Functional Whole Genome Screen of Nutrient-Starved Mycobacterium tuberculosis Identifies Genes Involved in Rifampin Tolerance
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains
2.2. Generation of Deletions
2.3. Media and Buffers
2.4. Generation of Transposon Mutant Library
2.5. Setup for Tn Screen in Nutrient-Rich Broth
2.6. Setup for Tn Screen in Nutrient Starvation
2.7. DNA Extraction
2.8. DNA Library Preparation
2.9. Hypothesis Testing for Antibiotic Hypersusceptibility
2.10. Oligonucleotides and Primers
2.11. Plasmids
2.12. Time-Kill Assays in Nutrient-Rich Broth
2.13. Time-Kill Assays in Nutrient Starvation Conditions
3. Results
3.1. Overview of Isoniazid and Rifampin Screens of Mtb Transposon Mutant Libraries
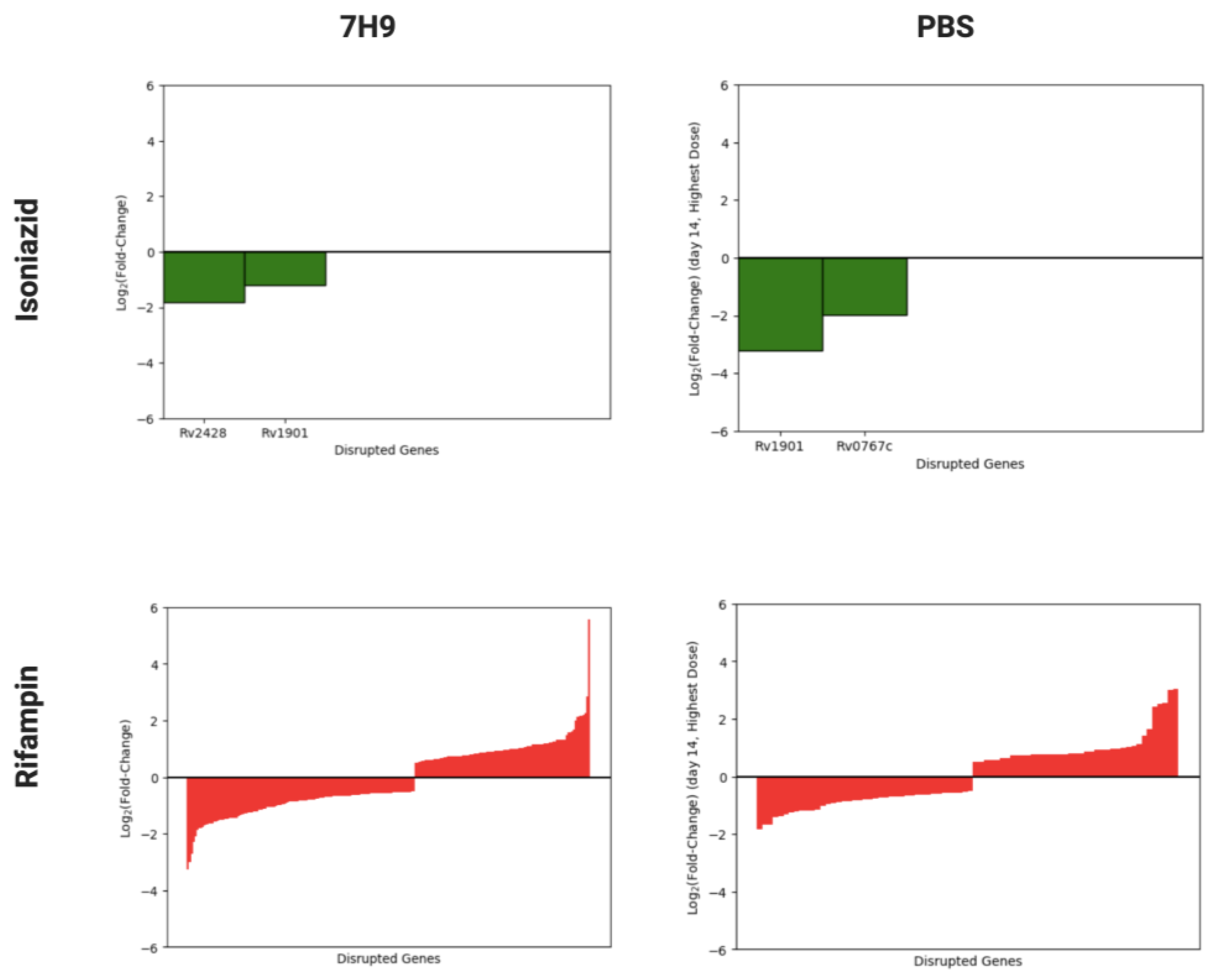
3.2. Identification of Mtb Genes Contributing to Rifampin Tolerance in Nutrient-Rich Broth and under Nutrient Starvation
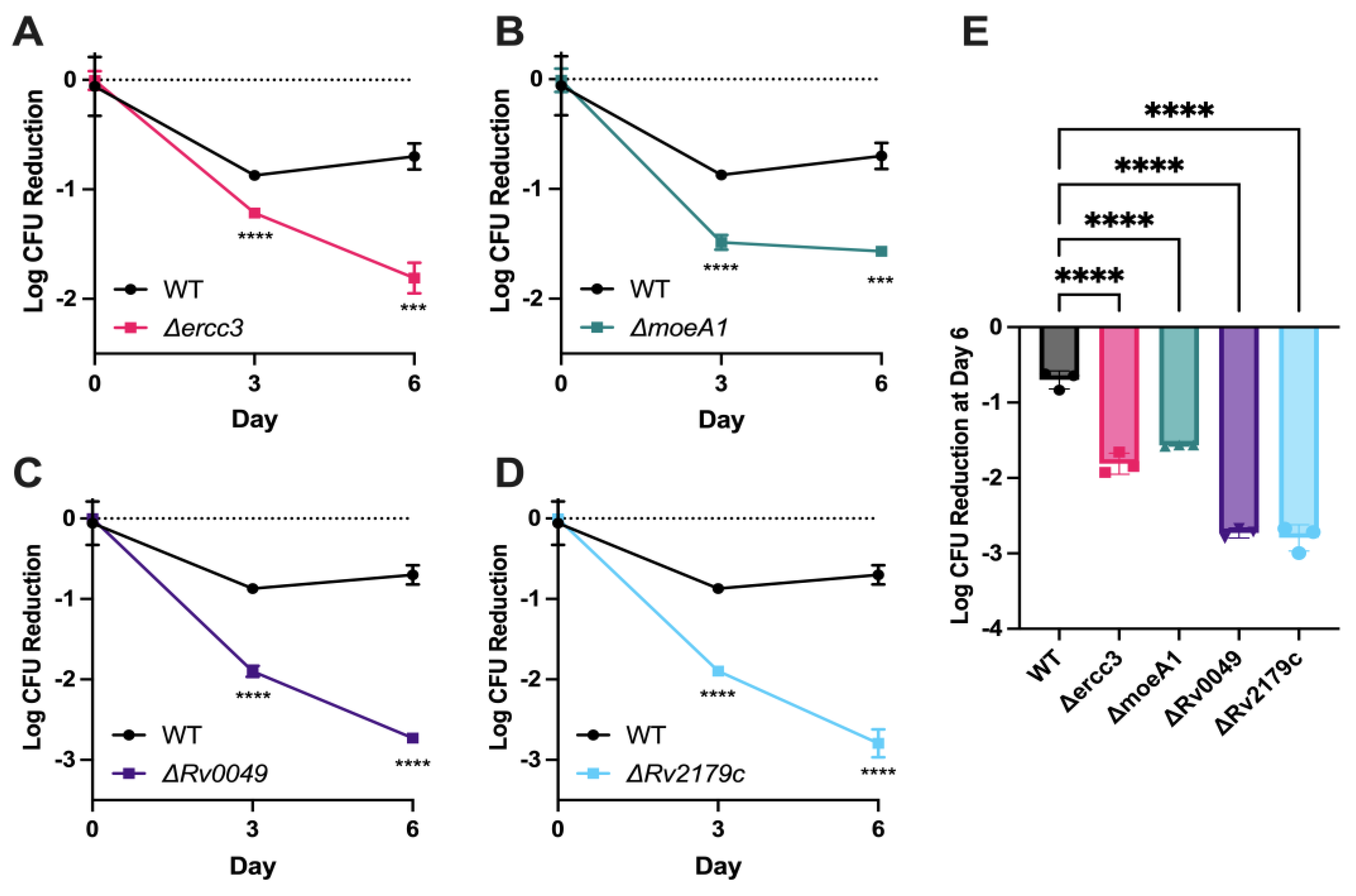
3.3. Validation of Specific Mtb Genetic Requirements for Rifampin Hypersusceptibility Using Targeted Mutagenesis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bagcchi, S. WHO’s Global Tuberculosis Report 2022. Lancet Microbe 2023, 4, e20. [Google Scholar] [CrossRef] [PubMed]
- Conradie, F.; Bagdasaryan, T.R.; Borisov, S.; Howell, P.; Mikiashvili, L.; Ngubane, N.; Samoilova, A.; Skornykova, S.; Tudor, E.; Variava, E.; et al. Bedaquiline-Pretomanid-Linezolid Regimens for Drug-Resistant Tuberculosis. N. Engl. J. Med. 2022, 387, 810–823. [Google Scholar] [CrossRef] [PubMed]
- Carr, W.; Kurbatova, E.; Starks, A.; Goswami, N.; Allen, L.; Winston, C. Interim Guidance: 4-Month Rifapentine-Moxifloxacin Regimen for the Treatment of Drug-Susceptible Pulmonary Tuberculosis—United States, 2022. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Strathdee, S.A.; Hatfull, G.F.; Mutalik, V.K.; Schooley, R.T. Phage therapy: From biological mechanisms to future directions. Cell 2023, 186, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Diacon, A.H.; Guerrero-Bustamante, C.A.; Rosenkranz, B.; Rubio Pomar, F.J.; Vanker, N.; Hatfull, G.F. Mycobacteriophages to Treat Tuberculosis: Dream or Delusion? Respiration 2022, 101, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Rhee, K.Y. Tuberculosis Drug Development: History and Evolution of the Mechanism-Based Paradigm. Cold Spring Harb. Perspect. Med. 2015, 5, a021147. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, J.; Thomas, A.; Levine, C.; Shrestha, R.; Levy, S.; Safi, H.; Pentakota, S.R.; Kumar, P.; Alland, D. Origin and Dynamics of Mycobacterium tuberculosis Subpopulations That Predictably Generate Drug Tolerance and Resistance. mBio 2022, 13, e0279522. [Google Scholar] [CrossRef]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef]
- Parker, H.; Lorenc, R.; Castillo, J.R.; Karakousis, P.C. Mechanisms of Antibiotic Tolerance in Mycobacterium avium Complex: Lessons From Related Mycobacteria. Front. Microbiol. 2020, 11, 573983. [Google Scholar] [CrossRef]
- Goossens, S.N.; Sampson, S.L.; Van Rie, A. Mechanisms of Drug-Induced Tolerance in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 2020, 34, e00141-20. [Google Scholar] [CrossRef]
- Wayne, L.G.; Hayes, L.G. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 1996, 64, 2062–2069. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Siddiqi, N.; Rubin, E.J. Differential antibiotic susceptibilities of starved Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 2005, 49, 4778–4780. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.J.; Abramovitch, R.B. Genetic and metabolic regulation of Mycobacterium tuberculosis acid growth arrest. Sci. Rep. 2018, 8, 4168. [Google Scholar] [CrossRef] [PubMed]
- Rifat, D.; Bishai, W.R.; Karakousis, P.C. Phosphate depletion: A novel trigger for Mycobacterium tuberculosis persistence. J. Infect. Dis. 2009, 200, 1126–1135. [Google Scholar] [CrossRef] [PubMed]
- DeJesus, M.A.; Zhang, Y.J.; Sassetti, C.M.; Rubin, E.J.; Sacchettini, J.C.; Ioerger, T.R. Bayesian analysis of gene essentiality based on sequencing of transposon insertion libraries. Bioinformatics 2013, 29, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Langridge, G.C.; Phan, M.-D.; Turner, D.J.; Perkins, T.T.; Parts, L.; Haase, J.; Charles, I.; Maskell, D.J.; Peters, S.E.; Dougan, G.; et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 2009, 19, 2308–2316. [Google Scholar] [CrossRef] [PubMed]
- Loebel, R.O.; Shorr, E.; Richardson, H.B. The Influence of Adverse Conditions upon the Respiratory Metabolism and Growth of Human Tubercle Bacilli. J. Bacteriol. 1933, 26, 167–200. [Google Scholar] [CrossRef]
- Gengenbacher, M.; Rao, S.P.S.; Pethe, K.; Dick, T. Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology 2010, 156 Pt 1, 81–87. [Google Scholar] [CrossRef]
- Betts, J.C.; Lukey, P.T.; Robb, L.C.; McAdam, R.A.; Duncan, K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 2002, 43, 717–731. [Google Scholar] [CrossRef]
- Loebel, R.O.; Shorr, E.; Richardson, H.B. The Influence of Foodstuffs upon the Respiratory Metabolism and Growth of Human Tubercle Bacilli. J. Bacteriol. 1933, 26, 139–166. [Google Scholar] [CrossRef]
- Dutta, N.K.; Pinn, M.L.; Karakousis, P.C. Reduced emergence of isoniazid resistance with concurrent use of thioridazine against acute murine tuberculosis. Antimicrob. Agents Chemother. 2014, 58, 4048–4053. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.C.; Nelson, S.J.; Nambi, S.; Papavinasasundaram, K.; Baer, C.E.; Sassetti, C.M. ORBIT: A New Paradigm for Genetic Engineering of Mycobacterial Chromosomes. mBio 2018, 9, e01467-18. [Google Scholar] [CrossRef] [PubMed]
- Long, J.E.; DeJesus, M.; Ward, D.; Baker, R.E.; Ioerger, T.; Sassetti, C.M. Identifying essential genes in Mycobacterium tuberculosis by global phenotypic profiling. Methods Mol. Biol. 2015, 1279, 79–95. [Google Scholar] [PubMed]
- Choudhri, S.H.; Hawken, M.; Gathua, S.; Minyiri, G.O.; Watkins, W.; Sahai, J.; Sitar, D.S.; Aoki, F.Y.; Long, R. Pharmacokinetics of antimycobacterial drugs in patients with tuberculosis, AIDS, and diarrhea. Clin. Infect. Dis. 1997, 25, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Matern, W.M.; Jenquin, R.L.; Bader, J.S.; Karakousis, P.C. Identifying the essential genes of Mycobacterium avium subsp. hominissuis with Tn-Seq using a rank-based filter procedure. Sci. Rep. 2020, 10, 1095. [Google Scholar] [CrossRef] [PubMed]
- Matern, W.M.; Parker, H.; Danchik, C.; Hoover, L.; Bader, J.S.; Karakousis, P.C. Genetic Determinants of Intrinsic Antibiotic Tolerance in Mycobacterium avium. Microbiol. Spectr. 2021, 9, e0024621. [Google Scholar] [CrossRef] [PubMed]
- Biswas, T.; Pero, J.M.; Joseph, C.G.; Tsodikov, O.V. DNA-dependent ATPase activity of bacterial XPB helicases. Biochemistry 2009, 48, 2839–2848. [Google Scholar] [CrossRef]
- Balasingham, S.V.; Zegeye, E.D.; Homberset, H.; Rossi, M.L.; Laerdahl, J.K.; Bohr, V.A.; Tønjum, T. Enzymatic activities and DNA substrate specificity of Mycobacterium tuberculosis DNA helicase XPB. PLoS ONE 2012, 7, e36960. [Google Scholar] [CrossRef]
- Williams, M.; Mizrahi, V.; Kana, B.D. Molybdenum cofactor: A key component of Mycobacterium tuberculosis pathogenesis? Crit. Rev. Microbiol. 2014, 40, 18–29. [Google Scholar] [CrossRef]
- McGuire, A.M.; Weiner, B.; Park, S.T.; Wapinski, I.; Raman, S.; Dolganov, G.; Peterson, M.; Riley, R.; Zucker, J.; Abeel, T.; et al. Comparative analysis of Mycobacterium and related Actinomycetes yields insight into the evolution of Mycobacterium tuberculosis pathogenesis. BMC Genom. 2012, 13, 120. [Google Scholar] [CrossRef]
- Rengarajan, J.; Bloom, B.R.; Rubin, E.J. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. USA 2005, 102, 8327–8332. [Google Scholar] [CrossRef] [PubMed]
- Gautam, U.S.; Mehra, S.; Kumari, P.; Alvarez, X.; Niu, T.; Tyagi, J.S.; Kaushal, D. Mycobacterium tuberculosis sensor kinase DosS modulates the autophagosome in a DosR-independent manner. Commun. Biol. 2019, 2, 349. [Google Scholar] [CrossRef]
- Abendroth, J.; Ollodart, A.; Andrews, E.S.; Myler, P.J.; Staker, B.L.; Edwards, T.E.; Arcus, V.L.; Grundner, C. Mycobacterium tuberculosis Rv2179c protein establishes a new exoribonuclease family with broad phylogenetic distribution. J. Biol. Chem. 2014, 289, 2139–2147. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peng, Z.; Chen, L.; Zhang, H. Serum proteomic analysis of Mycobacterium tuberculosis antigens for discriminating active tuberculosis from latent infection. J. Int. Med. Res. 2020, 48, 300060520910042. [Google Scholar] [CrossRef] [PubMed]
- Winther, K.; Tree, J.J.; Tollervey, D.; Gerdes, K. VapCs of Mycobacterium tuberculosis cleave RNAs essential for translation. Nucleic Acids Res. 2016, 44, 9860–9871. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, U.; Barth, V.C.; Woychik, N.A. tRNA(fMet) Inactivating Mycobacterium tuberculosis VapBC Toxin-Antitoxin Systems as Therapeutic Targets. Antimicrob. Agents Chemother. 2022, 66, e0189621. [Google Scholar] [CrossRef] [PubMed]
- Marmiesse, M.; Brodin, P.; Buchrieser, C.; Gutierrez, C.; Simoes, N.; Vincent, V.; Glaser, P.; Cole, S.T.; Brosch, R. Macro-array and bioinformatic analyses reveal mycobacterial ‘core’ genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology 2004, 150 Pt 2, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Sverrisson, F.; Feydy, J.; Correia, B.E.; Bronstein, M.M. Fast end-to-end learning on protein surfaces. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 19–25 June 2021; IEEE: Nashville, TN, USA. [Google Scholar]
- Gainza, P.; Sverrisson, F.; Monti, F.; Rodolà, E.; Boscaini, D.; Bronstein, M.M.; Correia, B.E. Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning. Nat. Methods 2020, 17, 184–192. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Namugenyi, S.B.; Aagesen, A.M.; Elliott, S.R.; Tischler, A.D. Mycobacterium tuberculosis PhoY Proteins Promote Persister Formation by Mediating Pst/SenX3-RegX3 Phosphate Sensing. mBio 2017, 8, e00494-17. [Google Scholar] [CrossRef]
- Brokaw, A.M.; Eide, B.J.; Muradian, M.; Boster, J.M.; Tischler, A.D. Mycobacterium smegmatis PhoU Proteins Have Overlapping Functions in Phosphate Signaling and Are Essential. Front. Microbiol. 2017, 8, 2523. [Google Scholar] [CrossRef]
- Newton, G.L.; Buchmeier, N.; Fahey, R.C. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol. Mol. Biol. Rev. 2008, 72, 471–494. [Google Scholar] [CrossRef] [PubMed]
- Rubin, E.J.; Akerley, B.J.; Novik, V.N.; Lampe, D.J.; Husson, R.N.; Mekalanos, J.J. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. USA 1999, 96, 1645–1650. [Google Scholar] [CrossRef]
- Xu, W.; DeJesus, M.A.; Rücker, N.; Engelhart, C.A.; Wright, M.G.; Healy, C.; Lin, K.; Wang, R.; Park, S.W.; Ioerger, T.R.; et al. Chemical Genetic Interaction Profiling Reveals Determinants of Intrinsic Antibiotic Resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2017, 61, e01334-17. [Google Scholar] [CrossRef]
- Block, A.M.; Namugenyi, S.B.; Palani, N.P.; Brokaw, A.M.; Zhang, L.; Beckman, K.B.; Tischler, A.D. Mycobacterium tuberculosis Requires the Outer Membrane Lipid Phthiocerol Dimycocerosate for Starvation-Induced Antibiotic Tolerance. mSystems 2023, 8, e0069922. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Poulton, N.C.; Chang, J.S.; Azadian, Z.A.; DeJesus, M.A.; Ruecker, N.; Zimmerman, M.D.; Eckartt, K.A.; Bosch, B.; Engelhart, C.A.; et al. CRISPRi chemical genetics and comparative genomics identify genes mediating drug potency in Mycobacterium tuberculosis. Nat. Microbiol. 2022, 7, 766–779. [Google Scholar] [CrossRef] [PubMed]
- Fenn, K.; Wong, C.T.; Darbari, V.C. Mycobacterium tuberculosis Uses Mce Proteins to Interfere with Host Cell Signaling. Front. Mol. Biosci. 2019, 6, 149. [Google Scholar] [CrossRef]
- Kreutzfeldt, K.M.; Jansen, R.S.; Hartman, T.E.; Gouzy, A.; Wang, R.; Krieger, I.V.; Zimmerman, M.D.; Gengenbacher, M.; Sarathy, J.P.; Xie, M.; et al. CinA mediates multidrug tolerance in Mycobacterium tuberculosis. Nat. Commun. 2022, 13, 2203. [Google Scholar] [CrossRef]

| Mtb Gene | Mav Gene | Mtb LFC | Mav LFC | Mtb Annotation | Mav Annotation |
|---|---|---|---|---|---|
| rv0049 | DFS55_00355 | −2.99 | −1.09 | hypothetical protein | hypothetical protein |
| rv0819 | DFS55_21365 | 1.33 | 1.23 | mycothiol acetyltransferase | mycothiol synthase |
| rv0820 | DFS55_21345 | −1.42 | −1.69 | phosphate ABC transporter ATP-binding protein PhoT | phosphate ABC transporter ATP-binding protein |
| rv0929 | DFS55_20215 | −1.23 | −1.87 | phosphate ABC transporter permease PstC | phosphate ABC transporter permease PstC |
| rv0930 | DFS55_20210 | −1.16 | −1.00 | phosphate ABC transporter permease PstA | phosphate ABC transporter permease PstA |
| rv1836c | DFS55_12730 | −1.45 | −1.44 | hypothetical protein | hypothetical protein |
| rv2179c | DFS55_14810 | −2.72 | −1.09 | 3′-5′ exoribonuclease A | hypothetical protein |
| rv2224c | DFS55_15065 | −1.80 | −2.17 | carboxylesterase A | alpha/beta hydrolase |
| rv3005c | DFS55_07355 | −1.25 | −1.06 | hypothetical protein | hypothetical protein |
| Gene | 7H9 6d LFC | PBS 7d LFC | PBS 14d LFC | Annotation |
|---|---|---|---|---|
| rv0458 | −0.11 | 2.30 | 1.05 | aldehyde dehydrogenase |
| rv0819 | 1.33 | −1.44 | −1.67 | mycothiolacetyl transferase |
| rv0989c | 0.03 | 2.77 | 1.01 | polyprenyl-diphosphate synthase GrcC |
| rv0998 | −0.22 | −1.24 | −1.25 | acetyltransferase Pat |
| rv1183 | 0.1 | 0.52 | 1.12 | transmembrane transport protein MmpL10 |
| rv1908c | 0.85 | −0.88 | −1.15 | catalase-peroxidase |
| rv2051c | −0.19 | −1.33 | −1.23 | polyprenol-monophosphomannose synthase |
| rv2199c | −0.5 | 2.53 | 2.42 | cytochrome c oxidase polypeptide 4 |
| rv2374c | −0.2 | −1.47 | −1.68 | heat-inducible transcription repressor HrcA |
| rv2392 | 0.95 | −1.29 | −1.17 | phosphoadenosine phosphosulfate reductase |
| rv2633c | 0.41 | 1.76 | 1.02 | hypothetical protein |
| rv2709 | 1.16 | −1.26 | −1.16 | transmembrane protein |
| rv2733c | 0.01 | −1.55 | −1.19 | (Dimethylallyl)adenosine tRNA methylthiotransferase |
| rv3680 | −0.26 | −1.12 | −1.40 | anion transporter ATPase |
| rv3923c | −0.17 | −1.1 | −1.36 | ribonuclease P protein component |
| Gene | 7H9 6d LFC | PBS 7d LFC | PBS 14d LFC | Annotation |
|---|---|---|---|---|
| rv0049 | −2.99 | −1.45 | −1.81 | hypothetical protein |
| rv0199 | 1.14 | 4.50 | 2.53 | membrane protein |
| rv0200 | 1.61 | 4.69 | 2.56 | transmembrane protein |
| rv0655 | 1.32 | 5.03 | 3.01 | ABC transporter ATP-binding protein |
| rv0819 | 1.33 | −1.44 | −1.67 | mycothiol acetyltransferase |
| rv0994 | −3.25 | −1.27 | −1.01 | molybdopterin molybdenumtransferase 1 |
| rv2179c | −2.72 | −1.32 | −1.30 | 3′-5′ exoribonuclease |
| rv2690c | 5.56 | 3.26 | 1.63 | integral membrane protein |
| rv2709 | 1.16 | −1.26 | −1.16 | transmembrane protein |
| rv3723 | 1.18 | 4.94 | 3.04 | transmembrane protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matern, W.M.; Harris, H.T.; Danchik, C.; McDonald, M.; Patel, G.; Srivastava, A.; Ioerger, T.R.; Bader, J.S.; Karakousis, P.C. Functional Whole Genome Screen of Nutrient-Starved Mycobacterium tuberculosis Identifies Genes Involved in Rifampin Tolerance. Microorganisms 2023, 11, 2269. https://doi.org/10.3390/microorganisms11092269
Matern WM, Harris HT, Danchik C, McDonald M, Patel G, Srivastava A, Ioerger TR, Bader JS, Karakousis PC. Functional Whole Genome Screen of Nutrient-Starved Mycobacterium tuberculosis Identifies Genes Involved in Rifampin Tolerance. Microorganisms. 2023; 11(9):2269. https://doi.org/10.3390/microorganisms11092269
Chicago/Turabian StyleMatern, William M., Harley T. Harris, Carina Danchik, Marissa McDonald, Gopi Patel, Aashish Srivastava, Thomas R. Ioerger, Joel S. Bader, and Petros C. Karakousis. 2023. "Functional Whole Genome Screen of Nutrient-Starved Mycobacterium tuberculosis Identifies Genes Involved in Rifampin Tolerance" Microorganisms 11, no. 9: 2269. https://doi.org/10.3390/microorganisms11092269
APA StyleMatern, W. M., Harris, H. T., Danchik, C., McDonald, M., Patel, G., Srivastava, A., Ioerger, T. R., Bader, J. S., & Karakousis, P. C. (2023). Functional Whole Genome Screen of Nutrient-Starved Mycobacterium tuberculosis Identifies Genes Involved in Rifampin Tolerance. Microorganisms, 11(9), 2269. https://doi.org/10.3390/microorganisms11092269






