Diversity and Network Relationship Construction of Soil Fungal Communities in Lactarius hatsudake Tanaka Orchard during Harvest
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plantation Overview
2.2. Experimental Design and Soil Sample Collection
2.3. DNA Extraction, PCR Amplification, and High-Throughput Sequencing
2.4. Data Analysis
2.4.1. Community Diversity Analysis
2.4.2. Analysis of Community Species Differences
2.4.3. Molecular Ecological Network Construction and Analysis
2.4.4. Soil Fungal-Community Assembly Process
2.4.5. Statistics and Analysis
3. Results
3.1. Overall Sequencing Results
3.2. Analysis of Soil Fungal Diversity in the L. hatsudake Orchard during the Harvest Period
3.2.1. Analysis of Fungal α-Diversity
3.2.2. Analysis of Fungal β-Diversity
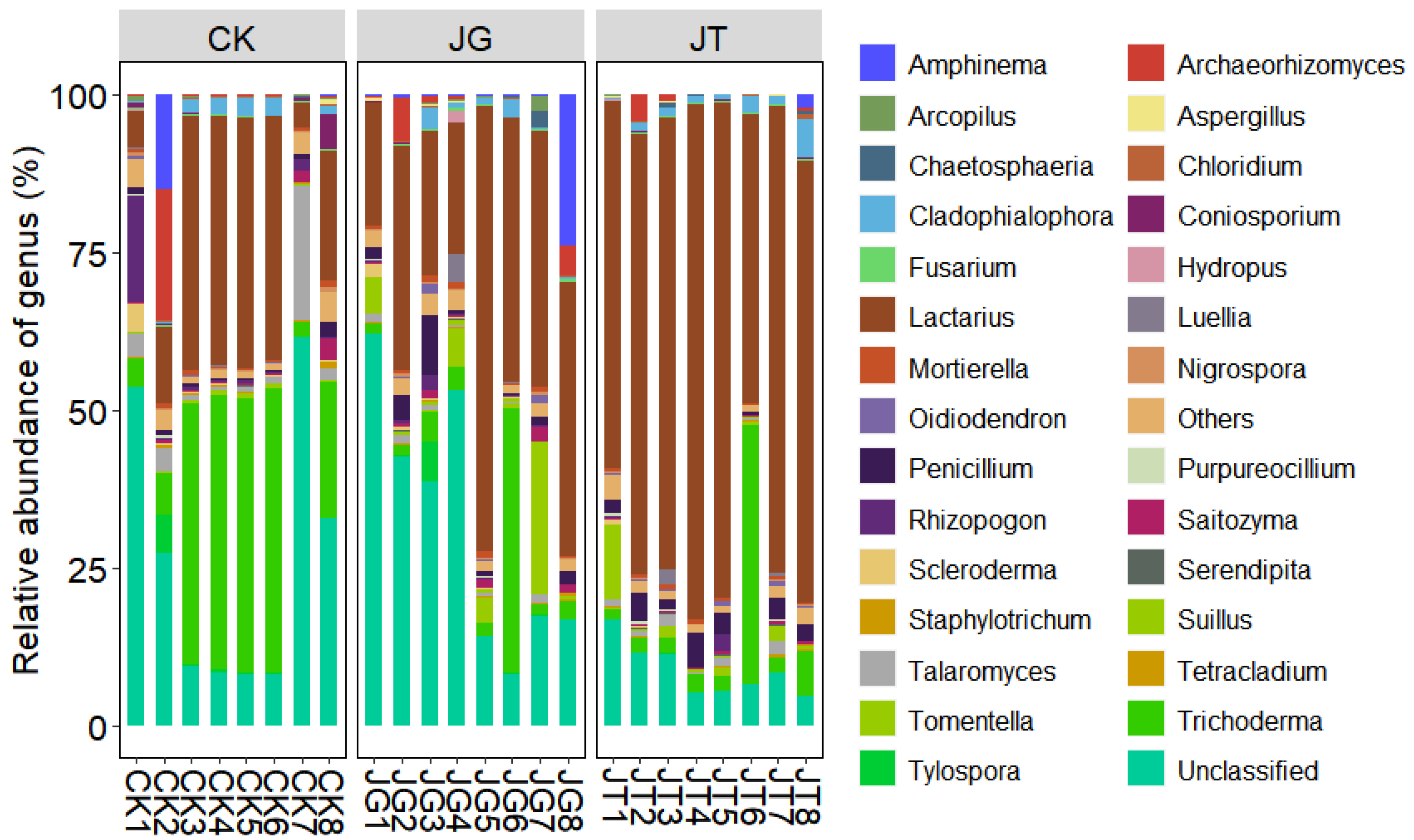
3.3. Soil Fungal-Community Structure in the L. hatsudake Orchard during the Harvest Period
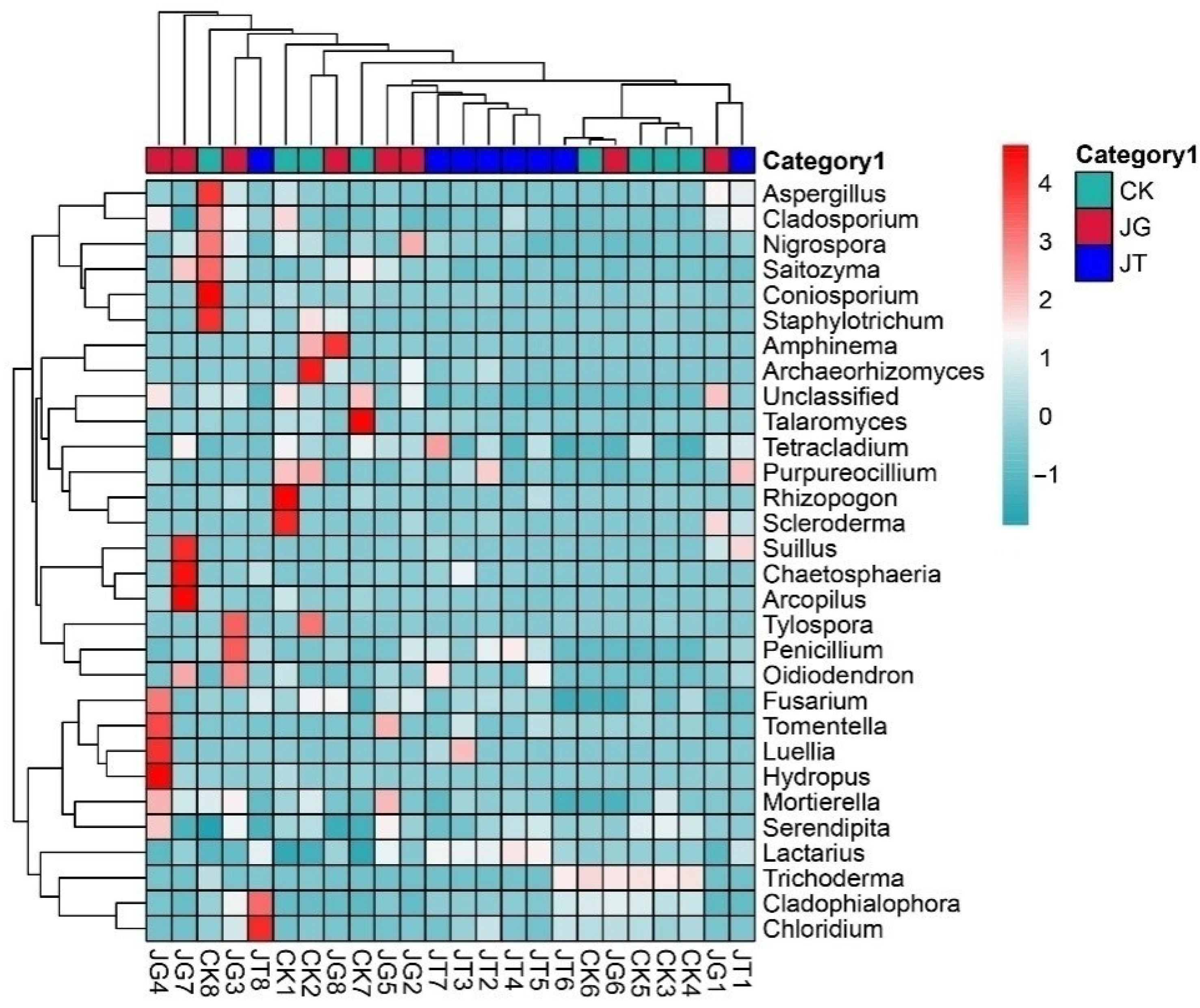
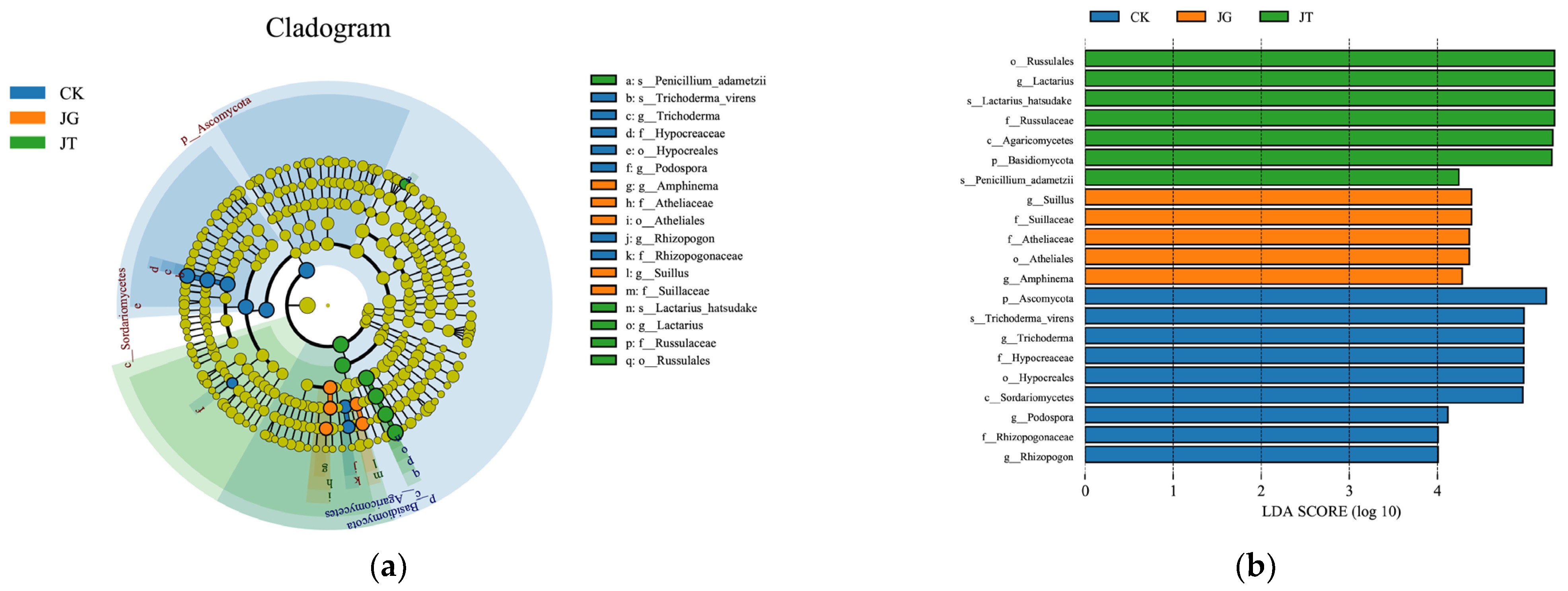
3.4. Analysis of the Differences in the Fungal Community of Different Sites in the L. hatsudake Orchard during the Harvesting Period
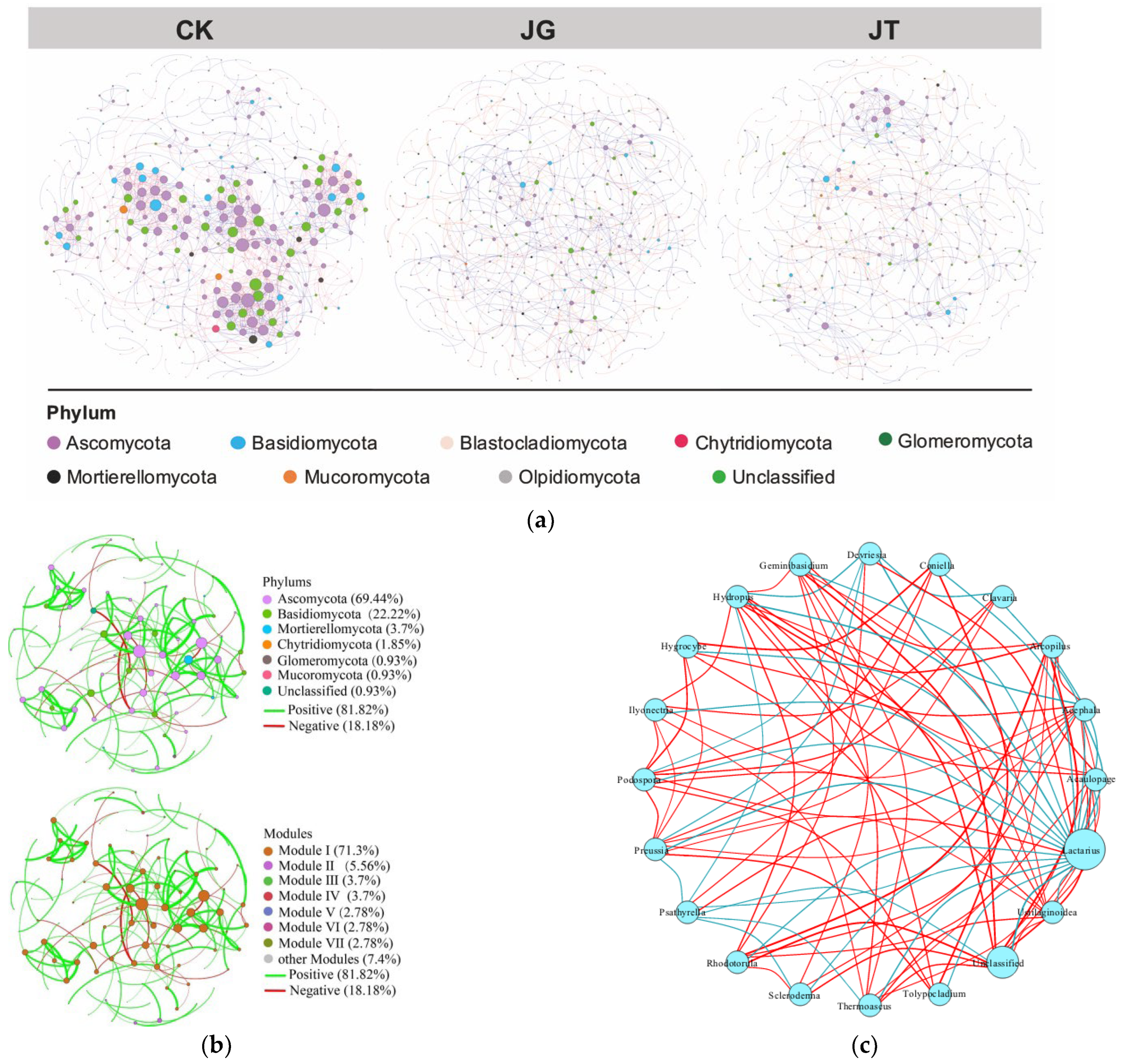
3.5. Molecular Ecological Network Structure Characteristics of Soil Fungi in the L. hatsudake Orchard during the Harvesting Period
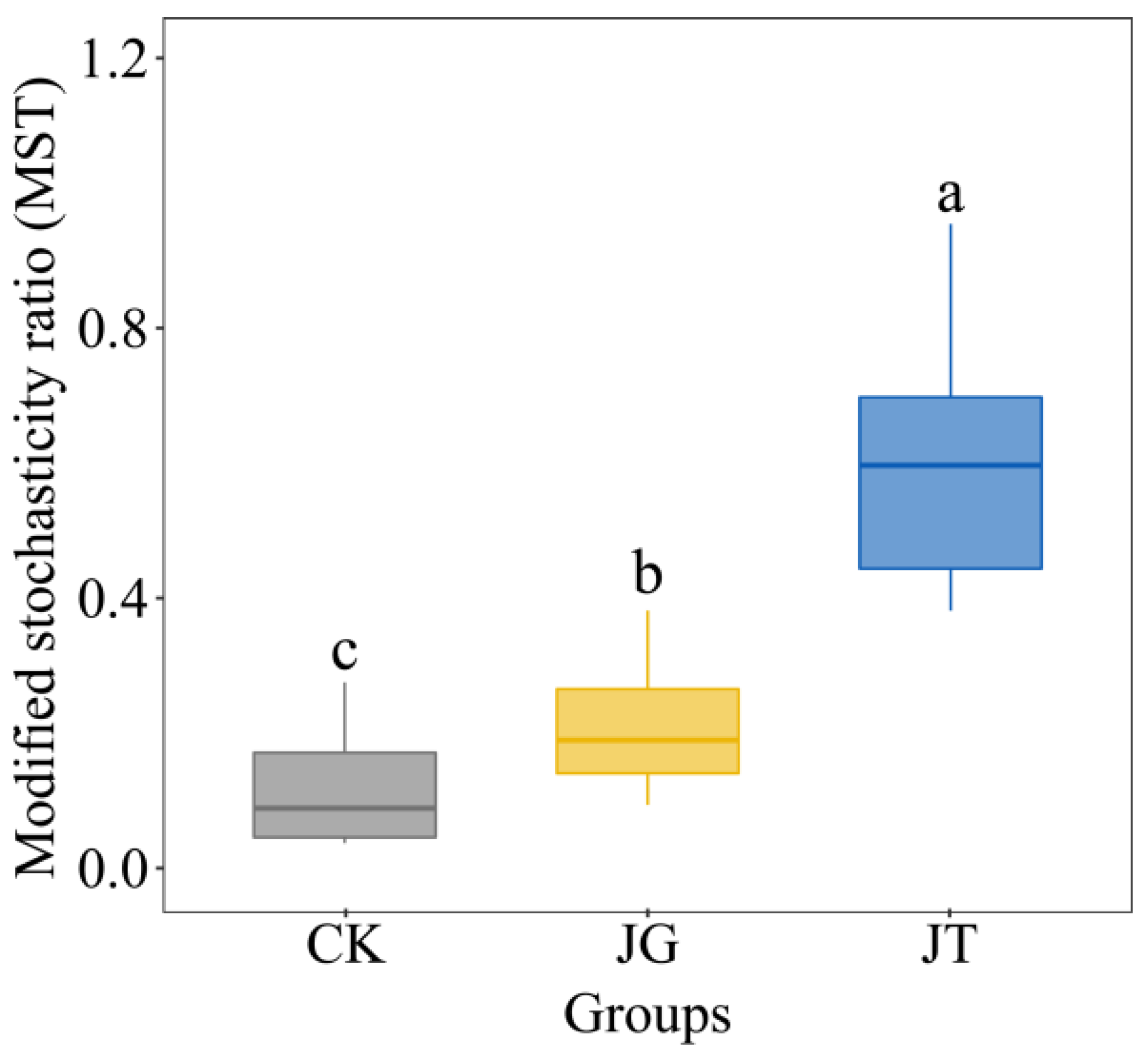
3.6. Microbial-Community Assembly Process
4. Conclusions and Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tan, Z.M. A Study on the Biological Characters of Lactarius hatsutake Tanaka and Its Semi-Artificaial Cultivation. Ph.D. Thesis, Hunan Agricultural University, Changsha, China, 2005. [Google Scholar]
- Wang, L.; Li, Z.; Zhu, M.; Meng, L.; Wang, H.; Ng, T.B. An acidic feruloyl esterase from the mushroom Lactarius hatsudake: A potential animal feed supplement. Int. J. Biol. Macromol. 2016, 93, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Song, F.; Jiang, L.; Liu, Z.; Tu, Y. Novel fluorescent nano carbon quantum dots derived from Lactarius hatsudake for high selective vitamin b12 detection. J. Aoac Int. 2022, 105, 1350–1359. [Google Scholar] [CrossRef]
- Zhang, A.L.; Liu, L.P.; Wang, M.; Gao, J.M. Bioactive ergosterol derivatives isolated from the fungus Lactarius hatsudake. Chem. Nat. Compd. 2007, 43, 637–638. [Google Scholar] [CrossRef]
- Zhang, X.F.; Cheng, M.Y.; Yang, J.; Chang, S.L.; Ren, J.L. Structural characteristics of polysaccharides from Lactarius hatsudake and its effect on intestinal. Food Mac. 2022, 38, 143–148. [Google Scholar] [CrossRef]
- Mitsuo, M.; Yumi, K.; Naoki, M. Character impact odorants from wild mushroom (Lactarius hatsudake) used in Japanese traditional food. Flavour Frag. J. 2010, 25, 197–201. [Google Scholar] [CrossRef]
- Yang, M.H.; Yang, X.M.; Chen, L.G. Studies on the relationship between Tricholoma matsutake and other rhizosphere microorganisms. Acta Agric. Univ. Jiangxiensis 1997, 14, 78–82. [Google Scholar]
- Perlińska-Lenart, U.; Pisyk, S.; Gryz, E.; Turo, J.; Kruszewska, J.S. Identification of bacteria and fungi inhabiting fruiting bodies of burgundy truffle (Tuber aestivumvittad.). Arch. Microbiol. 2020, 202, 2727–2738. [Google Scholar] [CrossRef]
- Mello, A.; Zampieri, E.; Zambonelli, A. Truffle Ecology: Genetic Diversity, Soil Interactions and Functioning. In Mycorrhiza—Function, Diversity, State of the Art; Varma, A., Prasad, R., Tuteja, N., Eds.; Springer: Berlin, Germany, 2017. [Google Scholar] [CrossRef]
- Vahdatzadeh, M.; Deveau, A.; Splivallo, R. The role of the microbiome of truffles in aroma formation: A meta-analysis approach. Appl. Environ. Microb. 2015, 81, 6946–6952. [Google Scholar] [CrossRef]
- Navarro-Ródenas, A.; Berná, L.M.; Lozano-Carrillo, C.; Andrino, A.; Morte, A. Beneficial native bacteria improve survival and mycorrhization of desert truffle mycorrhizal plants in nursery conditions. Mycorrhiza 2016, 26, 769–779. [Google Scholar] [CrossRef]
- Wang, T.; Meng, X.; Qinqin, M.A.; Yong, B.; Miao, Y. Functional diversity of culturable actinomycetes isolated from tuber inducum mycorrhizal soils in panzhihua, sichuan. Ecol. Sci. 2019, 38, 127–137. [Google Scholar] [CrossRef]
- Guo, X.; Li, Q.Q. Isolation and identification of a mycorrhiza helper bacteria strain from the rhizosphere soil of Boletus edulis and Pinus thunbergii. J. Forest Environ. 2019, 39, 315–319. [Google Scholar] [CrossRef]
- Guo, X.; Zheng, Y.H. Isolation and identification of mycorrhiza helper bacteria from the colonized soil of Boletus edulis. Mycosystema 2018, 37, 1802–1807. [Google Scholar] [CrossRef]
- Buzzini, P.; Gasparetti, C.; Turchetti, B.; Cramarossa, M.R.; Vaughan-Martini, A.; Martini, A.; Pagnoni, U.M.; Forti, L. Production of volatile organic compounds (vocs) by yeasts isolated from the ascocarps of black (Tuber melanosporum vitt.) and white (Tuber magnatum pico) truffles. Arch. Microbiol. 2005, 184, 187–193. [Google Scholar] [CrossRef]
- Miguel, A.M.; Águeda, B.; Sánchez, S.; Parladé, J. Ectomycorrhizal fungus diversity and community structure with natural and cultivated truffle hosts: Applying lessons learned to future truffle culture. Mycorrhiza 2014, 24, 5–18. [Google Scholar] [CrossRef]
- Kataoka, R.; Siddiqui, Z.A.; Kikuchi, J.; Ando, M.; Sriwati, R.; Nozaki, A.; Futai, K. Detecting nonculturable bacteria in the active mycorrhizal zone of the pine mushroom Tricholoma matsutake. J. Microbiol. 2012, 50, 199–206. [Google Scholar] [CrossRef]
- Napoli, C.; Mello, A.; Borra, A.; Vizzini, A.; Sourzat, P.; Bonfante, P. Tuber melanosporum, when dominant, affects fungal dynamics in truffle grounds. New Phytol. 2010, 185, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Arenas, F.; Navarro-Ródenas, A.; Marqués-Gálvez, J.E.; Ghignone, S.; Mello, A.; Morte, A. Different patterns in root and soil fungal diversity drive plant productivity of the desert truffle Terfezia claveryi in plantation. Environ. Microbiol. 2021, 23, 5917–5933. [Google Scholar] [CrossRef]
- Yamamoto, S.; Sato, H.; Tanabe, A.S.; Hidaka, A.; Kadowaki, K.; Toju, H. Spatial segregation and aggregation of ectomycorrhizal and root-endophytic fungi in the seedlings of two Quercus species. PLoS ONE 2014, 9, e96363. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.M. Morphological and structural characteristics of Lactarius hatsudake and artificial mycorrhizal fungi. Acta Edulis Fungi 2006, 13, 36–40. [Google Scholar] [CrossRef]
- Ministry of Civil Affairs of the People’s Republic of China; Li, L.G.; Yu, C.M.; Duan, L.Y. The People’s Republic of China Administrative District Grand Canon Hunan Volume; China Society Press: Beijing, China, 2015; pp. 246–247.
- Kim, M.; Yoon, H.; You, Y.H.; Kim, Y.E.; Kim, J.G. Metagenomic analysis of fungal communities inhabiting the fairy ring zone of Tricholoma matsutake. J. Microbiol. Biotechnol. 2013, 23, 1347. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Liu, Z.; Lin, Y.; Yang, J.; Chen, W.; Wei, G. Bacterial communities in oil contaminated soils: Biogeography and co-occurrence patterns. Soil Biol. Biochem. 2016, 98, 64–73. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Nguyen, N.; Song, Z.; Bates, S.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.; Kennedy, P. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Edgar, R. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2012, 10, 57–59. [Google Scholar] [CrossRef]
- Kõljalg, U.; Larsson, K.; Abarenkov, K.; Nilsson, R.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; et al. UNITE: A database providing web-based methods for the molecular identification of ectomy16corrhizal fungi. New Phytol. 2005, 166, 1063–1068. [Google Scholar] [CrossRef]
- Looft, T.; Johnson, T.A.; Allen, H.K.; Bayles, D.O.; Alt, D.P.; Stedtfeld, R.D.; Sul, W.J.; Stedtfeld, T.M.; Chai, B.; Cole, J.R.; et al. In-feed antibiotic effects on the swine intestinal microbiome. Pans 2012, 109, 1691–1696. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the International AAAI Conference on Web and Social Media, San Jose, CA, USA, 17–20 May 2009. [Google Scholar]
- Yang, Z. Study on Artificial Mycorrhizal Synthesis and Shiroes Soil Microbial Diversity of Thelephora ganbajun. Master’s Thesis, Yunnan University, Kunming, China, 2019. [Google Scholar]
- Ye, L. Study on the Correlation of Soil Fungal Community Structure and Edaphic Factor in Tuber indicum Producing Areas. Master’s Thesis, Sichuan Agricultural University, Chengdu, China, 2019. [Google Scholar]
- Ye, L.; Fu, Y.; Zhang, X.P.; Sun, Q.; Zhang, X.P.; Li, X.L. Structure and diversity of cultivable endophyte from hosts of truffle (Pinus armandii). J. Microbiol. 2018, 38, 21–28. [Google Scholar]
- Ye, L.; Fu, Y.; Zou, J.; Li, X.L. Isolation and identification of endophytic fungi from three commercial truffles in Sichuan province. Chin. Agric. Sci. Bull. 2018, 34, 57–63. [Google Scholar]
- Yang, J.Q. Isolation, Purification and Identification of 11 Species of Accompanying Fungi with Xerocomus spadiceus. Master’s Thesis, Yunnan University, Kunming, China, 2019. [Google Scholar]
- Hilszczańska, D.; Szmidla, H.; Horak, J.; Rosa-Gruszecka, A. Ectomycorrhizal communities in a Tuber aestivum vittad. orchard in poland. Cent. Eur. J. Biol. 2016, 11, 348–357. [Google Scholar] [CrossRef]
- Sánchez, S.; Ágreda, T.; Águeda, B.; María, M.; María de Miguel, A.; Barriuso, J. Persistence and detection of black truffle ectomycorrhizas in plantations: Comparison between two field detection methods. Mycorrhiza 2014, 24, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Salerni, E.; D’Aguanno, M.; Leonardi, P.; Perini, C. Ectomycorrhizal communities above and below ground and truffle productivity in a Tuber aestivum orchard. Forest Syst. 2014, 23, 329–338. [Google Scholar] [CrossRef]
- De Miguel, A.M.; Àgueda, B.; Sàez, R.; Sànchez, S.; Parladé, J. Diversity of ectomycorrhizal Thelephoraceae in Tuber melanosporum–cultivated orchards of Northern Spain. Mycorrhiza 2016, 6, 227–236. [Google Scholar] [CrossRef]
- Oliach, D.; Colinas, C.; Castao, C.; Fischer, C.R.; Oliva, J. The influence of forest surroundings on the soil fungal community of black truffle (Tuber melanosporum) plantations. Forest Ecol. Manag. 2020, 470–471, 118212. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Z.S.; Yang, F.C.; Cheng, B.; Liang, J.F.; Lu, J.K. Mycorrhizosphere microbial community structure of Castanopsis hystrix and Pinus massoniana mixed plantation in Pingxiang, Guangxi of South China. Mycosystema 2021, 40, 1343–1356. [Google Scholar] [CrossRef]
- Ren, K.Y.; Zhao, J.C.; Guo, S.; Wang, S.S.; Zhang, C.J.; Pang, S.J.; Yang, S.D. Characteristics of soil biological properties and fungal diversity of Russula vinosa in Castanopsis hystrix. Southwest China J. Agric. Sci. 2020, 33, 109–116. [Google Scholar] [CrossRef]
- Mello, A.; Lumini, E.; Napoli, C.; Bianciotto, V.; Bonfante, P. Arbuscular mycorrhizal fungal diversity in the Tuber melanosporum Arbuscular mycorrhizal fungal diversity in the Tuber melanosporum brule. Fungal Biol. 2015, 119, 518–527. [Google Scholar] [CrossRef]
- Li, Q.; Yan, L.; Ye, L.; Zhou, J.; Zhang, B.; Peng, W.; Zhang, X.; Li, X. Chinese black truffle (Tuber indicum) alters the ectomycorrhizosphere and endoectomycosphere microbiome and metabolic profiles of the host tree Quercus aliena. Front. Microbiol. 2018, 9, 2202. [Google Scholar] [CrossRef]
- Liu, D.; Pérez-Moreno, J.; He, X.; Garibay-Orijel, R.; Yu, F. Truffle microbiome is driven by fruit body compartmentalization rather than soils conditioned by different host trees. Msphere 2021, 6, e0003921. [Google Scholar] [CrossRef]
- Elisa, T.; Mathieu, S.; Adrien, T.; Javier, P.; Marc-Andre, S.; Franck, R. Whose truffle is this? Distribution patterns of ectomycorrhizal fungal diversity in Tuber melanosporum brules developed in multi-host Mediterranean plant communities. Environ. Microbiol. 2015, 17, 2747–2761. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, J.; Xiong, C.; Li, X.; Chen, Z.; Li, P.; Huang, W. Tuber indicum shapes the microbial communities of ectomycorhizosphere soil and ectomycorrhizae of an indigenous tree (Pinus armandii). PLoS ONE 2017, 12, e0175720. [Google Scholar] [CrossRef]
- Streiblová, E.; Gryndlerová, H.; Gryndler, M. Truffle brûlé: An efficient fungal life strategy. FEMS Microbiol. Ecol. 2012, 80, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zampieri, E.; Chiapello, M.; Daghino, S.; Bonfante, P.; Mello, A. Soil metaproteomics reveals an inter-kingdom stress response to the presence of black truffles. Sci. Rep. 2016, 6, 25773. [Google Scholar] [CrossRef] [PubMed]


| Sample ID | Richness | ACE | Chao1 | Simpson | Shannon |
|---|---|---|---|---|---|
| CK | 511 ± 8.01a | 595.88 ± 9.60a | 616.87 ± 10.89a | 0.78 ± 0.05a | 3.77 ± 0.34a |
| JG | 502 ± 8.90a | 594.35 ± 10.16a | 612.18 ± 13.62a | 0.77 ± 0.05a | 3.86 ± 0.44a |
| JT | 474 ± 6.61b | 569.76 ± 6.48b | 573.05 ± 4.53b | 0.49 ± 0.04b | 2.42 ± 0.15b |
| Network | Index | Non-Mushroom- Producing Area | Mushroom-Producing Area | |
|---|---|---|---|---|
| CK | JG | JT | ||
| Empirical | RMT threshold | 0.910 | 0.890 | 0.890 |
| Total nodes | 405 | 441 | 399 | |
| Total edges | 1360 | 647 | 586 | |
| R2 of power-law | 0.775 | 0.802 | 0.900 | |
| Average degree (avgK) | 6.716 | 2.934 | 2.937 | |
| Average clustering coefficient (avgCC) | 0.321 | 0.210 | 0.189 | |
| Average path distance (GD) | 5.592 | 9.850 | 9.260 | |
| Positive edges | 787 (57.9%) | 308 (47.6%) | 257 (43.9%) | |
| Negative edges | 573 (42.1%) | 339 (52.4%) | 329 (56.1%) | |
| Randomized | Average clustering coefficient (avgCC) | 0.017 ± 0.002 | 0.007 ± 0.003 | 0.007 ± 0.004 |
| Average path distance (GD) | 3.361 ± 0.008 | 5.569 ± 0.094 | 5.479 ± 0.090 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, A.; Shen, B.; Liu, L.; Tan, Y.; Zeng, L.; Tan, Z.; Li, J. Diversity and Network Relationship Construction of Soil Fungal Communities in Lactarius hatsudake Tanaka Orchard during Harvest. Microorganisms 2023, 11, 2279. https://doi.org/10.3390/microorganisms11092279
Shen A, Shen B, Liu L, Tan Y, Zeng L, Tan Z, Li J. Diversity and Network Relationship Construction of Soil Fungal Communities in Lactarius hatsudake Tanaka Orchard during Harvest. Microorganisms. 2023; 11(9):2279. https://doi.org/10.3390/microorganisms11092279
Chicago/Turabian StyleShen, Airong, Baoming Shen, Lina Liu, Yun Tan, Liangbin Zeng, Zhuming Tan, and Jilie Li. 2023. "Diversity and Network Relationship Construction of Soil Fungal Communities in Lactarius hatsudake Tanaka Orchard during Harvest" Microorganisms 11, no. 9: 2279. https://doi.org/10.3390/microorganisms11092279




