LPS-Induced Mortality in Zebrafish: Preliminary Characterisation of Common Fish Pathogens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. LPS Extraction and Quantification
2.3. Ethical Statement
2.4. Zebrafish Husbandry and Breeding
2.5. Challenge Tests with LPS and Live Bacteria on Zebrafish
2.6. Statistical Analysis
3. Results
3.1. Fish Pathogens A. hydrophila, V. harveyi, T. maritimum and P. damselae subsp. piscicida Have Different Lipopolysaccharides (LPS) Profiles
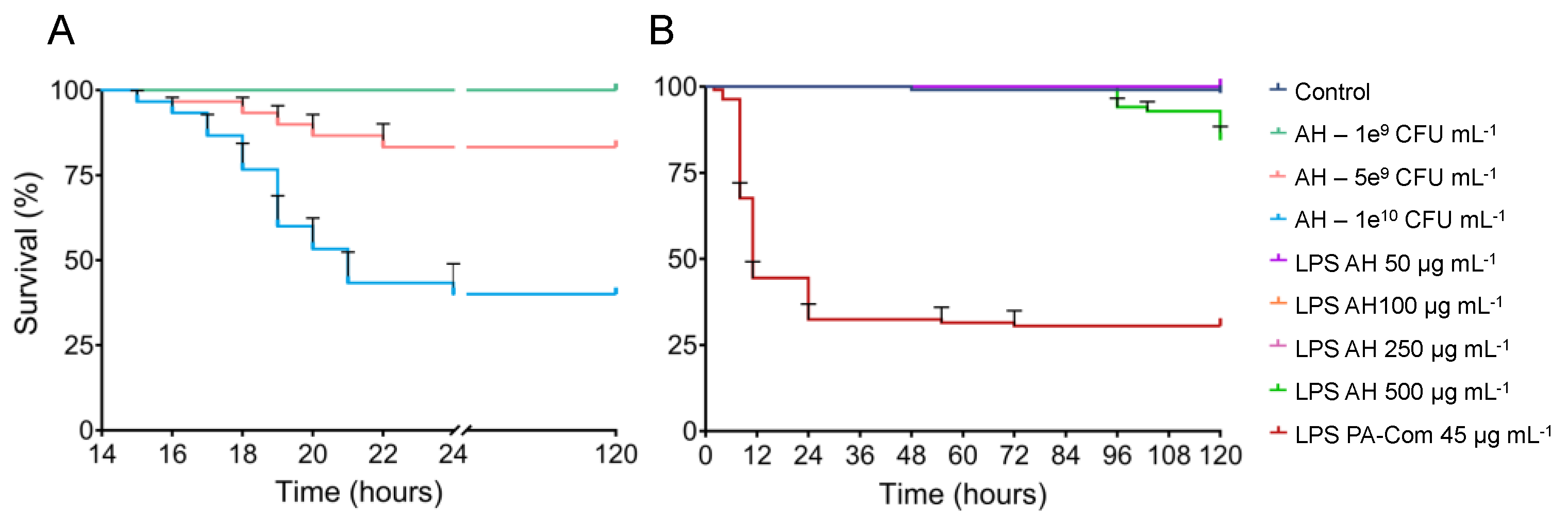
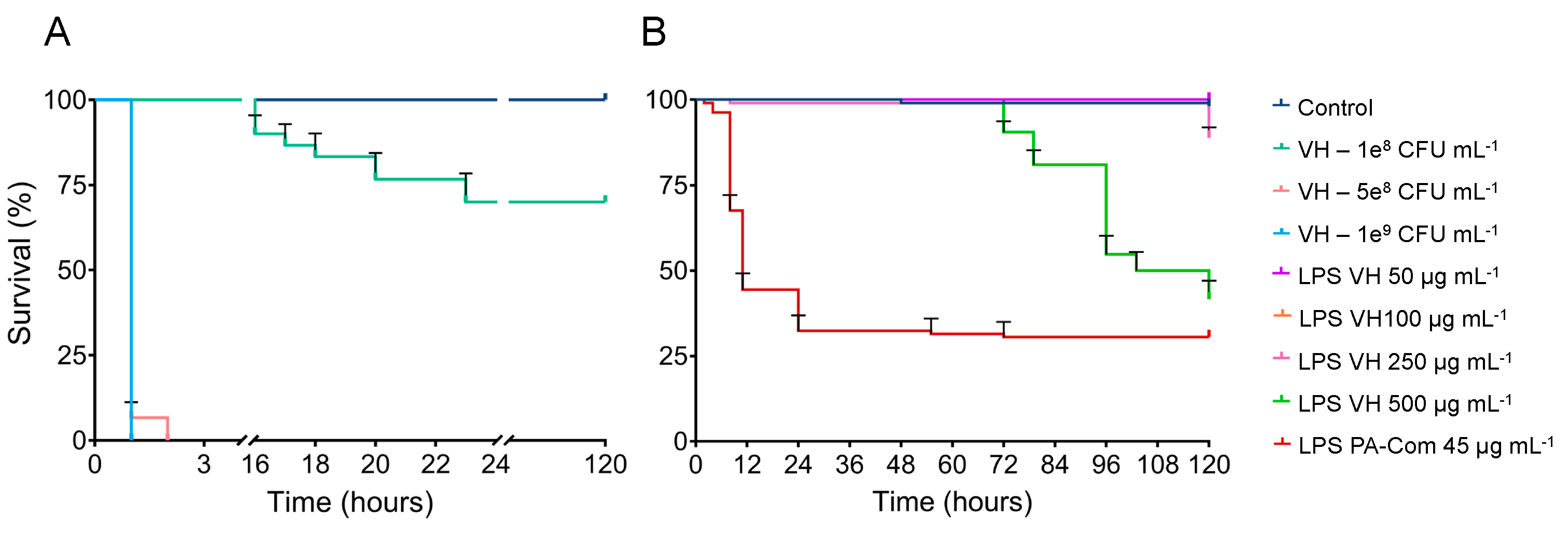
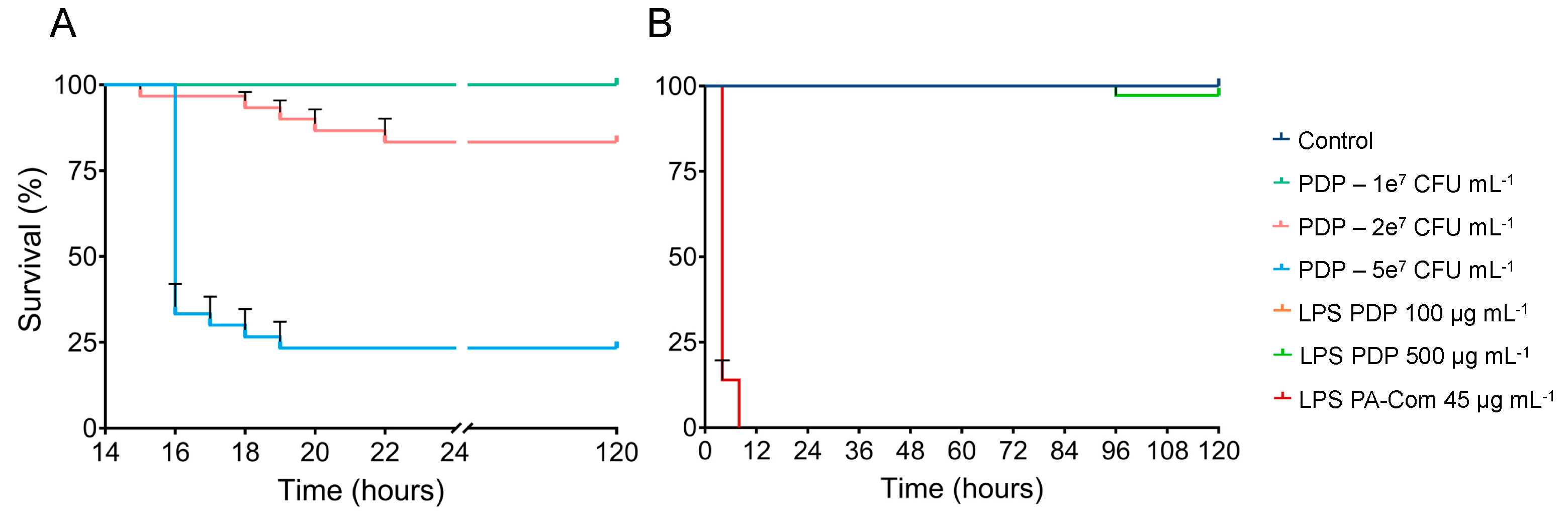
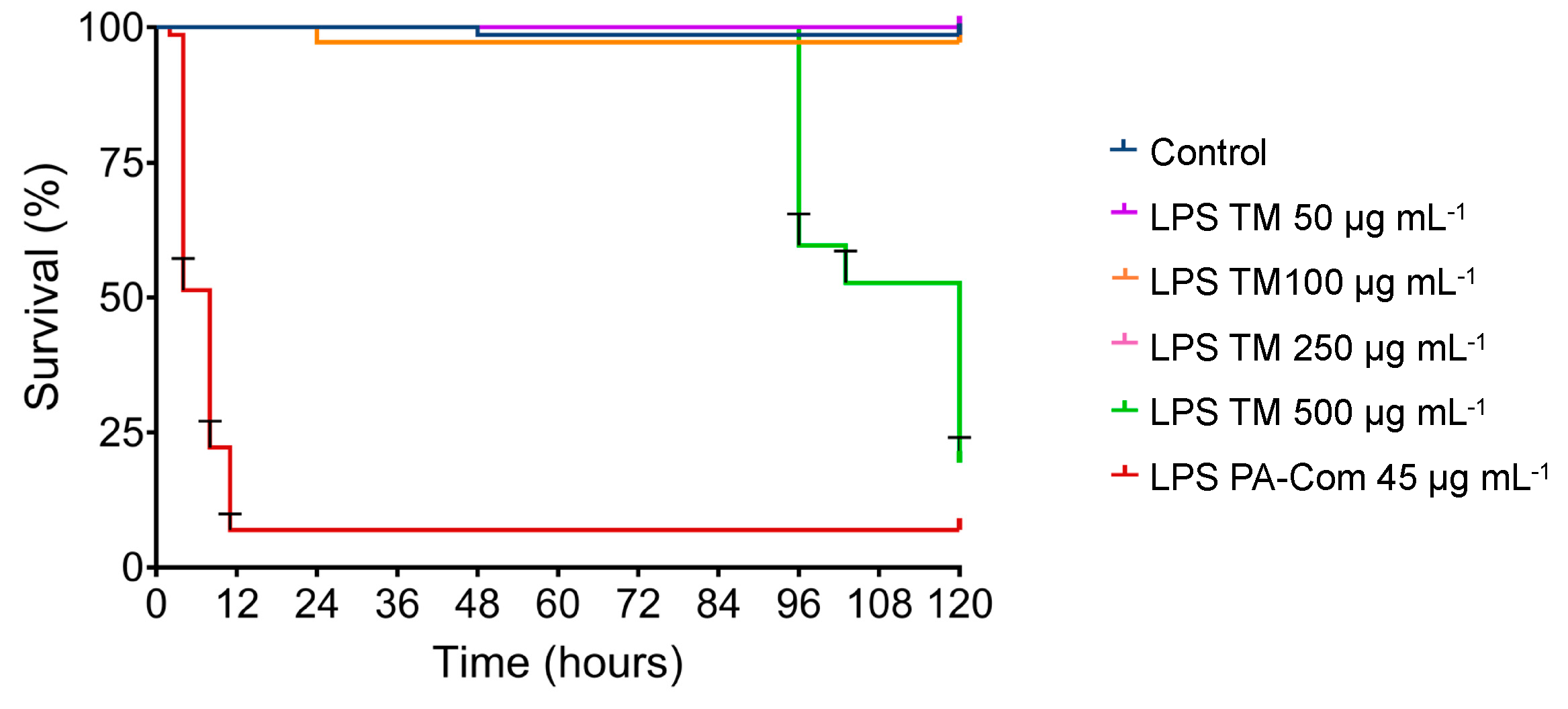
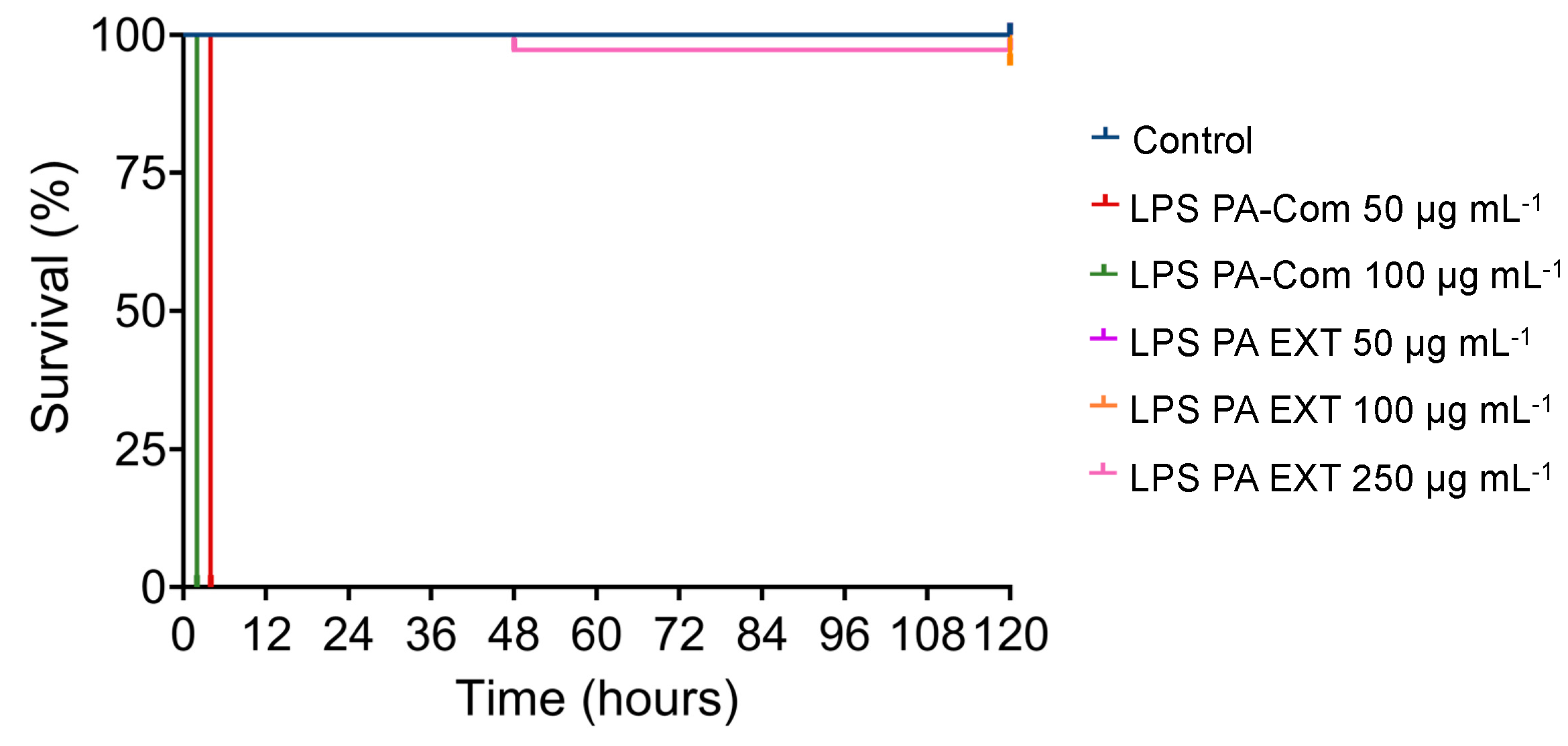
3.2. Virulence of A. hydrophila, V. harveyi, P. damselae subsp. piscicida and T. maritimum Cells and LPS Is Highly Variable and Strain Specific
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture 2020; Sustainability in Action; FAO: Rome, Italy, 2020. [Google Scholar]
- Ina-Salwany, M.Y.; Al-Saari, N.; Mohamad, A.; Mursidi, F.A.; Mohd-Aris, A.; Amal, M.N.A.; Kasai, H.; Mino, S.; Sawabe, T.; Zamri-Saad, M. Vibriosis in Fish: A Review on Disease Development and Prevention. J. Aquat. Anim. Health 2019, 31, 3–22. [Google Scholar] [CrossRef]
- Andreoni, F.; Magnani, M. Photobacteriosis: Prevention and Diagnosis. J. Immunol. Res. 2014, 2014, 7. [Google Scholar] [CrossRef]
- Fernandez-Bravo, A.; Figueras, M.J. An Update on the Genus Aeromonas: Taxonomy, Epidemiology, and Pathogenicity. Microorganisms 2020, 8, 129. [Google Scholar] [CrossRef]
- Fernandez-Alvarez, C.; Santos, Y. Identification and typing of fish pathogenic species of the genus Tenacibaculum. Appl. Microbiol. Biotechnol. 2018, 102, 9973–9989. [Google Scholar] [CrossRef]
- Simpson, B.W.; Trent, M.S. Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol. 2019, 17, 403–416. [Google Scholar] [CrossRef]
- Swain, P.; Nayak, S.K.; Nanda, P.K.; Dash, S. Biological effects of bacterial lipopolysaccharide (endotoxin) in fish: A review. Fish Shellfish. Immunol. 2008, 25, 191–201. [Google Scholar] [CrossRef]
- Novoa, B.; Bowman, T.V.; Zon, L.; Figueras, A. LPS response and tolerance in the zebrafish (Danio rerio). Fish Shellfish. Immunol. 2009, 26, 326–331. [Google Scholar] [CrossRef]
- Védrine, M.; Berthault, C.; Leroux, C.; Répérant-Ferter, M.; Gitton, C.; Barbey, S.; Rainard, P.; Gilbert, F.B.; Germon, P. Sensing of Escherichia coli and LPS by mammary epithelial cells is modulated by O-antigen chain and CD14. PLoS ONE 2018, 13, e0202664. [Google Scholar] [CrossRef]
- Farhana, A.; Khan, Y.S. Biochemistry, Lipopolysaccharide. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2023. [Google Scholar]
- Ruyra, A.; Cano-Sarabia, M.; García-Valtanen, P.; Yero, D.; Gibert, I.; Mackenzie, S.A.; Estepa, A.; Maspoch, D.; Roher, N. Targeting and stimulation of the zebrafish (Danio rerio) innate immune system with LPS/dsRNA-loaded nanoliposomes. Vaccine 2014, 32, 3955–3962. [Google Scholar] [CrossRef]
- Sepulcre, M.P.; Alcaraz-Perez, F.; Lopez-Munoz, A.; Roca, F.J.; Meseguer, J.; Cayuela, M.L.; Mulero, V. Evolution of lipopolysaccharide (LPS) recognition and signaling: Fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J. Immunol. 2009, 182, 1836–1845. [Google Scholar] [CrossRef]
- Nguyen, M.P.; Tran, L.V.H.; Namgoong, H.; Kim, Y.H. Applications of different solvents and conditions for differential extraction of lipopolysaccharide in Gram-negative bacteria. J. Microbiol. 2019, 57, 644–654. [Google Scholar] [CrossRef]
- Forn-Cuni, G.; Varela, M.; Pereiro, P.; Novoa, B.; Figueras, A. Conserved gene regulation during acute inflammation between zebrafish and mammals. Sci. Rep. 2017, 7, 41905. [Google Scholar] [CrossRef]
- Bi, D.; Wang, Y.; Gao, Y.; Li, X.; Chu, Q.; Cui, J.; Xu, T. Recognition of Lipopolysaccharide and Activation of NF-κB by Cytosolic Sensor NOD1 in Teleost Fish. Front. Immunol. 2018, 9, 1413. [Google Scholar] [CrossRef]
- Gauthier, A.E.; Rotjan, R.D.; Kagan, J.C. Lipopolysaccharide detection by the innate immune system may be an uncommon defence strategy used in nature. Open Biol. 2022, 12, 220146. [Google Scholar] [CrossRef]
- Li, Y.; Xia, P.; Wu, J.; Huang, A.; Bu, G.; Meng, F.; Kong, F.; Cao, X.; Han, X.; Yu, G.; et al. The potential sensing molecules and signal cascades for protecting teleost fishes against lipopolysaccharide. Fish Shellfish. Immunol. 2020, 97, 235–247. [Google Scholar] [CrossRef]
- Heiss, C.; Wang, Z.; Thurlow, C.M.; Hossain, M.J.; Sun, D.; Liles, M.R.; Saper, M.A.; Azadi, P. Structure of the capsule and lipopolysaccharide O-antigen from the channel catfish pathogen, Aeromonas hydrophila. Carbohydr. Res. 2019, 486, 107858. [Google Scholar] [CrossRef]
- Merino, S.; Canals, R.; Knirel, Y.A.; Tomás, J.M. Molecular and chemical analysis of the lipopolysaccharide from Aeromonas hydrophila strain AH-1 (Serotype O11). Mar. Drugs 2015, 13, 2233–2249. [Google Scholar] [CrossRef]
- Merino, S.; Gonzalez, V.; Tomás, J.M. The Polymerization of Aeromonas hydrophila AH-3 O-Antigen LPS: Concerted Action of WecP and Wzy. PLoS ONE 2015, 10, e0131905. [Google Scholar] [CrossRef]
- Diaz-Rosales, P.; Chabrillon, M.; Morinigo, M.A.; Balebona, M.C. Survival against exogenous hydrogen peroxide of Photobacterium damselae subsp. piscicida under different culture conditions. J. Fish Dis. 2003, 26, 305–308. [Google Scholar] [CrossRef]
- Rezania, S.; Amirmozaffari, N.; Tabarraei, B.; Jeddi-Tehrani, M.; Zarei, O.; Alizadeh, R.; Masjedian, F.; Zarnani, A.H. Extraction, Purification and Characterization of Lipopolysaccharide from Escherichia coli and Salmonella typhi. Avicenna J. Med. Biotechnol. 2011, 3, 3–9. [Google Scholar]
- Matinha-Cardoso, J.; Coutinho, F.; Lima, S.; Eufrasio, A.; Carvalho, A.P.; Oliva-Teles, A.; Bessa, J.; Tamagnini, P.; Serra, C.R.; Oliveira, P. Novel protein carrier system based on cyanobacterial nano-sized extracellular vesicles for application in fish. Microb. Biotechnol. 2022, 15, 2191–2207. [Google Scholar] [CrossRef]
- Santos, R.A.; Monteiro, M.; Rangel, F.; Jerusik, R.; Saavedra, M.J.; Carvalho, A.P.; Oliva-Teles, A.; Serra, C.R. Bacillus spp. Inhibit Edwardsiella tarda Quorum-Sensing and Fish Infection. Mar. Drugs 2021, 19, 602. [Google Scholar] [CrossRef] [PubMed]
- Novoa, B.; Figueras, A. Zebrafish: Model for the study of inflammation and the innate immune response to infectious diseases. Adv. Exp. Med. Biol. 2012, 946, 253–275. [Google Scholar] [CrossRef] [PubMed]
- Avendaño-Herrera, R.; Toranzo, A.E.; Magariños, B. Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: A review. Dis. Aquat. Org. 2006, 71, 255–266. [Google Scholar] [CrossRef]
- Huszczynski, S.M.; Lam, J.S.; Khursigara, C.M. The Role of Pseudomonas aeruginosa Lipopolysaccharide in Bacterial Pathogenesis and Physiology. Pathogens 2019, 9, 6. [Google Scholar] [CrossRef]
- Qin, Y.; Wang, J.; Wang, S.; Yan, Q. Antigenicity analysis of Vibrio harveyi TS-628 strain. Front. Biol. China 2007, 2, 263–267. [Google Scholar] [CrossRef]
- Montero, A.; Austin, B. Characterization of extracellular products from an isolate of Vibrio harveyi recovered from diseased post-larval Penaeus vannamei (Bonne). J. Fish Dis. 1999, 22, 377–386. [Google Scholar] [CrossRef]
- Avendaño-Herrera, R.; Magariños, B.; Moriñigo, M.A.; Romalde, J.L.; Toranzo1, A.E. A novel O-serotype in Tenacibaculum maritimum strains isolated from cultured sole (Solea senegalensis). Bull. Eur. Ass. Fish Pathol. 2005, 25, 70. [Google Scholar]
- Lopez, P.; Bridel, S.; Saulnier, D.; David, R.; Magariños, B.; Torres, B.S.; Bernardet, J.F.; Duchaud, E. Genomic characterization of Tenacibaculum maritimum O-antigen gene cluster and development of a multiplex PCR-based serotyping scheme. Transbound Emerg. Dis. 2022, 69, e2876–e2888. [Google Scholar] [CrossRef]
- Avendaño-Herrera, R.; Magariños, B.; López-Romalde, S.; Romalde, J.L.; Toranzo, A.E. Phenotypic characterization and description of two major O-serotypes in Tenacibaculum maritimum strains from marine fishes. Dis. Aquat. Organ. 2004, 58, 1–8. [Google Scholar] [CrossRef]
- Van Gelderen, R.; Carson, J.; Gudkovs, N.; Nowak, B. Physical characterisation of Tenacibaculum maritimum for vaccine development. J. Appl. Microbiol. 2010, 109, 1668–1676. [Google Scholar] [CrossRef] [PubMed]
- Merino, S.; Tomás, J.M. The Aeromonas salmonicida Lipopolysaccharide Core from Different Subspecies: The Unusual subsp. pectinolytica. Front. Microbiol. 2016, 7, 125. [Google Scholar] [CrossRef] [PubMed]
- Kuwae, T.; Sasaki, T.; Kurata, M. Chemical and biological properties of lipopolysaccharide from a marine bacterium, Photobacterium phosphoreum PJ-1. Microbiol. Immunol. 1982, 26, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Biedroń, R.; Peruń, A.; Józefowski, S. CD36 Differently Regulates Macrophage Responses to Smooth and Rough Lipopolysaccharide. PLoS ONE 2016, 11, e0153558. [Google Scholar] [CrossRef] [PubMed]
- Labella, A.M.; Rosado, J.J.; Balado, M.; Lemos, M.L.; Borrego, J.J. Virulence properties of three new Photobacterium species affecting cultured fish. J. Appl. Microbiol. 2020, 129, 37–50. [Google Scholar] [CrossRef]
- Zhang, X.H.; He, X.; Austin, B. Vibrio harveyi: A serious pathogen of fish and invertebrates in mariculture. Mar. Life Sci. Technol. 2020, 2, 231–245. [Google Scholar] [CrossRef]
- Mohamad, N.; Amal, M.N.A.; Yasin, I.S.M.; Saad, M.Z.; Nasruddin, N.S.; Al-saari, N.; Mino, S.; Sawabe, T. Vibriosis in cultured marine fishes: A review. Aquaculture 2019, 512, 734289. [Google Scholar] [CrossRef]
- Yazid, S.H.M.; Daud, H.M.; Azmai, M.N.A.; Mohamad, N.; Nor, N.M. Estimating the Economic Loss Due to Vibriosis in Net-Cage Cultured Asian Seabass (Lates calcarifer): Evidence From the East Coast of Peninsular Malaysia. Front. Vet. Sci. 2021, 8, 644009. [Google Scholar] [CrossRef]
- Defoirdt, T.; Boon, N.; Sorgeloos, P.; Verstraete, W.; Bossier, P. Quorum sensing and quorum quenching in Vibrio harveyi: Lessons learned from in vivo work. ISME J. 2008, 2, 19–26. [Google Scholar] [CrossRef]
- Ng, W.L.; Bassler, B.L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009, 43, 197–222. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Ye, X.; Emam, M.; Zhang, H.; Wang, H. Current advances in Vibrio harveyi quorum sensing as drug discovery targets. Eur. J. Med. Chem. 2020, 207, 112741. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, R.A.; Cardoso, C.; Pedrosa, N.; Gonçalves, G.; Matinha-Cardoso, J.; Coutinho, F.; Carvalho, A.P.; Tamagnini, P.; Oliva-Teles, A.; Oliveira, P.; et al. LPS-Induced Mortality in Zebrafish: Preliminary Characterisation of Common Fish Pathogens. Microorganisms 2023, 11, 2205. https://doi.org/10.3390/microorganisms11092205
Santos RA, Cardoso C, Pedrosa N, Gonçalves G, Matinha-Cardoso J, Coutinho F, Carvalho AP, Tamagnini P, Oliva-Teles A, Oliveira P, et al. LPS-Induced Mortality in Zebrafish: Preliminary Characterisation of Common Fish Pathogens. Microorganisms. 2023; 11(9):2205. https://doi.org/10.3390/microorganisms11092205
Chicago/Turabian StyleSantos, Rafaela A., Cláudia Cardoso, Neide Pedrosa, Gabriela Gonçalves, Jorge Matinha-Cardoso, Filipe Coutinho, António P. Carvalho, Paula Tamagnini, Aires Oliva-Teles, Paulo Oliveira, and et al. 2023. "LPS-Induced Mortality in Zebrafish: Preliminary Characterisation of Common Fish Pathogens" Microorganisms 11, no. 9: 2205. https://doi.org/10.3390/microorganisms11092205
APA StyleSantos, R. A., Cardoso, C., Pedrosa, N., Gonçalves, G., Matinha-Cardoso, J., Coutinho, F., Carvalho, A. P., Tamagnini, P., Oliva-Teles, A., Oliveira, P., & Serra, C. R. (2023). LPS-Induced Mortality in Zebrafish: Preliminary Characterisation of Common Fish Pathogens. Microorganisms, 11(9), 2205. https://doi.org/10.3390/microorganisms11092205






