Diversity and Abundance of Bacterial and Fungal Communities Inhabiting Camellia sinensis Leaf, Rhizospheric Soil, and Gut of Agriophara rhombata
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Site and Data Collection
2.2. DNA Extraction, Amplification, and Illumina Sequencing of ITS and 16S rRNA Amplicons
2.3. Metagenomic Sequence Data Analyses
2.4. Statistical Analysis
3. Results
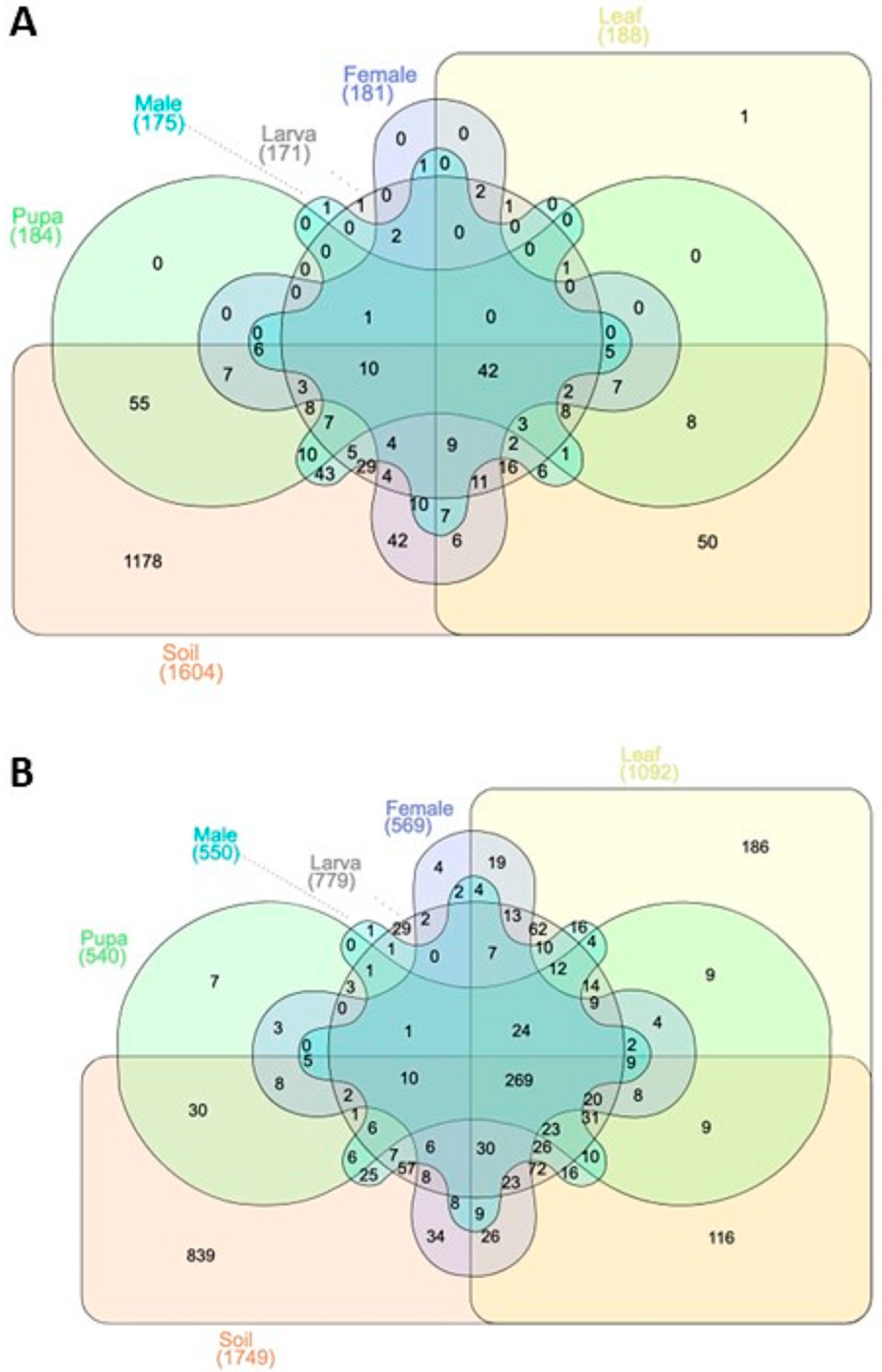
3.1. OTUs Annotation Results
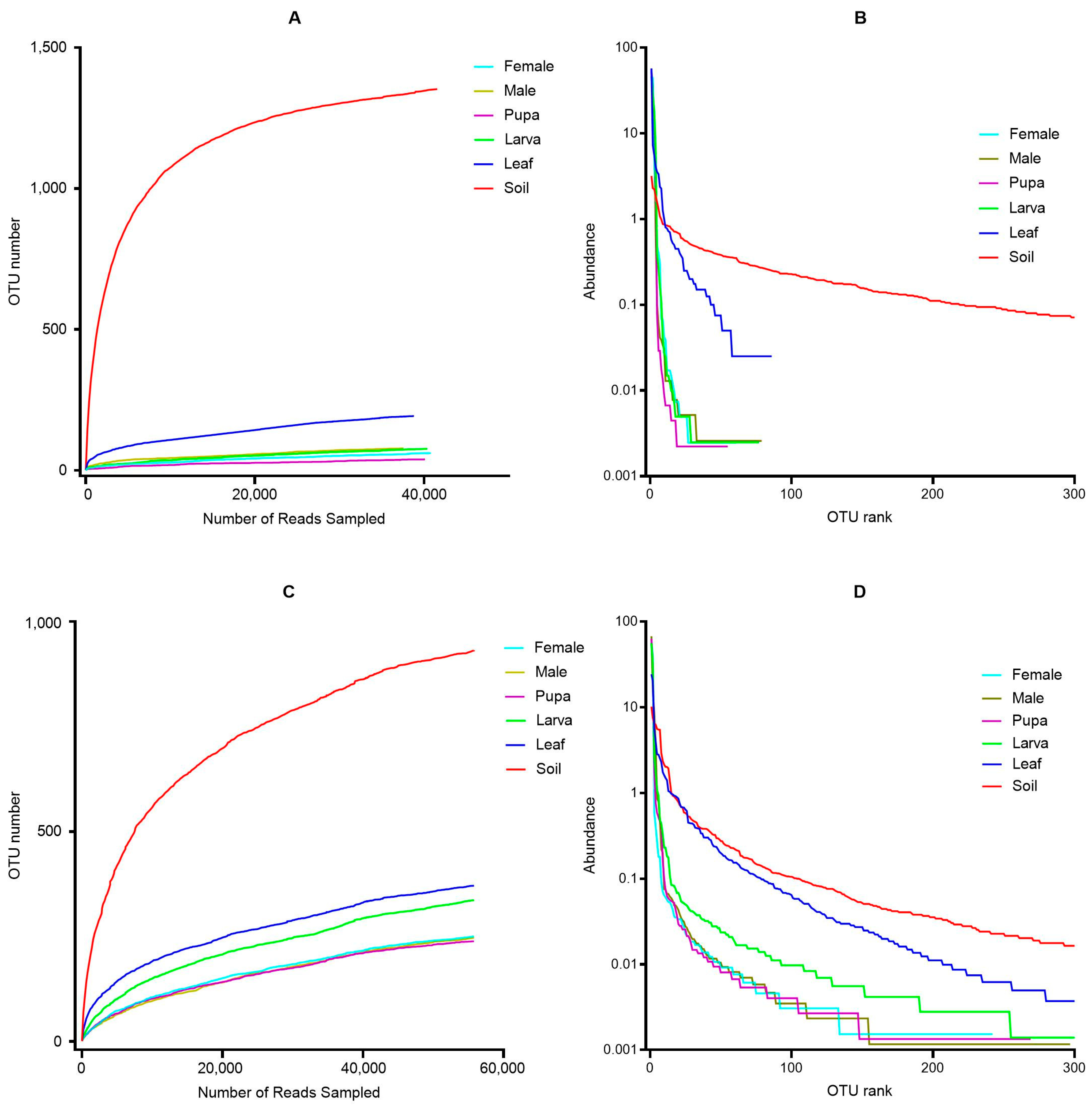
3.2. Alpha Diversity
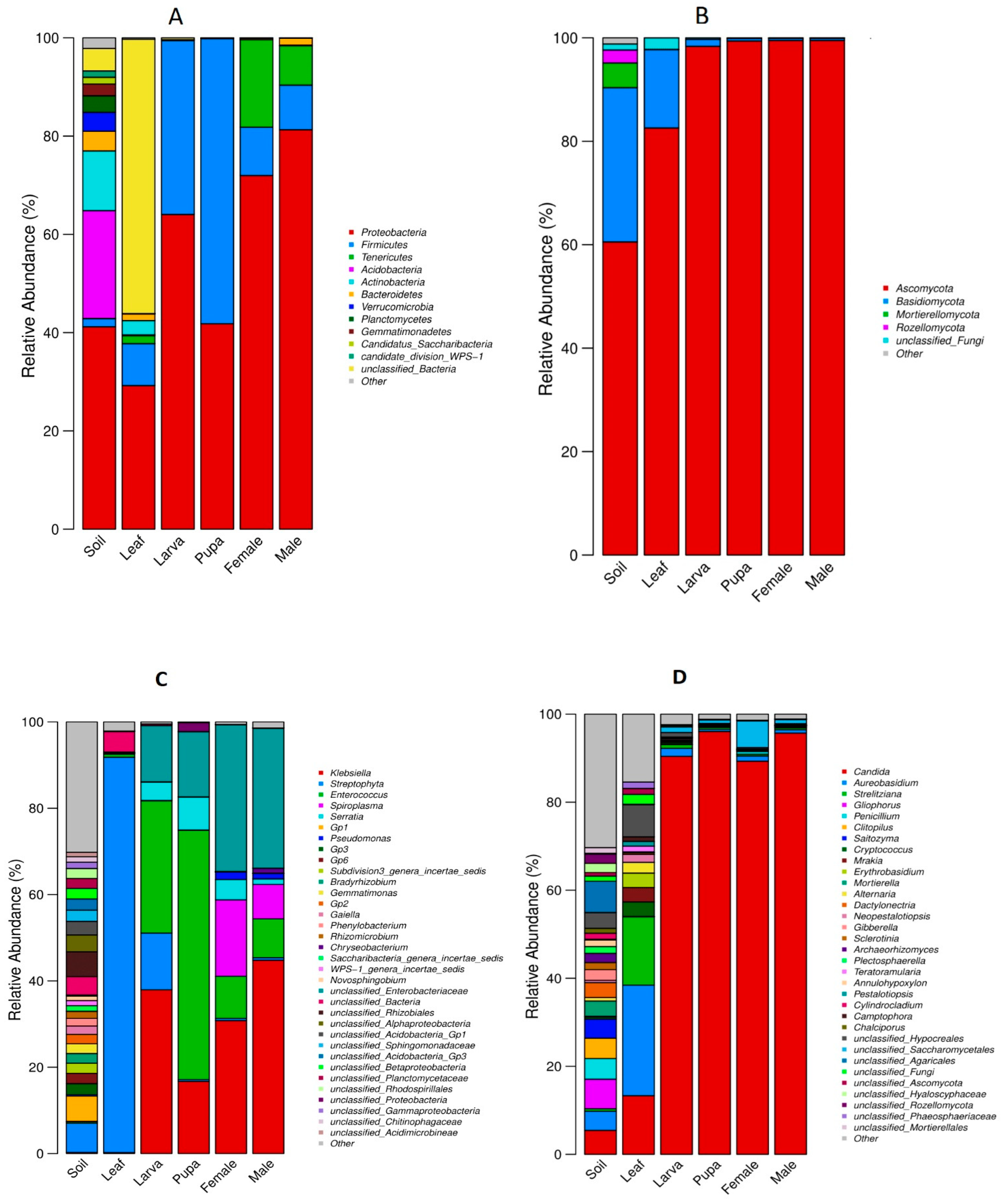
3.3. Relative Abundance of Bacterial and Fungal Communities
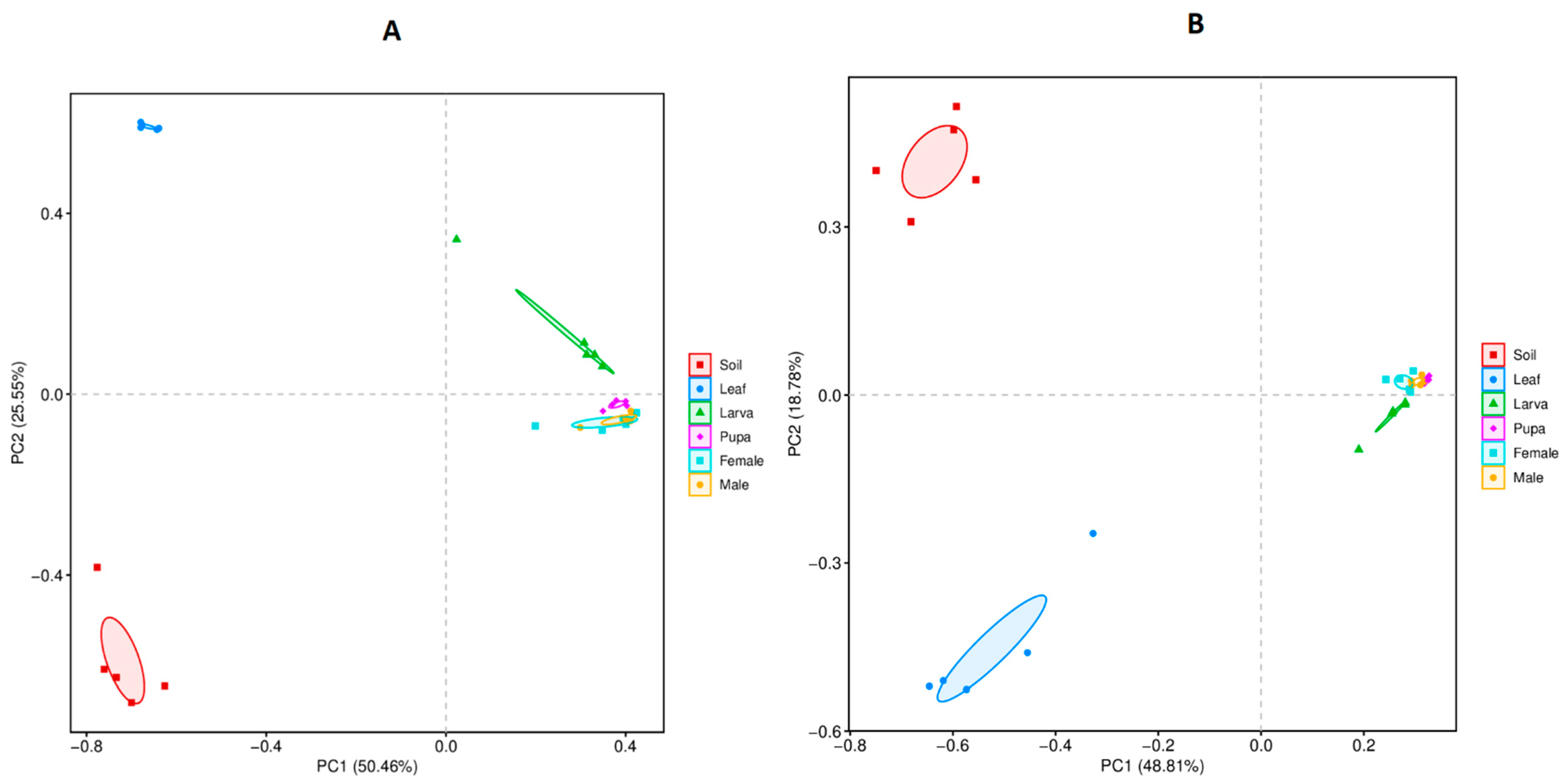
3.4. Beta Diversity
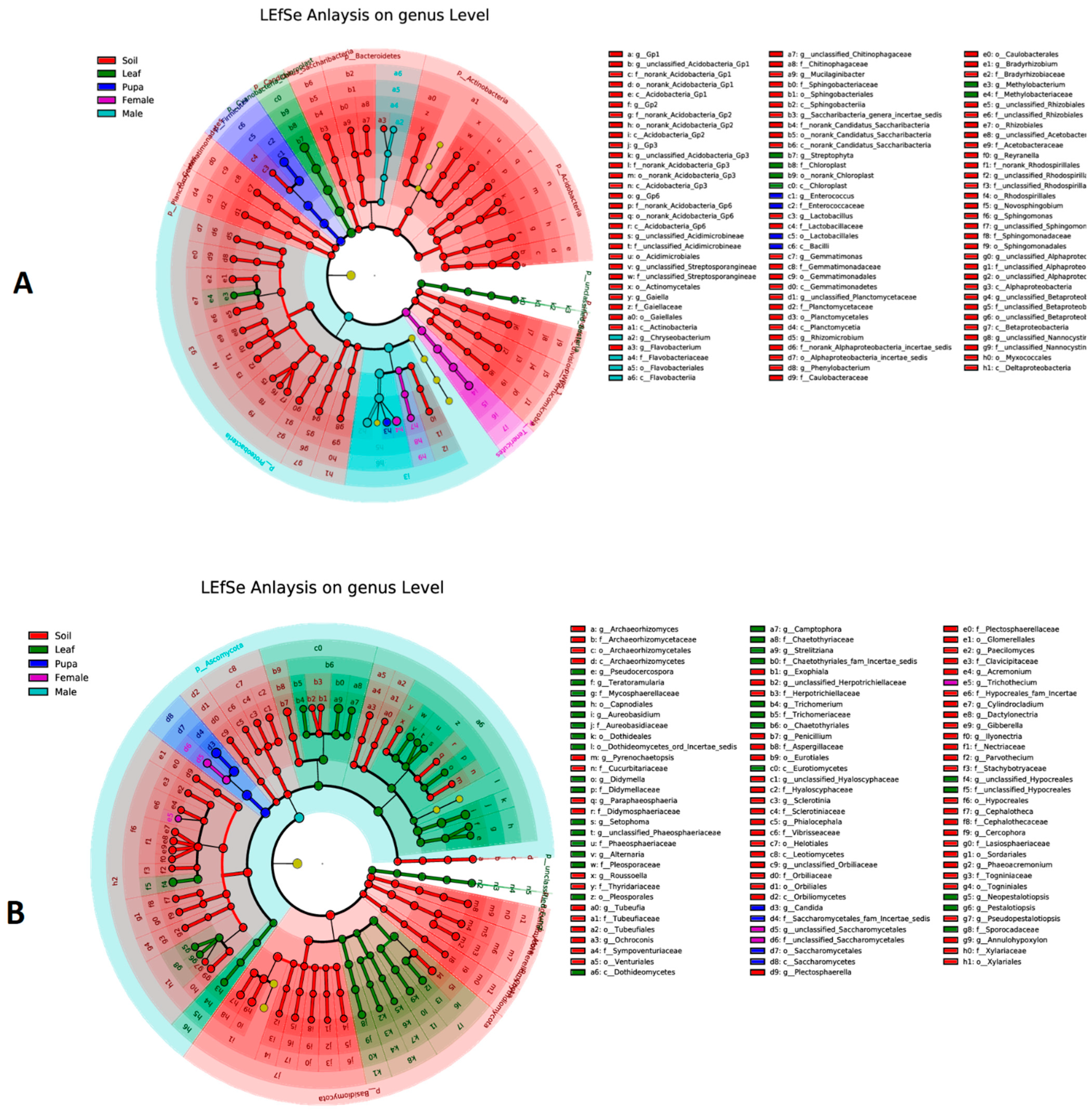
3.5. Biomarker Taxa of Bacterial and Fungal Communities
3.6. Functional Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wei, C.; Yang, H.; Wang, S.; Zhao, J.; Liu, C.; Gao, L.; Xia, E.; Lu, Y.; Tai, Y.; She, G. Draft Genome Sequence of Camellia sinensis Var. Sinensis Provides Insights into the Evolution of the Tea Genome and Tea Quality. Proc. Natl. Acad. Sci. USA 2018, 115, E4151–E4158. [Google Scholar] [CrossRef] [PubMed]
- Vishnoi, H.; Bodla, R.B.; Kant, R.; Bodla, R.B. Green Tea (Camellia sinensis) and Its Antioxidant Property: A Review. Int. J. Pharm. Sci. Res. 2018, 9, 1723–1736. [Google Scholar]
- Chan, E.W.C.; Lim, Y.Y.; Chew, Y.L. Antioxidant Activity of Camellia sinensis Leaves and Tea from a Lowland Plantation in Malaysia. Food Chem. 2007, 102, 1214–1222. [Google Scholar] [CrossRef]
- Jiang, H.; Yu, F.; Qin, L.I.; Zhang, N.; Cao, Q.; Schwab, W.; Li, D.; Song, C. Dynamic Change in Amino Acids, Catechins, Alkaloids, and Gallic Acid in Six Types of Tea Processed from the Same Batch of Fresh Tea (Camellia sinensis L.) Leaves. J. Food Compos. Anal. 2019, 77, 28–38. [Google Scholar] [CrossRef]
- Too, J.C.; Kinyanjui, T.; Wanyoko, J.K.; Wachira, F.N. Effect of Sunlight Exposure and Different Withering Durations on Theanine Levels in Tea (Camellia sinensis). Food Nutr. Sci. 2015, 6, 1014. [Google Scholar]
- Ji, H.-G.; Lee, Y.-R.; Lee, M.-S.; Hwang, K.H.; Kim, E.-H.; Park, J.S.; Hong, Y.-S. Metabolic Phenotyping of Various Tea (Camellia sinensis L.) Cultivars and Understanding of Their Intrinsic Metabolism. Food Chem. 2017, 233, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Sharangi, A.B. Medicinal and Therapeutic Potentialities of Tea (Camellia sinensis L.)—A Review. Food Res. Int. 2009, 42, 529–535. [Google Scholar] [CrossRef]
- Mondal, T.K.; Bhattacharya, A.; Laxmikumaran, M.; Singh Ahuja, P. Recent Advances of Tea (Camellia sinensis) Biotechnology. Plant Cell Tissue Organ Cult. 2004, 76, 195–254. [Google Scholar] [CrossRef]
- Liao, Y.; Zhou, X.; Zeng, L. How Does Tea (Camellia sinensis) Produce Specialized Metabolites Which Determine Its Unique Quality and Function: A Review. Crit. Rev. Food Sci. Nutr. 2022, 62, 3751–3767. [Google Scholar] [CrossRef]
- Wu, Q.; Sun, C.; Yang, J. Economic Analysis of the Change of Tea Production Layout in China. J. Phys. Conf. Ser. 2020, 1629, 12048. [Google Scholar] [CrossRef]
- Das, T.; Rahman, A. Lepidopteran Pests of Tea: Biology, Geographical Distribution, and Management. Phytoparasitica 2023, 51, 461–489. [Google Scholar] [CrossRef]
- Liu, Z.; Bashir, R.N.; Iqbal, S.; Shahid, M.M.A.; Tausif, M.; Umer, Q. Internet of Things (IoT) and Machine Learning Model of Plant Disease Prediction–Blister Blight for Tea Plant. IEEE Access 2022, 10, 44934–44944. [Google Scholar] [CrossRef]
- Ding, X.; Lu, Q.; Li, L.; Li, H.; Sarkar, A. Measuring the Impact of Relative Deprivation on Tea Farmers’ Pesticide Application Behavior: The Case of Shaanxi, Sichuan, Zhejiang, and Anhui Province, China. Horticulturae 2023, 9, 342. [Google Scholar] [CrossRef]
- Borah, A.; Hazarika, S.N.; Thakur, D. Potentiality of Actinobacteria to Combat against Biotic and Abiotic Stresses in Tea [Camellia sinensis (L) O. Kuntze]. J. Appl. Microbiol. 2022, 133, 2314–2330. [Google Scholar] [CrossRef] [PubMed]
- Ng’etich, W.K.; Bore, J.K.; Cheserek, B. Biotic and Abiotic Factors Affecting Tea: A Review. Tea 2006, 27, 29–35. [Google Scholar]
- Paul, S.K.; Ahmed, M.; Mamun, M.S.A.; Alam, M.J. Diversity of Insect, Mite and Nematode Species in Tea Ecosystem of Bangladesh. J. Biodivers. Conserv. Bioresour. Manag. 2017, 3, 31–44. [Google Scholar] [CrossRef][Green Version]
- Roy, S.; Prasad, A.K.; Pradhan, B.; Mukhopadhyay, A. Pestiferous Red Slug Caterpillars of Eterusia Aedea (Lepidoptera: Zygaenidae): Status, Bioecology and Management in Tea Plantations of India. J. Lepid. Soc. 2018, 72, 87–95. [Google Scholar] [CrossRef]
- Ye, G.-Y.; Xiao, Q.; Chen, M.; Chen, X.; Yuan, Z.; Stanley, D.W.; Hu, C. Tea: Biological Control of Insect and Mite Pests in China. Biol. Control 2014, 68, 73–91. [Google Scholar] [CrossRef]
- Long, Y.; Luo, Z.; Wang, X.; Long, L.; Yu, X.; Li, J.; Qu, H.; Wang, Y.; Chen, L. Antenna Transcriptome and Olfactory-Related Genes Analysis of Adult Tea Moth. J. Tea Sci. 2021, 41, 553–563. [Google Scholar]
- Lozupone, C.A.; Knight, R. Global Patterns in Bacterial Diversity. Proc. Natl. Acad. Sci. USA 2007, 104, 11436–11440. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A Global Atlas of the Dominant Bacteria Found in Soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef]
- Chi, F.; Shen, S.-H.; Cheng, H.-P.; Jing, Y.-X.; Yanni, Y.G.; Dazzo, F.B. Ascending Migration of Endophytic Rhizobia, from Roots to Leaves, inside Rice Plants and Assessment of Benefits to Rice Growth Physiology. Appl. Environ. Microbiol. 2005, 71, 7271–7278. [Google Scholar] [CrossRef] [PubMed]
- Hannula, S.E.; Zhu, F.; Heinen, R.; Bezemer, T.M. Foliar-Feeding Insects Acquire Microbiomes from the Soil Rather than the Host Plant. Nat. Commun. 2019, 10, 1254. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Moran, N.A. The Gut Microbiota of Insects–Diversity in Structure and Function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E. Multiorganismal Insects: Diversity and Function of Resident Microorganisms. Annu. Rev. Entomol. 2015, 60, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Serrat, X.; Esteban, R.; Guibourt, N.; Moysset, L.; Nogués, S.; Lalanne, E. EMS Mutagenesis in Mature Seed-Derived Rice Calli as a New Method for Rapidly Obtaining TILLING Mutant Populations. Plant Methods 2014, 10, 5. [Google Scholar] [CrossRef]
- Paniagua Voirol, L.R.; Frago, E.; Kaltenpoth, M.; Hilker, M.; Fatouros, N.E. Bacterial Symbionts in Lepidoptera: Their Diversity, Transmission, and Impact on the Host. Front. Microbiol. 2018, 9, 556. [Google Scholar] [CrossRef]
- Scoble, M.J. The Lepidoptera. Form, Function and Diversity; Oxford University Press: Oxford, UK, 1992; ISBN 0198540310. [Google Scholar]
- Tringe, S.G.; von Mering, C.; Kobayashi, A.; Salamov, A.A.; Chen, K.; Chang, H.W.; Podar, M.; Short, J.M.; Mathur, E.J.; Detter, J.C.; et al. Comparative metagenomics of microbial communities. Science 2005, 308, 554–557. [Google Scholar] [CrossRef]
- Llewellyn, S.; Fitzpatrick, R.; Kenny, D.A.; Murphy, J.J.; Scaramuzzi, R.J.; Wathes, D.C. Effect of Negative Energy Balance on the Insulin-like Growth Factor System in Pre-Recruitment Ovarian Follicles of Post Partum Dairy Cows. Reproduction 2007, 133, 627–639. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M. Ultra-High-Throughput Microbial Community Analysis on the Illumina HiSeq and MiSeq Platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Tedersoo, L.; Anslan, S.; Bahram, M.; Põlme, S.; Riit, T.; Liiv, I.; Kõljalg, U.; Kisand, V.; Nilsson, H.; Hildebrand, F. Shotgun Metagenomes and Multiple Primer Pair-Barcode Combinations of Amplicons Reveal Biases in Metabarcoding Analyses of Fungi. MycoKeys 2015, 10, 1–43. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and Clustering Orders of Magnitude Faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- De Hollander, M. Nioo-Knaw/Hydra: 1.3. 3. Zenodo. 2017, 10. Available online: http://www.arb-silva.de/ (accessed on 15 July 2022).
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.-H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; Pennanen, T. The UNITE Database for Molecular Identification of Fungi–Recent Updates and Future Perspectives. New Phytol. 2010, 186, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Marco-Ramell, A.; Palau-Rodriguez, M.; Alay, A.; Tulipani, S.; Urpi-Sarda, M.; Sanchez-Pla, A.; Andres-Lacueva, C. Evaluation and Comparison of Bioinformatic Tools for the Enrichment Analysis of Metabolomics Data. BMC Bioinform. 2018, 19, 1. [Google Scholar] [CrossRef]
- Das, T.; Bhattacharyya, A.; Bhar, A. Linking Phyllosphere and Rhizosphere Microbiome to the Plant–Insect Interplay: The New Dimension of Tripartite Interaction. Physiologia 2023, 3, 129–144. [Google Scholar] [CrossRef]
- Jiang, N.; Liu, S.; Xue, D.; Tang, M.; Xiao, Q.; Han, H. External Morphology and Molecular Identification of Two Tea Geometrid Moth from Southern China. Chin. J. Appl. Entomol. 2014, 51, 987–1002. [Google Scholar]
- Hammer, T.J.; Janzen, D.H.; Hallwachs, W.; Jaffe, S.P.; Fierer, N. Caterpillars Lack a Resident Gut Microbiome. Proc. Natl. Acad. Sci. USA 2017, 114, 9641–9646. [Google Scholar] [CrossRef]
- Šigut, M.; Pyszko, P.; Šigutová, H.; Višňovská, D.; Kostovčík, M.; Kotásková, N.; Dorňák, O.; Kolařík, M.; Drozd, P. Fungi Are More Transient than Bacteria in Caterpillar Gut Microbiomes. Sci. Rep. 2022, 12, 15552. [Google Scholar] [CrossRef]
- González-Serrano, F.; Pérez-Cobas, A.E.; Rosas, T.; Baixeras, J.; Latorre, A.; Moya, A. The Gut Microbiota Composition of the Moth Brithys Crini Reflects Insect Metamorphosis. Microb. Ecol. 2020, 79, 960–970. [Google Scholar] [CrossRef]
- Xia, X.; Gurr, G.M.; Vasseur, L.; Zheng, D.; Zhong, H.; Qin, B.; Lin, J.; Wang, Y.; Song, F.; Li, Y. Metagenomic Sequencing of Diamondback Moth Gut Microbiome Unveils Key Holobiont Adaptations for Herbivory. Front. Microbiol. 2017, 8, 663. [Google Scholar] [CrossRef] [PubMed]
- MsangoSoko, K.; Gandotra, S.; Chandel, R.K.; Sharma, K.; Ramakrishinan, B.; Subramanian, S. Composition and Diversity of Gut Bacteria Associated with the Eri Silk Moth, Samia Ricini,(Lepidoptera: Saturniidae) as Revealed by Culture-Dependent and Metagenomics Analysis. J. Microbiol. Biotechnol. 2020, 30, 1367–1378. [Google Scholar] [CrossRef] [PubMed]
- Brumfield, K.D.; Raupp, M.J.; Haji, D.; Simon, C.; Graf, J.; Cooley, J.R.; Janton, S.T.; Meister, R.C.; Huq, A.; Colwell, R.R. Gut Microbiome Insights from 16S RRNA Analysis of 17-Year Periodical Cicadas (Hemiptera: Magicicada Spp.) Broods II, VI, and X. Sci. Rep. 2022, 12, 16967. [Google Scholar] [CrossRef]
- Chen, J.; Ma, Y.; Huang, S.; Li, J.; Zhang, Y.; Wang, H.; Qi, G.; Shi, Q.; Zhang, Z.; Yang, M. The Dynamics of the Microbial Community in Fall Armyworm Spodoptera Frugiperda during a Life Cycle. Entomol. Exp. Appl. 2023, 171, 502–513. [Google Scholar] [CrossRef]
- Chen, B.; Teh, B.-S.; Sun, C.; Hu, S.; Lu, X.; Boland, W.; Shao, Y. Biodiversity and Activity of the Gut Microbiota across the Life History of the Insect Herbivore Spodoptera Littoralis. Sci. Rep. 2016, 6, 29505. [Google Scholar] [CrossRef]
- Reid, N.M.; Addison, S.L.; Macdonald, L.J.; Lloyd-Jones, G. Biodiversity of Active and Inactive Bacteria in the Gut Flora of Wood-Feeding Huhu Beetle Larvae (Prionoplus reticularis). Appl. Environ. Microbiol. 2011, 77, 7000–7006. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Gu, Y.; Zhou, Z.; Liu, Z.; Yin, H.; Qin, C.; Yi, T.; Tao, J. Ecological Strategies of Biological and Chemical Control Agents on Wildfire Disease of Tobacco (Nicotiana tabacum L.). BMC Microbiol. 2021, 21, 184. [Google Scholar] [CrossRef]
- Teh, B.-S.; Apel, J.; Shao, Y.; Boland, W. Colonization of the Intestinal Tract of the Polyphagous Pest Spodoptera Littoralis with the GFP-Tagged Indigenous Gut Bacterium Enterococcus Mundtii. Front. Microbiol. 2016, 7, 928. [Google Scholar] [CrossRef]
- Zheng, Y.; Xiao, G.; Zhou, W.; Gao, Y.; Li, Z.; Du, G.; Chen, B. Midgut Microbiota Diversity of Potato Tuber Moth Associated with Potato Tissue Consumed. BMC Microbiol. 2020, 20, 58. [Google Scholar] [CrossRef]
- Porter, T.M.; Schadt, C.W.; Rizvi, L.; Martin, A.P.; Schmidt, S.K.; Scott-Denton, L.; Vilgalys, R.; Moncalvo, J.-M. Widespread Occurrence and Phylogenetic Placement of a Soil Clone Group Adds a Prominent New Branch to the Fungal Tree of Life. Mol. Phylogenet. Evol. 2008, 46, 635–644. [Google Scholar] [CrossRef]
- Liu, H.; Yao, J.; Liu, B.; Li, M.; Liu, J.; Jiang, S.; Yu, W.; Zhao, Y.; Duran, R. Active Tailings Disturb the Surrounding Vegetation Soil Fungal Community: Diversity, Assembly Process and Co-Occurrence Patterns. Sci. Total Environ. 2023, 865, 161133. [Google Scholar] [CrossRef]
- Sumbula, V.; Kurian, P.S.; Girija, D.; Cherian, K.A. Impact of Foliar Application of Fungicides on Tomato Leaf Fungal Community Structure Revealed by Metagenomic Analysis. Folia Microbiol. 2022, 67, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, A.; Nicosia, M.G.L.D.; Mosca, S.; Cacciola, S.O.; Schena, L. Metagenomic Analysis of the Fungal Microbiome in the Olive Canopy. Order 2014, 1, 3. [Google Scholar]
- Deng, X.; Yang, J.; Wan, Y.; Han, Y.; Tong, H.; Chen, Y. Characteristics of Leaf Spot Disease Caused by Didymella Species and the Influence of Infection on Tea Quality. Phytopathology 2023, 113, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Duan, C.; Sang, Y.; Wu, S.; Ru, J.; Cui, X. Effects of Graphene on Bacterial Community Diversity and Soil Environments of Haplic Cambisols in Northeast China. Forests 2018, 9, 677. [Google Scholar] [CrossRef]
- Sun, Y.; Fan, M.; He, Y. DNA Methylation Analysis of the Citrullus Lanatus Response to Cucumber Green Mottle Mosaic Virus Infection by Whole-Genome Bisulfite Sequencing. Genes 2019, 10, 344. [Google Scholar] [CrossRef]
- Li, T.; Zhou, C.; Gong, J.; Xi, Z.; Hong, X. Recently Introduced Wolbachia Reduces Bacterial Species Richness and Reshapes Bacterial Community Structure in Nilaparvata Lugens. Pest Manag. Sci. 2022, 78, 1881–1894. [Google Scholar] [CrossRef]
- Mazumdar, T.; Teh, B.S.; Murali, A.; Schmidt-Heck, W.; Schlenker, Y.; Vogel, H.; Boland, W. Transcriptomics Reveal the Survival Strategies of Enterococcus Mundtii in the Gut of Spodoptera Littoralis. J. Chem. Ecol. 2021, 47, 227–241. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, H.; Long, Y.; Wang, X.; Wang, K.; Chen, L.; Yang, Y.; Chen, L. Diversity and Abundance of Bacterial and Fungal Communities Inhabiting Camellia sinensis Leaf, Rhizospheric Soil, and Gut of Agriophara rhombata. Microorganisms 2023, 11, 2188. https://doi.org/10.3390/microorganisms11092188
Qu H, Long Y, Wang X, Wang K, Chen L, Yang Y, Chen L. Diversity and Abundance of Bacterial and Fungal Communities Inhabiting Camellia sinensis Leaf, Rhizospheric Soil, and Gut of Agriophara rhombata. Microorganisms. 2023; 11(9):2188. https://doi.org/10.3390/microorganisms11092188
Chicago/Turabian StyleQu, Hao, Yaqin Long, Xuesong Wang, Kaibo Wang, Long Chen, Yunqiu Yang, and Linbo Chen. 2023. "Diversity and Abundance of Bacterial and Fungal Communities Inhabiting Camellia sinensis Leaf, Rhizospheric Soil, and Gut of Agriophara rhombata" Microorganisms 11, no. 9: 2188. https://doi.org/10.3390/microorganisms11092188
APA StyleQu, H., Long, Y., Wang, X., Wang, K., Chen, L., Yang, Y., & Chen, L. (2023). Diversity and Abundance of Bacterial and Fungal Communities Inhabiting Camellia sinensis Leaf, Rhizospheric Soil, and Gut of Agriophara rhombata. Microorganisms, 11(9), 2188. https://doi.org/10.3390/microorganisms11092188





