Comparison of Antibiofilm Activity of Pseudomonas aeruginosa Phages on Isolates from Wounds of Diabetic and Non-Diabetic Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. Bacteriophages
2.3. Lytic Activity of Phages
2.4. Antibiofilm Activity of Phages
2.5. Activity of Phages on Biofilms Formed on Different Materials
3. Results
3.1. Characteristics of Bacterial Isolates
3.2. Characterization of the Bacteriophages
3.3. Lytic Activity of Phages
3.4. Antibiofilm Activity of Phages
3.5. Activity of Phages on Biofilms Formed on Different Materials
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nagoba, B.; Davane, M.; Gandhi, R.; Wadher, B.; Suryawanshi, N.; Selkar, S. Treatment of skin and soft tissue infections caused by Pseudomonas aeruginosa—A review of our experiences with citric acid over the past 20 years. Wound Med. 2017, 19, 5–9. [Google Scholar] [CrossRef]
- Ballok, A.E.; O’Toole, G.A. Pouring salt on a wound: Pseudomonas aeruginosa virulence factors alter Na+ and Cl− flux in the lung. J. Bacteriol. 2013, 195, 4013–4019. [Google Scholar] [CrossRef] [PubMed]
- Jneid, J.; Lavigne, J.P.; La Scola, B.; Cassir, N. The diabetic foot microbiota: A review. Hum. Microbiome J. 2017, 5, 1–6. [Google Scholar] [CrossRef]
- World Health Organizations. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 13 July 2023).
- Tamma, P.D.; Aitken, S.L.; Bonomo, R.A.; Mathers, A.J.; van Duin, D.; Clancy, C.J. Infectious Diseases Society of America Guidance on the Treatment of Extended-Spectrum βlactamase Producing Enterobacterales (ESBL-E), Carbapenem-Resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with Difficult-to-Treat Resistance (DTR-P. aeruginosa). Clin. Infect. Dis. 2021, 72, e169–e183. [Google Scholar] [PubMed]
- Sulakvelidge, A.; Alavidze, Z.; Morris, J.G. Bacteriophage therapy. Antimicrob. Agents Chemother. 2001, 45, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Aziz, A.M.; Elgaml, A.; Ali, Y.M. Bacteriophage therapy increases complement-mediated lysis of bacteria and enhances bacterial clearance after acute lung infection with multidrug-resistant Pseudomonas aeruginosa. J. Infect. Dis. 2018, 219, 1439–1447. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Turner, P.E.; Kim, S.; Mojibian, H.R.; Elefteriades, J.A.; Narayan, D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evolut. Med. Public Health 2018, 2018, 60–66. [Google Scholar] [CrossRef]
- Ferry, T.; Boucher, F.; Fevre, C.; Perpoint, T.; Chateau, J.; Petitjean, C.; Josse, J.; Chidiac, C.; L’hostis, G.; Leboucher, G.; et al. Innovations for the treatment of a complex bone and joint infection due to XDR Pseudomonas aeruginosa including local application of a selected cocktail of bacteriophages. J. Antimicrob. Chemother. 2018, 73, 2901–2903. [Google Scholar] [CrossRef]
- Jennes, S.; Merabishvili, M.; Soentjens, P.; Pang, K.W.; Rose, T.; Keersebilck, E.; Soete, O.; Francois, P.-M.; Teodorescu, S.; Verween, G.; et al. Use of bacteriophages in the treatment of colistin-only-sensitive Pseudomonas aeruginosa septicaemia in a patient with acute kidney injury—A case report. Crit. Care 2017, 21, 129. [Google Scholar] [CrossRef]
- Jault, P.; Leclerc, T.; Jennes, S.; Pirnay, J.P.; Que, Y.-A.; Resch, G.; Rosseau, A.F.; Ravat, F.; Carsin, H.; Le Floch, R.; et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): A randomized, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 2019, 19, 35–45. [Google Scholar] [CrossRef]
- Soothill, J.S. Bacteriophage prevents destruction of skin grafts by Pseudomonas aeruginosa. Burns 1994, 20, 209–211. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.J.; Levin, B.R.; DeRouin, T.; Walker, N.; Bloch, C.A. Dynamics of success and failure in phage and antibiotic therapy in experimental infections. BMC Microbiol. 2002, 2, 35. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.; Vittal, R.V.; Raj, J.R.M. Isolation, Characterisation, and Comparison of Efficiencies of Bacteriophages to Reduce Planktonic and Biofilm-Associated Staphylococcus aureus. J. Health Allied Sci. 2020, 10, 102–108. [Google Scholar]
- Goldufsky, J.; Wood, S.J.; Jayaraman, V.; Majdobeh, O.; Chen, L.; Qin, S.; Zhang, C.; DiPietro, L.A.; Shafikhani, S.H. Pseudomonas aeruginosa uses T3SS to inhibit diabetic wound healing. Wound Repair. Regen. 2015, 23, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Phan, S.; Feng, C.H.; Huang, R.; Lee, Z.X.; Moua, Y.; Phung, O.J.; Lenhard, J.R. Relative Abundance and Detection of Pseudomonas aeruginosa from Chronic Wound Infections Globally. Microorganisms 2023, 11, 1210. [Google Scholar] [CrossRef] [PubMed]
- Grosso-Becerra, M.V.; Santos-Medellín, C.; González-Valdez, A.; Jose-Luiz, M.; Delgado, G.; Morales-Espinosa, R.; Servin-Gonzalez, L.; Alcaraz, L.-D.; Soberon-Chavez, G. Pseudomonas aeruginosa clinical and environmental isolates constitute a single population with high phenotypic diversity. BMC Genom. 2014, 15, 318. [Google Scholar] [CrossRef] [PubMed]
- Pirnay, J.P.; Bilocq, F.; Pot, B.; Cornelis, P.; Zizi, M.; Van Eldere, J.; Deschaght, P.; Vaneechoutte, M.; Jennes, S.; Pitt, T.; et al. Pseudomonas aeruginosa population structure revisited. PLoS ONE 2009, 4, e7740. [Google Scholar] [CrossRef]
- Vos, M.; Birkett, P.J.; Birch, E.; Griffiths, R.I.; Buckling, A. Local adaptation of bacteriophages to their bacterial hosts in soil. Science 2009, 325, 833. [Google Scholar] [CrossRef]
- De Vos, D.; Lim, A., Jr.; Pirnay, J.P.; Struelens, M.; Vandenvelde, C.; Duinslaeger, L.; Vanderkelen, A.; Cornelis, P. Direct detection and identification of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectorations by multiplex PCR based on two outer membrane lipoprotein genes, oprI and oprL. J. Clin. Microbiol. 1997, 35, 1295–1299. [Google Scholar] [CrossRef]
- Storms, Z.J.; Teel, M.R.; Mercurio, K.; Sauvageau, D. The virulence index: A metric for quantitative analysis of phage virulence. Phage 2020, 1, 27–36. [Google Scholar] [CrossRef]
- Fang, Q.; Feng, Y.; McNally, A.; Zong, Z. Characterization of phage resistance and phages capable of intestinal decolonization of carbapenem-resistant Klebsiella pneumoniae in mice. Commun. Biol. 2022, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011, 47, 2437. [Google Scholar]
- Stepanovic, S.; Vukovic, D.; Dakic, I.; Savic, B.; Svabic-Vlahovic, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Köhler, C.D.; Dobrindt, U. What defines extraintestinal pathogenic Escherichia coli? Int. J. Med. Microbiol. 2011, 301, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Khurana, S.; Bhardwaj, N.; Malhotra, R.; Mathur, P. Pathogen burden & associated antibiogram of Pseudomonas spp. in a tertiary care hospital of India. Indian J. Med. Res. 2019, 149, 295–298. [Google Scholar] [PubMed]
- Gandra, S.; Tseng, K.K.; Arora, A.; Bhowmik, B.; Robinson, M.L.; Panigrahi, B.; Laxminarayan, R.; Klein, E.Y. The mortality burden of multidrug-resistant pathogens in India: A retrospective, observational study. Clin. Infect. Dis. 2019, 69, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Javiya, V.A.; Ghatak, S.B.; Patel, K.R.; Patel, J.A. Antibiotic susceptibility patterns of Pseudomonas aeruginosa at a tertiary care hospital in Gujarat, India. Indian J. Pharmacol. 2008, 40, 230–234. [Google Scholar] [PubMed]
- Biswal, I.; Arora, B.S.; Kasana, D. Incidence of multidrug resistant Pseudomonas aeruginosa isolated from burn patients and environment of teaching institution. J. Clin. Diagnos Res. 2014, 8, 26–29. [Google Scholar]
- Suresh, S.; Prithvisagar, K.S.; Kumar, B.K.; Premanath, R. Unravelling the Distinctive Virulence Traits and Clonal Relationship among the Pseudomonas aeruginosa Isolates from Diabetic Patients. J. Pure Appl. Microbiol. 2022, 16, 1893–1908. [Google Scholar] [CrossRef]
- McVay, C.S.; Velasquez, M.; Fralick, J.A. Phage therapy of Pseudomonas aeruginosa infection in a mouse burn wound model. Antimicrob. Agents Chemother. 2007, 51, 1934–1938. [Google Scholar] [CrossRef]
- Shivu, M.M.; Rajeeva, B.C.; Girisha, S.K.; Karunasagar, I.; Krohne, G.; Karunasagar, I. Molecular characterization of Vibrio harveyi bacteriophages isolated from aquaculture environments along the coast of India. Environ. Microbiol. 2007, 9, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Jurczak-Kurek, A.; Gąsior, T.; Nejman-Faleńczyk, B.; Bloch, S.; Dydecka, A.; Topka, G.; Necel, A.; Jakubowska-Deredas, M.; Narajczyk, M.; Richert, M.; et al. Biodiversity of bacteriophages: Morphological and biological properties of a large group of phages isolated from urban sewage. Sci. Res. 2016, 6, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef] [PubMed]
- Drulis-Kawa, Z.; Majkowska-Skrobek, G.; Maciejewska, B. Bacteriophages and phage-derived proteins–application approaches. Curr. Med. Chem. 2015, 22, 1757–1773. [Google Scholar] [CrossRef] [PubMed]
- Sillankorva, S.; Oliveira, R.; Vieira, M.J.; Sutherland, I.; Azeredo, J. Bacteriophage Φ S1 infection of Pseudomonas fluorescens planktonic cells versus biofilms. Biofouling 2004, 20, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Chadha, J.; Harjai, K.; Chhibber, S. Revisiting the virulence hallmarks of Pseudomonas aeruginosa: A chronicle through the perspective of quorum sensing. Environ. Microbiol. 2022, 24, 2630–2656. [Google Scholar] [CrossRef] [PubMed]
- Cornelissen, A.; Ceyssens, P.J.; T'syen, J.; Van Praet, H.; Noben, J.P.; Shaburova, O.V. The T7-related Pseudomonas putida phage φ15 displays virion-associated biofilm degradation properties. PLoS ONE 2011, 6, e18597. [Google Scholar] [CrossRef] [PubMed]
- Fong, S.A.; Drilling, A.; Morales, S.; Cornet, M.E.; Woodworth, B.A.; Fokkens, W.J.; Psaltis, A.J.; Vreugde, S.; Wormald, P.J. Activity of Bacteriophages in Removing Biofilms of Pseudomonas aeruginosa Isolates from Chronic Rhinosinusitis Patients. Front. Cell Infect. Microbiol. 2017, 7, 418. [Google Scholar] [CrossRef]
- Latz, S.; Krüttgen, A.; Häfner, H.; Buhl, E.M.; Ritter, K.; Horz, H.P. Differential effect of newly isolated phages belonging to Pb1-like, Phikz-like and Luz24-like viruses against multidrug resistant Pseudomonas aeruginosa under varying growth conditions. Viruses 2017, 9, 315. [Google Scholar] [CrossRef]
- Oliveira, V.C.; Bim, F.L.; Monteiro, R.M.; Macedo, A.P.; Santos, E.S.; Silva-Lovato, C.H.; Paranhos, H.F.O.; Melo, L.D.R.; Santos, S.B.; Watanabe, E. Identification and Characterization of New Bacteriophages to Control Multidrug-Resistant Pseudomonas aeruginosa Biofilm on Endotracheal Tubes. Front. Microbiol. 2020, 11, 580779. [Google Scholar] [CrossRef]
- Spagnolo, A.M.; Sartini, M.C.; Maria, L. Pseudomonas aeruginosa in the healthcare facility setting. Rev. Med. Microbiol. 2021, 32, 169–175. [Google Scholar] [CrossRef]
- Adnan, M.; Ali Shah, M.R.; Jamal, M.; Jalil, F.; Andleeb, S.; Nawaz, M.A.; Pervez, S.; Hussain, T.; Shah, I.; Imran, M.; et al. Isolation and characterization of bacteriophage to control multidrug-resistant Pseudomonas aeruginosa planktonic cells and biofilm. Biologicals 2020, 63, 89–96. [Google Scholar] [CrossRef]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.H.; Webb, J.S.; Rehm, B.H.A. The role of polyhydroxyalkanoate biosynthesis by Pseudomonas aeruginosa in rhamnolipid and alginate production as well as stress tolerance and biofilm formation. Microbiol 2004, 150, 3405–3413. [Google Scholar] [CrossRef]
- Kebriaei, R.; Lehman, S.M.; Shah, R.M.; Stamper, K.C.; Kunz Coyne, A.J.; Holger, D.; El Ghali, A.; Rybak, M.J. Optimization of phage-antibiotic combinations against Staphylococcus aureus biofilms. Microbiol. Spectr. 2023, 11, e04918-22. [Google Scholar] [CrossRef] [PubMed]
- Ryan, E.M.; Alkawareek, M.Y.; Donnelly, R.F.; Gilmore, B.F. Synergistic phage-antibiotic combinations for the control of Escherichia coli biofilms in vitro. FEMS Immunol. Med. Microbiol. 2012, 65, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.; Wu, S.M.; Soni, I.; Wang-Crocker, C.; Matern, T.; Beck, J.P.; Loc-Carrillo, C. Phage and antibiotic combinations reduce Staphylococcus aureus in static and dynamic biofilms grown on an implant material. Viruses 2023, 15, 460. [Google Scholar] [CrossRef]
- Akturk, E.; Oliveira, H.; Santos, S.B.; Costa, S.; Kuyumcu, S.; Melo, L.D.R.; Azeredo, J. Synergistic Action of Phage and Antibiotics: Parameters to enhance the killing efficacy against mono and dual-species biofilms. Antibiotics 2019, 8, 103. [Google Scholar] [CrossRef]
- Chaudhry, W.N.; Concepción-Acevedo, J.; Park, T.; Andleeb, S.; Bull, J.J.; Levin, B.R. Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms. PLoS ONE 2017, 12, e0168615. [Google Scholar] [CrossRef]
 indicates that the host is susceptible to that phage, while
indicates that the host is susceptible to that phage, while  represents resistance.
represents resistance.
 indicates that the host is susceptible to that phage, while
indicates that the host is susceptible to that phage, while  represents resistance.
represents resistance.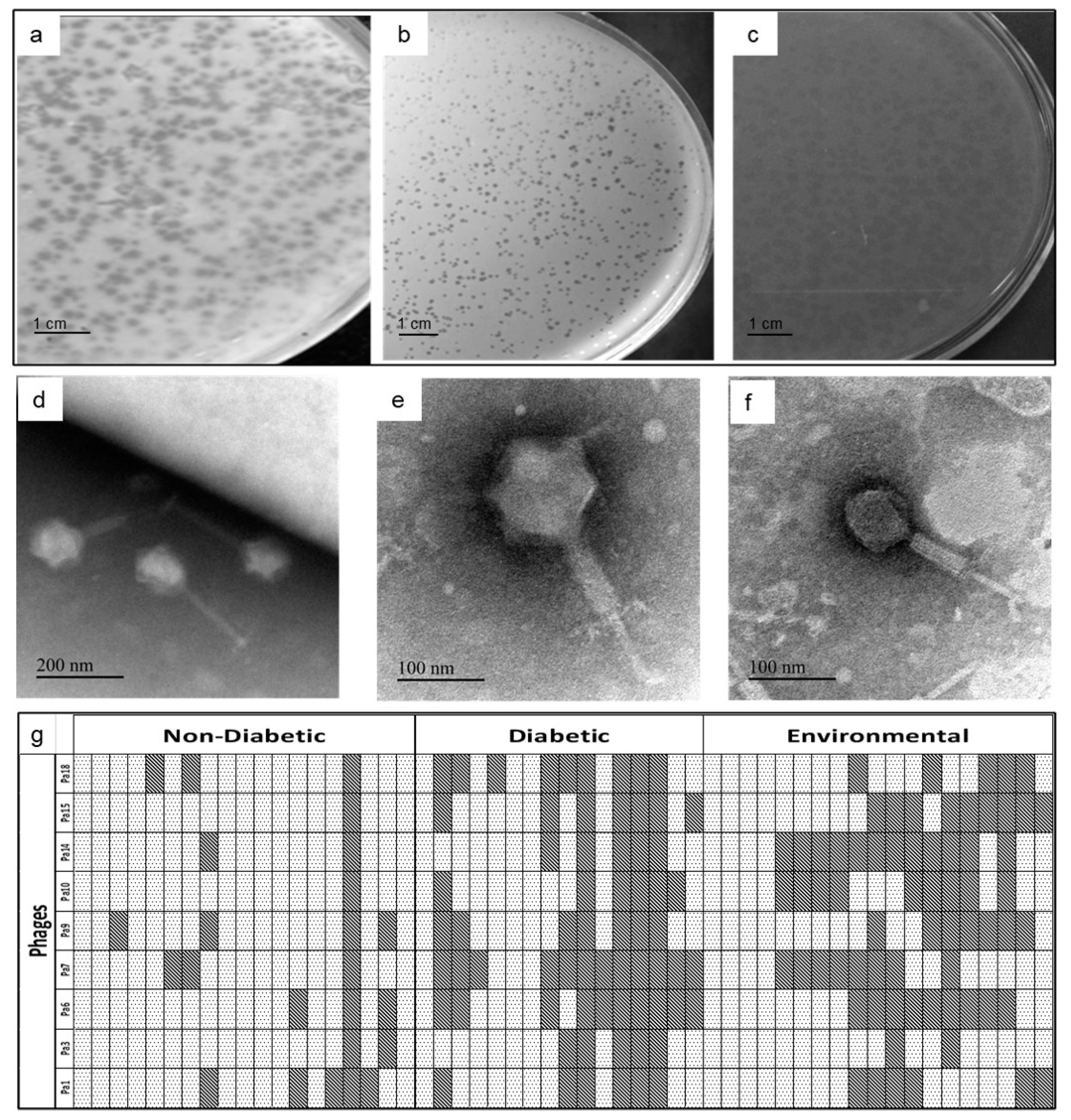



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suresh, S.; Saldanha, J.; Bhaskar Shetty, A.; Premanath, R.; Akhila, D.S.; Mohan Raj, J.R. Comparison of Antibiofilm Activity of Pseudomonas aeruginosa Phages on Isolates from Wounds of Diabetic and Non-Diabetic Patients. Microorganisms 2023, 11, 2230. https://doi.org/10.3390/microorganisms11092230
Suresh S, Saldanha J, Bhaskar Shetty A, Premanath R, Akhila DS, Mohan Raj JR. Comparison of Antibiofilm Activity of Pseudomonas aeruginosa Phages on Isolates from Wounds of Diabetic and Non-Diabetic Patients. Microorganisms. 2023; 11(9):2230. https://doi.org/10.3390/microorganisms11092230
Chicago/Turabian StyleSuresh, Sarika, Joylin Saldanha, Ashwini Bhaskar Shetty, Ramya Premanath, D. S. Akhila, and Juliet Roshini Mohan Raj. 2023. "Comparison of Antibiofilm Activity of Pseudomonas aeruginosa Phages on Isolates from Wounds of Diabetic and Non-Diabetic Patients" Microorganisms 11, no. 9: 2230. https://doi.org/10.3390/microorganisms11092230




