Evaluation of Bacillus spp. as Potent Probiotics with Reduction in AHPND-Related Mortality and Facilitating Growth Performance of Pacific White Shrimp (Litopenaeus vannamei) Farms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacillus spp. Isolation
2.2. VPAHPND Isolation and Identification
2.3. Phenotypic Characterization of Bacillus spp.
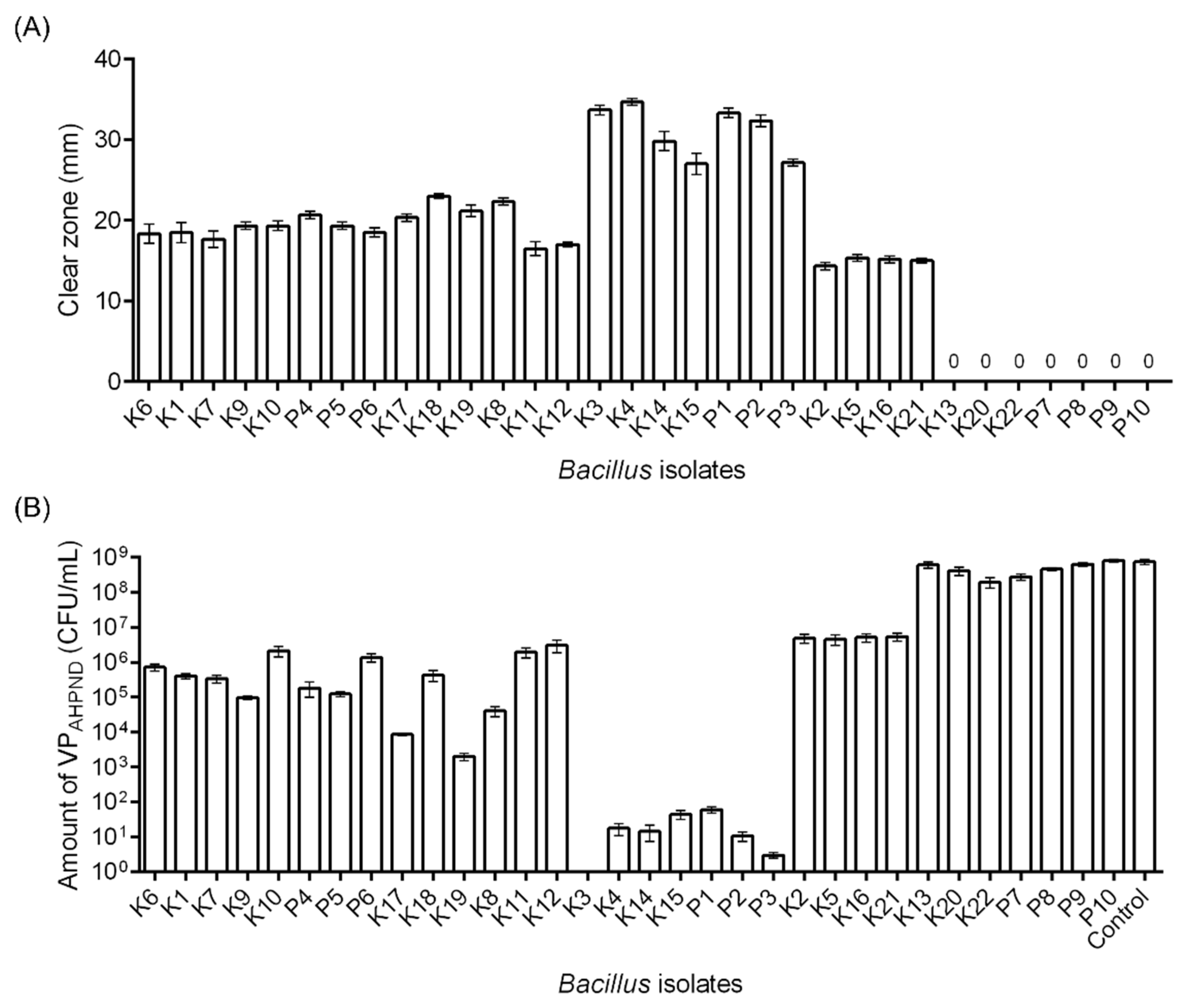
2.3.1. In Vitro Inhibition Assay: Solid Medium
2.3.2. In Vitro Inhibition Assay: Liquid Medium
2.3.3. Antibiotic Susceptibility of Bacillus spp.
2.4. Genotypic Characterization of Bacillus spp.
2.4.1. Species Identification
2.4.2. Antimicrobial Peptide (AMP)-Related Gene Determination
2.5. In Vivo AHPND Challenge Test and Efficacy Analysis
2.5.1. Experimental Shrimp
2.5.2. Preparation of Bacillus spp. and VPAHPND
2.5.3. Pathogenicity Analysis of Isolated VPAHPND in Shrimp
2.5.4. Efficiency of Isolated Bacillus in Controlling AHPND: Laboratory Level
2.5.5. Evaluation of AHPND Control Efficiency at Different Salinities
2.5.6. Validation of Disease Control Efficiency against Different Strains of VPAHPND
2.5.7. Efficiency of Isolated Bacillus in Controlling AHPND: Field Level
2.5.8. Statistical Analysis
3. Results
3.1. Bacillus spp. Isolation
3.2. VPAHPND Isolation, Identification, and Pathogenicity of Isolated VPAHPND
3.3. Phenotypic Characterization of Bacillus spp.
3.3.1. In Vitro Inhibition Assay: Solid Medium and Liquid Medium
3.3.2. Antibiotic Susceptibility of B. subtilis (K3)
3.4. Genotypic Characterization of Bacillus spp.
3.4.1. Species Identification
3.4.2. Antimicrobial Peptide (AMP)-Related Gene Determination
3.5. In Vivo Efficacy Analysis of Bacillus against AHPND
3.5.1. Efficiency of Isolated Bacillus in Controlling AHPND: Laboratory Level
3.5.2. Evaluation of AHPND Control Efficiency at Different Salinities
3.5.3. Validation of Disease Control Efficiency against Different Strains of VPAHPND
3.5.4. Efficiency of Isolated Bacillus in Controlling AHPND: Field Level
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kumar, R.; Ng, T.H.; Wang, H.C. Acute hepatopancreatic necrosis disease in penaeid shrimp. Rev. Aquac. 2020, 12, 1867–1880. [Google Scholar] [CrossRef]
- Lee, C.T.; Chen, I.T.; Yang, Y.T.; Ko, T.P.; Huang, Y.T.; Huang, J.Y.; Huang, M.H.; Lin, S.J.; Chen, C.Y.; Lin, S.S.; et al. The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc. Natl. Acad. Sci. USA 2015, 112, 10798–10803. [Google Scholar] [CrossRef] [PubMed]
- Schryver, P.D.; Defoirdt, T.; Sorgeloos, P. Early mortality syndrome outbreaks: A microbial management issue in shrimp farming. PLoS Pathog. 2014, 10, e1003919. [Google Scholar] [CrossRef] [PubMed]
- Decamp, O.; Moriarty, D.J.; Lavens, P. Probiotics for shrimp larviculture: Review of field data from Asia and Latin America. Aquac. Res. 2008, 39, 334–338. [Google Scholar] [CrossRef]
- FAO/WHO Joint Expert Consultation Report. In Evaluations of Health and Nutritional Properties of Probiotics in Food Including Powder Milk and Live Lactic Acid Bacteria; FAO/WHO: Cordoba, Argentina, 2001.
- Verschuere, L.; Rombaut, G.; Sorgeloos, P.; Verstraete, W. Probiotic bacteria as biological control agents in aquaculture. Microbiol. Mol. Biol. Rev. 2000, 64, 655–671. [Google Scholar] [CrossRef] [PubMed]
- Tseng, D.Y.; Ho, P.L.; Huang, S.Y.; Cheng, S.C.; Shiu, Y.L.; Chiu, C.S.; Liu, C.H. Enhancement of immunity and disease resistance in the white shrimp, Litopenaeus vannamei, by the probiotic, Bacillus subtilis E20. Fish ShellFish Immunol. 2009, 26, 339–344. [Google Scholar] [CrossRef]
- Liu, K.F.; Chiu, C.H.; Shiu, Y.L.; Cheng, W.; Liu, C.H. Effects of the probiotic, Bacillus subtilis E20, on the survival, development, stress tolerance, and immune status of white shrimp, Litopenaeus vannamei larvae. Fish ShellFish Immunol. 2010, 28, 837–844. [Google Scholar] [CrossRef]
- Knipe, H.; Temperton, B.; Lange, A.; Bass, D.; Tyler, C.R. Probiotics and competitive exclusion of pathogens in shrimp aquaculture. Rev. Aquac. 2021, 13, 324–352. [Google Scholar] [CrossRef]
- Vaseeharan, B.A.R.P.; Ramasamy, P. Control of pathogenic Vibrio spp. by Bacillus subtilis BT23, a possible probiotic treatment for black tiger shrimp Penaeus monodon. Lett. Appl. Microbiol. 2003, 36, 83–87. [Google Scholar] [CrossRef]
- Kuebutornye, F.K.; Abarike, E.D.; Lu, Y. A review on the application of Bacillus as probiotics in aquaculture. Fish ShellFish Immunol. 2019, 87, 820–828. [Google Scholar] [CrossRef]
- Balcázar, J.L.; Rojas-Luna, T. Inhibitory activity of probiotic Bacillus subtilis UTM 126 against Vibrio species confers protection against vibriosis in juvenile shrimp (Litopenaeus vannamei). Curr. Microbiol. 2007, 55, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Balcázar, J.L.; Rojas-Luna, T.; Cunningham, D.P. Effect of the addition of four potential probiotic strains on the survival of pacific white shrimp (Litopenaeus vannamei) following immersion challenge with Vibrio parahaemolyticus. J. Invertebr. Pathol. 2007, 96, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.E.; Baiano, J.C.F.; Barnes, A.C. Isolation of a novel strain of Bacillus pumilus from penaeid shrimp that is inhibitory against marine pathogens. J. Fish Dis. 2009, 32, 1007–1016. [Google Scholar] [CrossRef]
- Kewcharoen, W.; Srisapoome, P. Probiotic effects of Bacillus spp. from Pacific white shrimp (Litopenaeus vannamei) on water quality and shrimp growth, immune responses, and resistance to Vibrio parahaemolyticus (AHPND strains). Fish ShellFish Immunol. 2019, 94, 175–189. [Google Scholar] [CrossRef]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions: Bacillus subtilis antibiotics. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Sumi, C.D.; Yang, B.W.; Yeo In-Cheol Hahm, Y.T. Antimicrobial peptides of the genus Bacillus: A new era for antibiotics. Can. J. Microbiol. 2015, 61, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.; Blecha, F. Antimicrobial peptides and bacteriocins: Alternatives to traditional antibiotics. Anim. Health Res. Rev. 2008, 9, 227–235. [Google Scholar] [CrossRef]
- Lai, H.C.; Ng, T.H.; Ando, M.; Lee, C.T.; Chen, I.T.; Chuang, J.C.; Mavichak, R.; Chang, S.H.; Yeh, M.D.; Chiang, Y.A.; et al. Pathogenesis of acute hepatopancreatic necrosis disease (AHPND) in shrimp. Fish ShellFish Immunol. 2015, 47, 1006–1014. [Google Scholar] [CrossRef]
- de la Peña, L.D.; Nakai, T.; Muroga, K. Dynamics of Vibrio sp. PJ in organs of orally infected kuruma prawn, Penaeus japonicus. Fish Pathol. 1995, 30, 39–45. [Google Scholar] [CrossRef]
- Imaizumi, K.; Tinwongger, S.; Kondo, H.; Hirono, I. Analysis of microbiota in the stomach and midgut of two penaeid shrimps during probiotic feeding. Sci. Rep. 2021, 11, 9936. [Google Scholar] [CrossRef]
- Imaizumi, K.; Molex, W.; Jitnavee, C.; Direkbusarakom, S.; Kondo, H.; Hirono, I. Bacterial and eukaryotic communities in pond water of whiteleg shrimp Litopenaeus vannamei and the bacterial communities of their stomach and midgut. Aquaculture 2022, 554, 738139. [Google Scholar] [CrossRef]
- Restrepo, L.; Domínguez-Borbor, C.; Bajaña, L.; Betancourt, I.; Rodríguez, J.; Bayot, B.; Reyes, A. Microbial community characterization of shrimp survivors to AHPND challenge test treated with an effective shrimp probiotic (Vibrio diabolicus). Microbiome 2021, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Thitamadee, S.; Prachumwat, A.; Srisala, J.; Jaroenlak, P.; Salachan, P.V.; Sritunyalucksana, K.; Flegel, T.W.; Itsathitphaisarn, O. Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture 2016, 452, 69–87. [Google Scholar] [CrossRef]
- Sookchaiyaporn, N.; Srisapoome, P.; Unajak, S.; Areechon, N. Efficacy of Bacillus spp. isolated from Nile tilapia Oreochromis niloticus Linn. on its growth and immunity, and control of pathogenic bacteria. Fish Sci. 2020, 86, 353–365. [Google Scholar] [CrossRef]
- Tran, L.; Nunan, L.; Redman, R.M.; Mohney, L.L.; Pantoja, C.R.; Fitzsimmons, K.; Lightner, D.V. Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis. Aquat. Organ. 2013, 105, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Prompamorn, P.; Sithigorngul, P.; Rukpratanporn, S.; Longyant, S.; Sridulyakul, P.; Chaivisuthangkura, P. The development of loop-mediated isothermal amplification combined with lateral flow dipstick for detection of Vibrio parahaemolyticus. Lett. Appl. Microbiol. 2011, 52, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Tinwongger, S.; Proespraiwong, P.; Thawonsuwan, J.; Sriwanayos, P.; Kongkumnerd, J.; Chaweepack, T.; Mavichak, R.; Unajak, S.; Nozaki, R.; Kondo, H.; et al. Development of PCR diagnosis for shrimp acute hepatopancreatic necrosis disease (AHPND) strain of Vibrio parahaemolyticus. Fish Pathol. 2014, 49, 159–164. [Google Scholar] [CrossRef]
- Hockett, K.L.; Baltrus, D.A. Use of the soft-agar overlay technique to screen for bacterially produced inhibitory compounds. J. Vis. Exp. 2017, 119, 55064. [Google Scholar]
- Weinstein, M.P. Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021. [Google Scholar]
- Mora, I.; Cabrefiga, J.; Montesinos, E. Antimicrobial peptide genes in Bacillus strains from plant environments. Int. Microbiol. 2011, 14, 213–223. [Google Scholar]
- Jaroenlak, P.; Sanguanrut, P.; Williams, B.A.P.; Stentiford, G.D.; Flegel, T.W.; Sritunyalucksana, K.; Itsathitphaisarn, O. A nested PCR assay to avoid false positive detection of the microsporidian Enterocytozoon hepatopenaei (EHP) in environmental samples in shrimp farms. PLoS ONE 2016, 11, e0166320. [Google Scholar] [CrossRef]
- Durand, S.V.; Lightner, D.V. Quantitative real time PCR for the measurement of white spot syndrome virus in shrimp. J. Fish Dis. 2002, 25, 381–389. [Google Scholar] [CrossRef]
- Tang, K.F.; Navarro, S.A.; Lightner, D.V. PCR assay for discriminating between infectious hypodermal and hematopoietic necrosis virus (IHHNV) and virus-related sequences in the genome of Penaeus monodon. Dis. Aquat. Org. 2007, 74, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.F.; Wang, J.; Lightner, D.V. Quantitation of Taura syndrome virus by real-time RT-PCR with a TaqMan assay. J. Virol. Methods 2004, 115, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Wijegoonawardane, P.K.; Cowley, J.A.; Walker, P.J. A consensus real-time RT-PCR for detection of all genotypic variants of yellow head virus of penaeid shrimp. J. Virol. Methods 2010, 167, 5–9. [Google Scholar] [CrossRef]
- Dangtip, S.; Sirikharin, R.; Sanguanrut, P.; Thitamadee, S.; Sritunyalucksana, K.; Taengchaiyaphum, S.; Mavichak, R.; Proespraiwong, P.; Flegel, T.W. AP4 method for two-tube nested PCR detection of AHPND isolates of Vibrio parahaemolyticus. Aquac. Rep. 2015, 2, 158–162. [Google Scholar] [CrossRef]
- Kim, K.M.; Lee, J.Y.; Kim, C.K.; Kang, J.S. Isolation and characterization of surfactin produced by Bacillus polyfermenticus KJS-2. Arch. Pharm. Res. 2009, 32, 711–715. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.M.; Rong, Y.J.; Zhao, M.X.; Song, B.; Chi, Z.M. Antibacterial activity of the lipopetides produced by Bacillus amyloliquefaciens M1 against multidrug-resistant Vibrio spp. isolated from diseased marine animals. Appl. Microbiol. Biotechnol. 2014, 98, 127–136. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, S.A.; Mou, H.; Ma, Y.; Li, M.; Hu, X. Characterization of lipopeptide biosurfactants produced by Bacillus licheniformis MB01 from marine sediments. Front. Microbiol. 2017, 8, 871. [Google Scholar] [CrossRef]
- Liu, C.H.; Chiu, C.S.; Ho, P.L.; Wang, S.W. Improvement in the growth performance of white shrimp, Litopenaeus vannamei, by a protease-producing probiotic, Bacillus subtilis E20, from natto. J. Appl. Microbiol. 2009, 107, 1031–1041. [Google Scholar] [CrossRef]
- Ziaei-Nejad, S.; Rezaei, M.H.; Takami, G.A.; Lovett, D.L.; Mirvaghefi, A.R.; Shakouri, M. The effect of Bacillus spp. bacteria used as probiotics on digestive enzyme activity, survival and growth in the Indian white shrimp Fenneropenaeus indicus. Aquaculture 2006, 252, 516–524.4444. [Google Scholar] [CrossRef]
- López-Cervantes, G.; Álvarez-Ruiz, P.; Luna-Suárez, S.; Luna-González, A.; Esparza-Leal, H.M.; Castro-Martínez, C.; Gámez-Jiménez, C.; Soto-Alcalá, J. Temperature and salinity modulate virulence and PirA gene expression of Vibrio parahaemolyticus, the causative agent of AHPND. Aquac. Int. 2021, 29, 743–756. [Google Scholar] [CrossRef]
- Schofield, P.J.; Noble, B.L.; Caro, L.F.A.; Mai, H.N.; Padilla, T.J.; Millabas, J.; Dhar, A.K. Pathogenicity of acute hepatopancreatic necrosis disease (AHPND) on the freshwater prawn, Macrobrachium rosenbergii, and Pacific white shrimp, Penaeus vannamei, at various salinities. Aquac. Res. 2021, 52, 1480–1489. [Google Scholar] [CrossRef]
- Charmantier, G.; Charmantier-Daures, M.; Bouaricha, N.; Thuet, P.; Trilles, J.P.; Aiken, D.E. Ontogeny of osmoregulation and salinity tolerance in two decapod crustaceans: Homarus americanus and Penaeus japonicus. Biol. Bull. 1988, 175, 102–110. [Google Scholar] [CrossRef]
- Roy, L.A.; Davis, D.A.; Saoud, I.P.; Boyd, C.A.; Pine, H.J.; Boyd, C.E. Shrimp culture in inland low salinity waters. Rev. Aquac. 2010, 2, 191–208. [Google Scholar] [CrossRef]
- Li, E.; Wang, X.; Chen, K.; Xu, C.; Qin, J.G.; Chen, L. Physiological change and nutritional requirement of Pacific white shrimp Litopenaeus vannamei at low salinity. Rev. Aquac. 2017, 9, 57–75. [Google Scholar] [CrossRef]
- Chai, P.C.; Song, X.L.; Chen, G.F.; Xu, H.; Huang, J. Dietary supplementation of probiotic Bacillus PC465 isolated from the gut of Fenneropenaeus chinensis improves the health status and resistance of Litopenaeus vannamei against white spot syndrome virus. Fish ShellFish Immunol. 2016, 54, 602–611. [Google Scholar] [CrossRef]
- Holt, C.C.; Bass, D.; Stentiford, G.D.; van der Giezen, M. Understanding the role of the shrimp gut microbiome in health and disease. J. Invertebr. Pathol. 2021, 186, 107387. [Google Scholar] [CrossRef]
- Piewngam, P.; Zheng, Y.; Nguyen, T.H.; Dickey, S.W.; Joo, H.S.; Villaruz, A.E.; Glose, K.A.; Fisher, E.L.; Hunt, R.L.; Li, B.; et al. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 2018, 562, 532–537. [Google Scholar] [CrossRef]



| Gene | AMP | Primer | Sequence (5′ → 3′) | Product Size (bp) |
|---|---|---|---|---|
| fenD | Fengycin | fenD_F | GGCCCGTTCTCTAAATCCAT | 269 |
| fenD_R | GTCATGCTGACGAGAGCAAA | |||
| bmyB | Bacillomycin | bmyB_F | GAATCCCGTTGTTCTCCAAA | 370 |
| bmyB_R | GCGGGTATTGAATGCTTGTT | |||
| ituC | Iturin | ituC_F | GGCTGCTGCAGATGCTTTAT | 423 |
| ituC_R | TCGCAGATAATCGCAGTGAG | |||
| srfAA | Surfactin | srfAA_F | TCGGGACAGGAAGACATCAT | 201 |
| srfAA_R | CCACTCAAACGGATAATCCTGA | |||
| bacA | Bacilysin | bacA_F | CAGCTCATGGGAATGCTTTT | 498 |
| bacA_R | CTCGGTCCTGAAGGGACAAG | |||
| spaS | Subtilin | spaS_F | GGTTTGTTGGATGGAGCTGT | 375 |
| spaS_R | GCAAGGAGTCAGAGCAAGGT |
| Antibiotic | Disc Potency (µg) | Bacillus sp. Isolate K3 | |
|---|---|---|---|
| Zone Diameter (mm) | Interpretation | ||
| Amoxicillin | 10 | 15.0 | I |
| Ceftriaxone | 30 | 38.5 | S |
| Doxycycline | 30 | 27.0 | S |
| Gentamycin | 10 | 16.5 | S |
| Enrofloxacin | 5 | 27.5 | S |
| Erythromycin | 15 | 21.0 | S |
| Norfloxacin | 10 | 31.0 | S |
| Oxytetracycline | 30 | 26.5 | I |
| Streptomycin | 10 | 14.0 | I |
| Sulfa-trimethoprim | 25 | 28.5 | S |
| Tetracycline | 30 | 28.0 | S |
| Isolate | Origin of Isolation | 16S rRNA | BacA | srfAA | ituC | fenD | spaS | bmyB |
|---|---|---|---|---|---|---|---|---|
| K1 | Shrimp | B. methylotrophicus | + | + | − | + | − | + |
| K2 | Shrimp | B. licheniformis | − | + | − | − | − | − |
| K3 | Shrimp | B. subtilis | + | + | − | − | − | − |
| K4 | Shrimp | B. subtilis | + | + | − | − | − | − |
| K5 | Shrimp | B. licheniformis | − | + | − | − | − | − |
| K6 | Shrimp | B. amyloliquefaciens | + | + | + | + | − | + |
| K7 | Shrimp | B. amyloliquefaciens | + | + | − | + | − | + |
| K8 | Shrimp | B. subtilis | + | + | − | − | − | + |
| K9 | Shrimp | B. amyloliquefaciens | + | + | − | + | − | + |
| K10 | Shrimp | B. subtilis | + | + | − | + | − | + |
| K11 | Shrimp | B. vallismortis | − | + | − | − | − | + |
| K12 | Shrimp | B. subtilis | − | + | − | + | − | − |
| K13 | Shrimp | B. subtilis | − | − | − | − | − | − |
| K14 | Shrimp | B. subtilis | + | + | − | − | − | − |
| K15 | Shrimp | B. subtilis | + | + | − | − | − | − |
| K16 | Shrimp | B. licheniformis | − | + | − | − | − | − |
| K17 | Shrimp | B. subtilis | + | + | − | + | − | + |
| K18 | Shrimp | B. subtilis | + | + | − | − | + | − |
| K19 | Shrimp | B. subtilis | + | + | − | − | − | + |
| K20 | Shrimp | B. cereus | − | − | − | − | − | − |
| K21 | Shrimp | B. licheniformis | − | + | − | − | − | − |
| K22 | Shrimp | B. flexus | − | − | − | − | − | − |
| P1 | Mangrove | B. tequilensis | + | + | − | − | − | − |
| P2 | Mangrove | B. amyloliquefaciens | + | + | − | − | − | − |
| P3 | Mangrove | B. tequilensis | + | + | − | − | − | − |
| P4 | Mangrove | B. amyloliquefaciens | + | + | − | + | − | − |
| P5 | Mangrove | B. velezensis | + | + | − | + | − | + |
| P6 | Mangrove | B. velezensis | + | + | − | + | − | + |
| P7 | Mangrove | B. methylotrophicus | − | − | − | − | − | − |
| P8 | Mangrove | B. velezensis | − | − | − | − | − | − |
| P9 | Mangrove | B. firmus | − | − | − | − | − | − |
| P10 | Mangrove | B. velezensis | − | − | − | − | − | − |
| Parameters | Control | B. subtilis (K3) |
|---|---|---|
| No. of ponds | 30 | 30 |
| No. of shrimp stocked/pond | 600,000 | 600,000 |
| No. of AHPND-positive ponds | 10 | 2 |
| Average survival rate (%) | 68.1 | 94.6 |
| Average FCR | 1.67 | 1.45 |
| Number of drain ponds | 5 | 0 |
| Final size (pcs/kg) | 942.48 | 783.53 |
| Total shrimp weight (kg)/pond | 433.75 | 724.53 |
| Total feed (kg)/pond | 725.75 | 1053.90 |
| No. of surviving shrimp/pond | 408,801 | 567,604 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Proespraiwong, P.; Mavichak, R.; Imaizumi, K.; Hirono, I.; Unajak, S. Evaluation of Bacillus spp. as Potent Probiotics with Reduction in AHPND-Related Mortality and Facilitating Growth Performance of Pacific White Shrimp (Litopenaeus vannamei) Farms. Microorganisms 2023, 11, 2176. https://doi.org/10.3390/microorganisms11092176
Proespraiwong P, Mavichak R, Imaizumi K, Hirono I, Unajak S. Evaluation of Bacillus spp. as Potent Probiotics with Reduction in AHPND-Related Mortality and Facilitating Growth Performance of Pacific White Shrimp (Litopenaeus vannamei) Farms. Microorganisms. 2023; 11(9):2176. https://doi.org/10.3390/microorganisms11092176
Chicago/Turabian StyleProespraiwong, Porranee, Rapeepat Mavichak, Kentaro Imaizumi, Ikuo Hirono, and Sasimanas Unajak. 2023. "Evaluation of Bacillus spp. as Potent Probiotics with Reduction in AHPND-Related Mortality and Facilitating Growth Performance of Pacific White Shrimp (Litopenaeus vannamei) Farms" Microorganisms 11, no. 9: 2176. https://doi.org/10.3390/microorganisms11092176





