerase Mutations after Remdesivir Treatment for Chronic COVID-19 in Two Transplant Recipients: Case Report and Intra-Host Viral Genomic Investigation
Abstract
:1. Introduction
2. Methods
2.1. SARS-CoV-2 PCR
2.2. SARS-CoV-2 Sequencing and Mutation Analysis
2.3. Viral Load Monitoring
3. Results
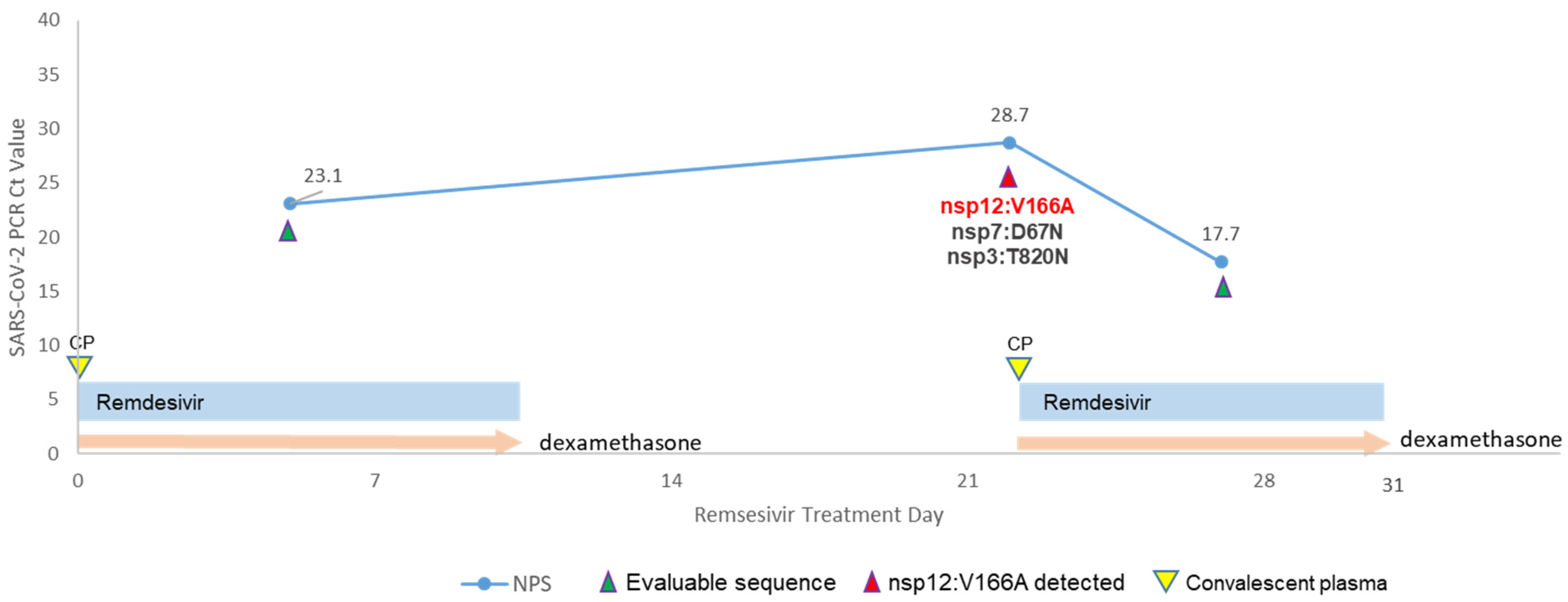
3.1. Clinical Course and Virologic Dynamics
3.2. Intra-Host Viral Evolution after Remdesivir Treatment
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Case | Collection Day | Specimen Source | Ct Value | Amino Acid Substitutions | GISAID ID | GenBank ID | SRA ID |
|---|---|---|---|---|---|---|---|
| 1 | Day −1 | NPS | 17.1 | none | EPI_ISL_962506 | OR387920 | SRR25496336 |
| 1 | Day 13 | NPS | 15.8 | none | EPI_ISL_962507 | OR387921 | SRR25496335 |
| 1 | Day 14 | plasma | 30.7 | nsp12:A449V (minor allele) | EPI_ISL_962514 | OR387927 | SRR25496328 |
| 1 | Day 17 | NPS | 23 | nsp12:A449V (minor allele) | EPI_ISL_962508 | OR387922 | SRR25496333 |
| 1 | Day 23 | NPS | 20.1 | nsp12:A449V | EPI_ISL_962509 | OR387923 | SRR25496332 |
| 1 | Day 29 | NPS | 27.2 | nsp12:A449V | EPI_ISL_962510 | OR387924 | SRR25496331 |
| 1 | Day 36 | NPS | 28.1 | nsp5:L24F | EPI_ISL_962512 | OR387925 | SRR25496330 |
| 1 | Day 42 | NPS | 26.9 | none | EPI_ISL_962513 | OR387926 | SRR25496329 |
| 1 | Day 50 | NPS | 19.5 | none | EPI_ISL_1191129 | OR387929 | SRR25496326 |
| 1 | Day 64 | NPS | 25.3 | none | EPI_ISL_1191131 | OR387930 | SRR25496334 |
| 1 | Day 68 | NPS | 24.2 | none | EPI_ISL_1191110 | OR387928 | SRR25496327 |
| 2 | Day 5 | NPS | 23 | none | EPI_ISL_17636781 | OR397702 | SRR25506571 |
| 2 | Day 22 | NPS | 28.7 | nsp3:T820N, nsp7:D67N, nsp12:V166A | EPI_ISL_17799305 | OR393372 | SRR25506336 |
| 2 | Day 27 | NPS | 17.7 | none | EPI_ISL_17636782 | OR397703 | SRR25506570 |
References
- Fitero, A.; Bungau, S.G.; Tit, D.M.; Endres, L.; Khan, S.A.; Bungau, A.F.; Romanul, I.; Vesa, C.M.; Radu, A.F.; Tarce, A.G.; et al. Comorbidities, Associated Diseases, and Risk Assessment in COVID-19—A Systematic Review. Int. J. Clin. Pract. 2022, 2022, 1571826. [Google Scholar] [CrossRef]
- Fung, M.; Babik, J.M. COVID-19 in Immunocompromised Hosts: What We Know So Far. Clin. Infect. Dis. 2021, 72, 340–350. [Google Scholar] [CrossRef]
- Sharma, A.; Bhatt, N.S.; St Martin, A.; Abid, M.B.; Bloomquist, J.; Chemaly, R.F.; Dandoy, C.; Gauthier, J.; Gowda, L.; Perales, M.A.; et al. Clinical characteristics and outcomes of COVID-19 in haematopoietic stem-cell transplantation recipients: An observational cohort study. Lancet Haematol. 2021, 8, e185–e193. [Google Scholar] [CrossRef]
- Schaenman, J.; Byford, H.; Grogan, T.; Motwani, Y.; Beaird, O.E.; Kamath, M.; Lum, E.; Meneses, K.; Sayah, D.; Vucicevic, D.; et al. Impact of solid organ transplant status on outcomes of hospitalized patients with COVID-19 infection. Transpl. Infect. Dis. 2022, 24, e13853. [Google Scholar] [CrossRef] [PubMed]
- NIH. Therapeutic Management of Hospitalized Adults with COVID-19. COVID-19 Treatment Guidelines 2021. Available online: https://www.covid19treatmentguidelines.nih.gov/management/clinical-management/hospitalized-adults--therapeutic-management/ (accessed on 25 February 2022).
- Mari, A.; Roloff, T.; Stange, M.; Sogaard, K.K.; Asllanaj, E.; Tauriello, G.; Alexander, L.T.; Schweitzer, M.; Leuzinger, K.; Gensch, A.; et al. Global Genomic Analysis of SARS-CoV-2 RNA Dependent RNA Polymerase Evolution and Antiviral Drug Resistance. Microorganisms 2021, 9, 1094. [Google Scholar] [CrossRef] [PubMed]
- Negru, P.A.; Radu, A.F.; Vesa, C.M.; Behl, T.; Abdel-Daim, M.M.; Nechifor, A.C.; Endres, L.; Stoicescu, M.; Pasca, B.; Tit, D.M.; et al. Therapeutic dilemmas in addressing SARS-CoV-2 infection: Favipiravir versus Remdesivir. Biomed. Pharmacother. 2022, 147, 112700. [Google Scholar] [CrossRef]
- Shannon, A.; Canard, B. Kill or corrupt: Mechanisms of action and drug-resistance of nucleotide analogues against SARS-CoV-2. Antivir. Res. 2023, 210, 105501. [Google Scholar] [CrossRef]
- Agostini, M.L.; Andres, E.L.; Sims, A.C.; Graham, R.L.; Sheahan, T.P.; Lu, X.; Smith, E.C.; Case, J.B.; Feng, J.Y.; Jordan, R.; et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Goldswain, H.; Dong, X.; Penrice-Randal, R.; Alruwaili, M.; Shawli, G.T.; Prince, T.; Williamson, M.K.; Raghwani, J.; Randle, N.; Jones, B.; et al. The P323L substitution in the SARS-CoV-2 polymerase (NSP12) confers a selective advantage during infection. Genome Biol. 2023, 24, 47. [Google Scholar] [CrossRef]
- Martinot, M.; Jary, A.; Fafi-Kremer, S.; Leducq, V.; Delagreverie, H.; Garnier, M.; Pacanowski, J.; Mekinian, A.; Pirenne, F.; Tiberghien, P.; et al. Emerging RNA-Dependent RNA Polymerase Mutation in a Remdesivir-Treated B-cell Immunodeficient Patient With Protracted Coronavirus Disease 2019. Clin. Infect. Dis. 2021, 73, e1762–e1765. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Zhou, S.; Graham, R.L.; Pruijssers, A.J.; Agostini, M.L.; Leist, S.R.; Schafer, A.; Dinnon, K.H., 3rd; Stevens, L.J.; et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020, 12, eabb5883. [Google Scholar] [CrossRef] [PubMed]
- Hogan, J.I.; Duerr, R.; Dimartino, D.; Marier, C.; Hochman, S.E.; Mehta, S.; Wang, G.; Heguy, A. Remdesivir Resistance in Transplant Recipients With Persistent Coronavirus Disease 2019. Clin. Infect. Dis. 2023, 76, 342–345. [Google Scholar] [CrossRef]
- Price, T.K.; Bowland, B.C.; Chandrasekaran, S.; Garner, O.B.; Yang, S. Performance Characteristics of Severe Acute Respiratory Syndrome Coronavirus 2 RT-PCR Tests in a Single Health System: Analysis of >10,000 Results from Three Different Assays. J. Mol. Diagn. 2021, 23, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Hemarajata, P.; Hilt, E.E.; Price, T.K.; Garner, O.B.; Green, N.M. Investigation of SARS-CoV-2 Epsilon Variant and Hospitalization Status by Genomic Surveillance in a Single Large Health System During the 2020-2021 Winter Surge in Southern California. Am. J. Clin. Pathol. 2021, 157, 649–652. [Google Scholar] [CrossRef]
- Gorzalski, A.J.; Boyles, C.; Sepcic, V.; Verma, S.; Sevinsky, J.; Libuit, K.; Van Hooser, S.; Pandori, M.W. Rapid repeat infection of SARS-CoV-2 by two highly distinct delta-lineage viruses. Diagn. Microbiol. Infect. Dis. 2022, 104, 115747. [Google Scholar] [CrossRef] [PubMed]
- Libuit, K.G.; Doughty, E.L.; Otieno, J.R.; Ambrosio, F.; Kapsak, C.J.; Smith, E.A.; Wright, S.M.; Scribner, M.R.; Petit Iii, R.A.; Mendes, C.I.; et al. Accelerating bioinformatics implementation in public health. Microb. Genom. 2023, 9. [Google Scholar] [CrossRef]
- Katz, K.S.; Shutov, O.; Lapoint, R.; Kimelman, M.; Brister, J.R.; O’Sullivan, C. STAT: A fast, scalable, MinHash-based k-mer tool to assess Sequence Read Archive next-generation sequence submissions. Genome Biol. 2021, 22, 270. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Gangavarapu, K.; Quick, J.; Matteson, N.L.; De Jesus, J.G.; Main, B.J.; Tan, A.L.; Paul, L.M.; Brackney, D.E.; Grewal, S.; et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019, 20, 8. [Google Scholar] [CrossRef]
- Schaffer, A.A.; Hatcher, E.L.; Yankie, L.; Shonkwiler, L.; Brister, J.R.; Karsch-Mizrachi, I.; Nawrocki, E.P. VADR: Validation and annotation of virus sequence submissions to GenBank. BMC Bioinform. 2020, 21, 211. [Google Scholar] [CrossRef]
- Aksamentov, I.; Roemer, C.; Hodcroft, E.; Neher, R. Nextclade: Clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021, 6, 3773. [Google Scholar] [CrossRef]
- Fajnzylber, J.; Regan, J.; Coxen, K.; Corry, H.; Wong, C.; Rosenthal, A.; Worrall, D.; Giguel, F.; Piechocka-Trocha, A.; Atyeo, C.; et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 2020, 11, 5493. [Google Scholar] [CrossRef]
- Jacobs, J.L.; Bain, W.; Naqvi, A.; Staines, B.; Castanha, P.M.S.; Yang, H.; Boltz, V.F.; Barratt-Boyes, S.; Marques, E.T.A.; Mitchell, S.L.; et al. SARS-CoV-2 Viremia is Associated with COVID-19 Severity and Predicts Clinical Outcomes. Clin. Infect. Dis. 2021, 74, 1525–1533. [Google Scholar] [CrossRef]
- Yuan, F.; Wang, L.; Fang, Y.; Wang, L. Global SNP analysis of 11,183 SARS-CoV-2 strains reveals high genetic diversity. Transbound. Emerg. Dis. 2021, 68, 3288–3304. [Google Scholar] [CrossRef]
- Williamson, M.K.; Hamilton, F.; Hutchings, S.; Pymont, H.; Hackett, M.; Arnold, D.; Maskell, N.; MacGowan, A.; Albur, M.; Jenkins, M.; et al. Chronic SARS-CoV-2 infection and viral evolution in a hypogammaglobulinaemic individual. medRxiv 2021. [Google Scholar] [CrossRef]
- Zheng, Y.; Deng, J.; Han, L.; Zhuang, M.W.; Xu, Y.; Zhang, J.; Nan, M.L.; Xiao, Y.; Zhan, P.; Liu, X.; et al. SARS-CoV-2 NSP5 and N protein counteract the RIG-I signaling pathway by suppressing the formation of stress granules. Signal Transduct. Target. Ther. 2022, 7, 22. [Google Scholar] [CrossRef]
- Stevens, L.J.; Pruijssers, A.J.; Lee, H.W.; Gordon, C.J.; Tchesnokov, E.P.; Gribble, J.; George, A.S.; Hughes, T.M.; Lu, X.; Li, J.; et al. Mutations in the SARS-CoV-2 RNA-dependent RNA polymerase confer resistance to remdesivir by distinct mechanisms. Sci. Transl. Med. 2022, 14, eabo0718. [Google Scholar] [CrossRef]
- Hillen, H.S.; Kokic, G.; Farnung, L.; Dienemann, C.; Tegunov, D.; Cramer, P. Structure of replicating SARS-CoV-2 polymerase. Nature 2020, 584, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Mao, C.; Luan, X.; Shen, D.-D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M.; et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020, 368, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Osipiuk, J.; Azizi, S.-A.; Dvorkin, S.; Endres, M.; Jedrzejczak, R.; Jones, K.A.; Kang, S.; Kathayat, R.S.; Kim, Y.; Lisnyak, V.G.; et al. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021, 12, 743. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Multani, A.; Garrigues, J.M.; Oh, M.S.; Hemarajata, P.; Burleson, T.; Green, N.M.; Oliai, C.; Gaynor, P.T.; Beaird, O.E.; et al. Transient SARS-CoV-2 RNA-Dependent RNA Polymerase Mutations after Remdesivir Treatment for Chronic COVID-19 in Two Transplant Recipients: Case Report and Intra-Host Viral Genomic Investigation. Microorganisms 2023, 11, 2096. https://doi.org/10.3390/microorganisms11082096
Yang S, Multani A, Garrigues JM, Oh MS, Hemarajata P, Burleson T, Green NM, Oliai C, Gaynor PT, Beaird OE, et al. Transient SARS-CoV-2 RNA-Dependent RNA Polymerase Mutations after Remdesivir Treatment for Chronic COVID-19 in Two Transplant Recipients: Case Report and Intra-Host Viral Genomic Investigation. Microorganisms. 2023; 11(8):2096. https://doi.org/10.3390/microorganisms11082096
Chicago/Turabian StyleYang, Shangxin, Ashrit Multani, Jacob M. Garrigues, Michael S. Oh, Peera Hemarajata, Taylor Burleson, Nicole M. Green, Caspian Oliai, Pryce T. Gaynor, Omer E. Beaird, and et al. 2023. "Transient SARS-CoV-2 RNA-Dependent RNA Polymerase Mutations after Remdesivir Treatment for Chronic COVID-19 in Two Transplant Recipients: Case Report and Intra-Host Viral Genomic Investigation" Microorganisms 11, no. 8: 2096. https://doi.org/10.3390/microorganisms11082096






