The Deciphering of Growth-Dependent Strategies for Quorum-Sensing Networks in Pseudomonas aeruginosa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms and Growth Media
2.2. Planktonic Culture under Dynamic Conditions
2.3. Dual Biofilm Formation in Liquid-Medium Cultures
2.4. Quantitative PCR Analysis
2.5. Scanning Electron Microscopy (SEM)
2.6. Metabolite Extraction
2.7. Quantitative High-Performance Liquid Chromatography (HPLC) with Tandem Mass Spectrometric (MS/MS) Analysis
2.8. Relative Quantitation of Pyocyanin via Matrix-Assisted Laser Desorption Ionization MS
3. Results and Discussion
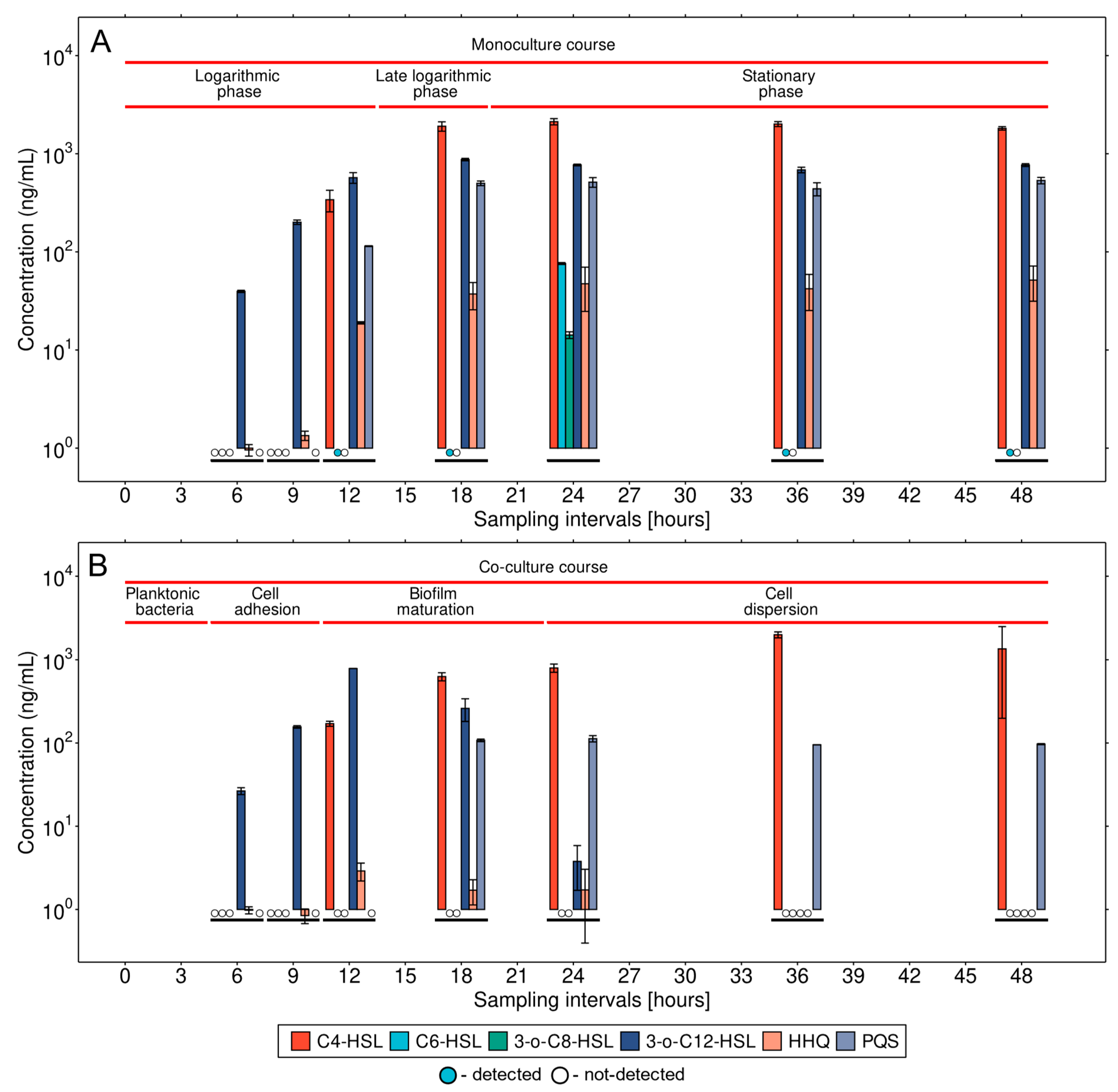
3.1. QS Expression Dynamics in a Monoculture of P. aeruginosa
3.2. Detailed Analysis of QS Molecules in the Co-Culture
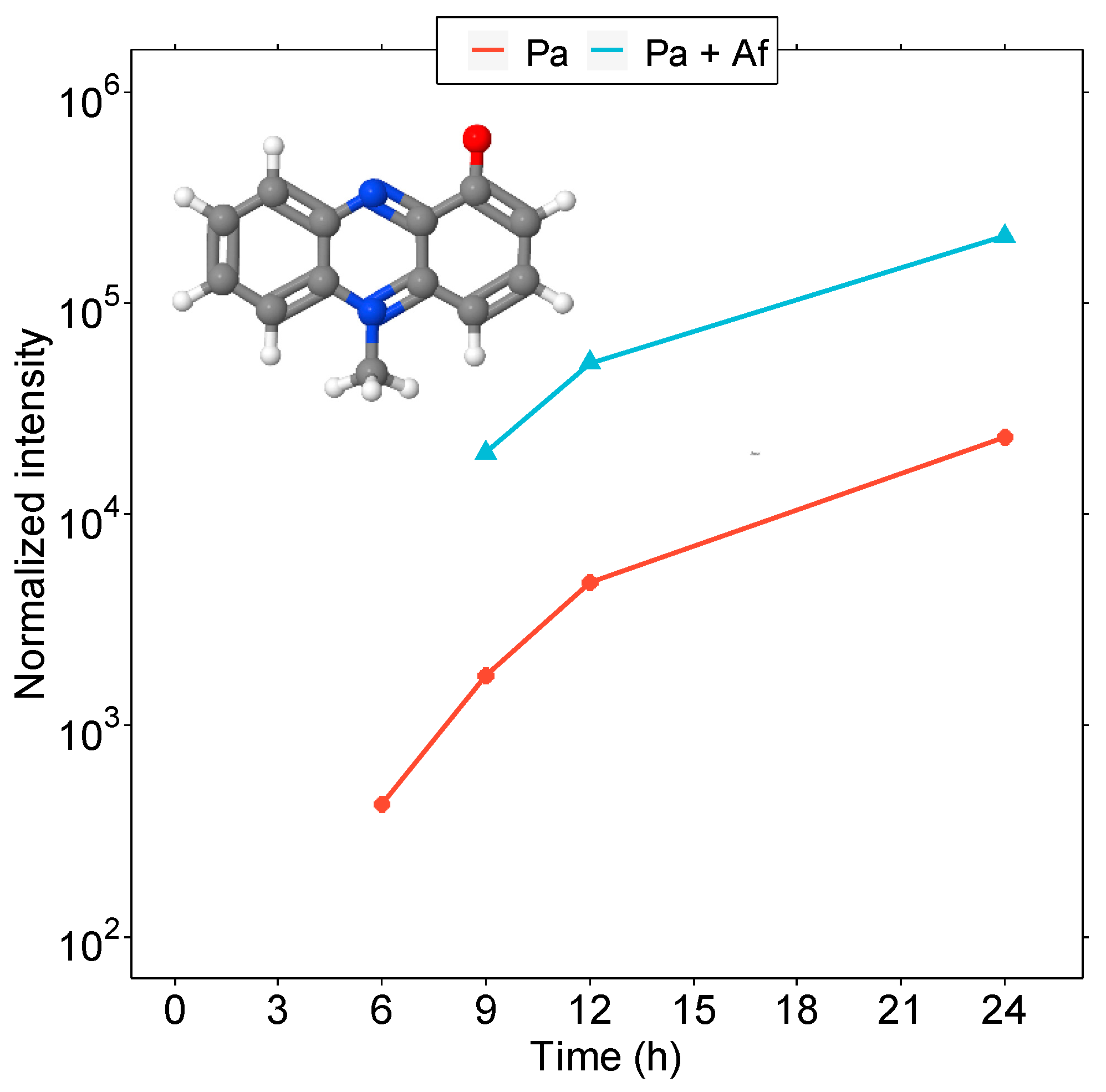
3.3. Microbial Interaction in the Co-Culture
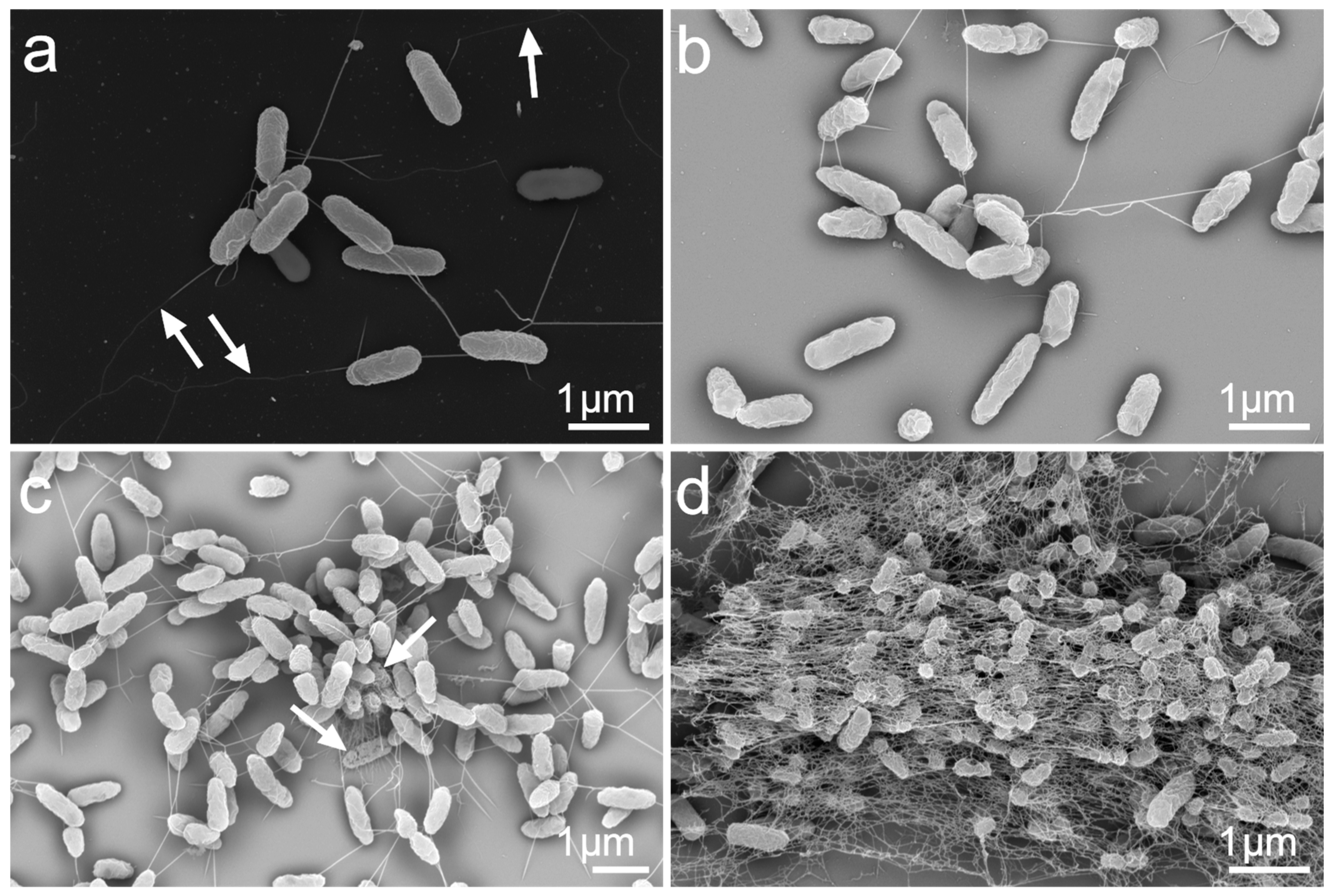
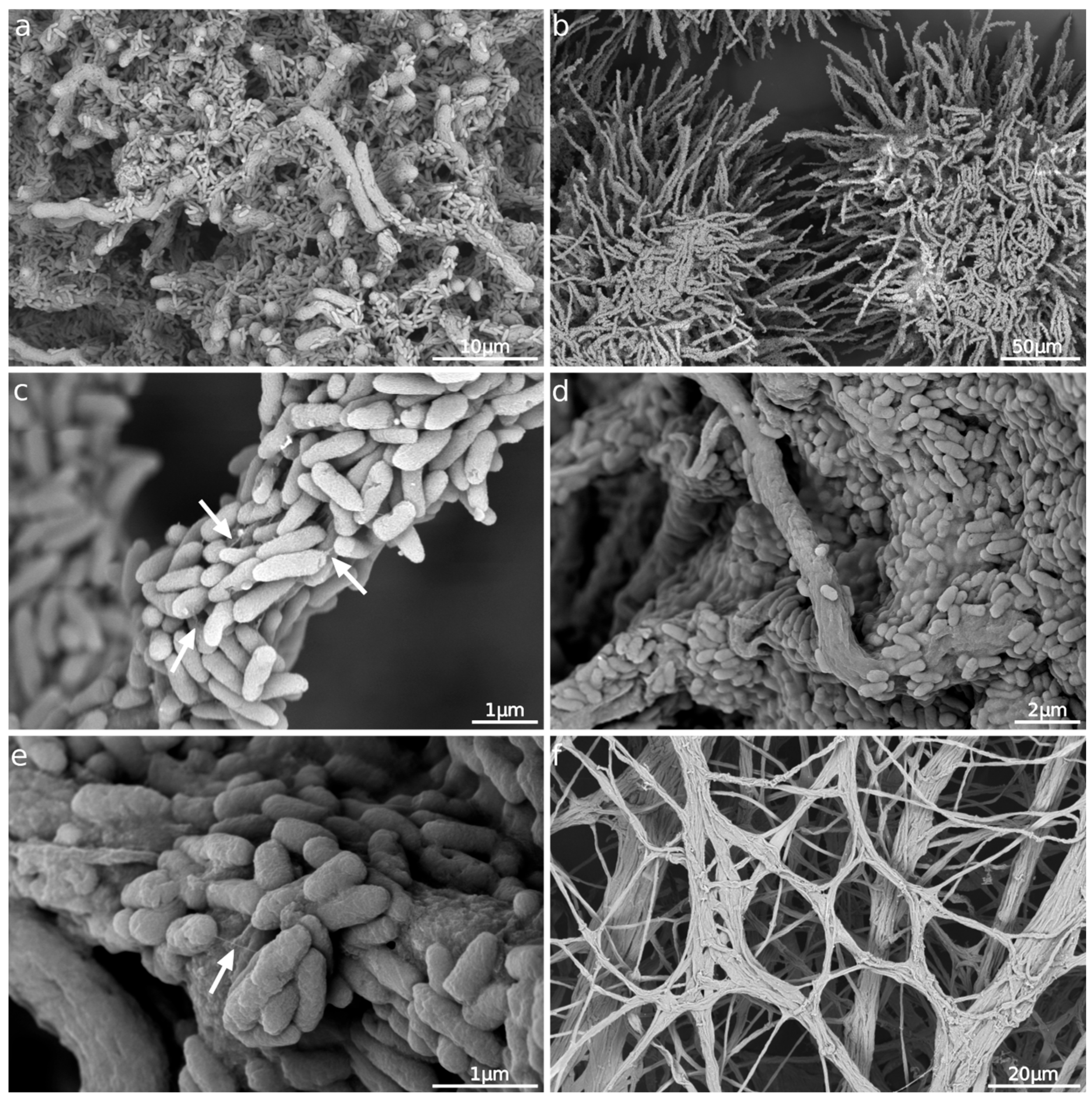
3.4. Morphological Characterization of the P. aeruginosa Monoculture and Biofilm Formation in the Presence of A. fumigatus
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Douglas, A.E. The microbial exometabolome: Ecological resource and architect of microbial communities. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190250. [Google Scholar] [CrossRef] [PubMed]
- Schroter, L.; Dersch, P. Phenotypic diversification of microbial pathogens—Cooperating and preparing for the future. J. Mol. Biol. 2019, 431, 4645–4655. [Google Scholar] [CrossRef] [PubMed]
- Spratt, M.R.; Lane, K. Navigating environmental transitions: The role of phenotypic variation in bacterial responses. mBio 2022, 13, e0221222. [Google Scholar] [CrossRef] [PubMed]
- Assefa, M.; Amare, A. Biofilm-associated multi-drug resistance in hospital-acquired infections: A review. Infect. Drug Resist. 2022, 31, 5061–5068. [Google Scholar] [CrossRef] [PubMed]
- Sauer, K.; Camper, A.K.; Ehrlich, G.D.; Costerton, J.W.; Davies, D.G. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 2002, 184, 1140–1154. [Google Scholar] [CrossRef]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The biofilm life cycle: Expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Sig. Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef]
- WHO/EMP/IAU/2017.12; Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis. World Health Organization: Geneva, Switzerland, 2017.
- Tashiro, Y.; Yawata, Y.; Toyofuku, M.; Uchiyama, H.; Nomura, N. Interspecies interaction between Pseudomonas aeruginosa and other microorganisms. Microbes Environ. 2013, 28, 13–24. [Google Scholar] [CrossRef]
- Hector, A.; Kirn, T.; Ralhan, A.; Graepler-Mainka, U.; Berenbrinker, S.; Riethmueller, J.; Hogardt, M.; Wagner, M.; Pfleger, A.; Hartl, D.; et al. Microbial colonization and lung function in adolescents with cystic fibrosis. J. Cyst. Fibros. 2016, 15, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Baxter, C.G.; Dunn, G.; Jones, A.M.; Webb, K.; Gore, R.; Richardson, M.D.; Denning, D.W. Novel immunologic classification of aspergillosis in adult cystic fibrosis. J. Allergy Clin. Immunol. 2013, 132, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Moss, A.; Juarez-Colunga, E.; Nathoo, F.; Wagner, B.; Sagel, S. A comparison of change point models with application to longitudinal lung function measurements in children with cystic fibrosis. Stat. Med. 2016, 35, 2058–2073. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.P.; Chamilos, G. Aspergillus fumigatus and aspergillosis in 2019. Clin. Microbiol. Rev. 2019, 33, e00140-18. [Google Scholar] [CrossRef]
- Agarwal, R.; Sehgal, I.S.; Dhooria, S.; Aggarwal, A.N. Developments in the diagnosis and treatment of allergic bronchopulmonary aspergillosis. Expert Rev. Respir. Med. 2016, 10, 1317–1334. [Google Scholar] [CrossRef]
- Brandt, C.; Roehmel, J.; Rickerts, V.; Melichar, V.; Niemann, N.; Schwarz, C. Aspergillus bronchitis in patients with cystic fibrosis. Mycopathologia 2018, 183, 61–69. [Google Scholar] [CrossRef]
- Reece, E.; Doyle, S.; Greally, P.; Renwick, J.; McClean, S. Aspergillus fumigatus inhibits Pseudomonas aeruginosa in co-culture: Implications of a mutually antagonistic relationship on virulence and inflammation in the CF Airway. Front. Microbiol. 2018, 5, 1205. [Google Scholar] [CrossRef]
- Bastos, R.W.; Akiyama, D.; Dos Reis, T.F.; Colabardini, A.C.; Luperini, R.S.; de Castro, P.A.; Baldini, R.L.; Fill, T.; Goldman, G.H. Secondary metabolites produced during Aspergillus fumigatus and Pseudomonas aeruginosa biofilm formation. mBio 2022, 30, e0185022. [Google Scholar] [CrossRef]
- Mowat, E.; Rajendran, R.; Williams, C.; McCulloch, E.; Jones, B.; Lang, S.; Ramage, G. Pseudomonas aeruginosa and their small diffusible extracellular molecules inhibit Aspergillus fumigatus biofilm formation. FEMS Microbiol. Lett. 2010, 313, 96–102. [Google Scholar] [CrossRef]
- Sass, G.; Nazik, H.; Penner, J.; Shah, H.; Ansari, S.R.; Clemons, K.V.; Groleau, M.C.; Dietl, A.M.; Visca, P.; Stevens, D.A.; et al. Studies of Pseudomonas aeruginosa mutants indicate pyoverdine as the central factor in inhibition of Aspergillus fumigatus biofilm. J. Bacteriol. 2018, 200, e00345-17. [Google Scholar] [CrossRef]
- Briard, B.; Mislin, G.L.A.; Latgé, J.P.; Beauvais, A. Interactions between Aspergillus fumigatus and pulmonary bacteria: Current state of the field, new data, and future perspective. J. Fungi 2019, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Sass, G.; Ansari, S.R.; Dietl, A.M.; Déziel, E.; Haas, H.; Stevens, D.A. Intermicrobial interaction: Aspergillus fumigatus siderophores protect against competition by Pseudomonas aeruginosa. PLoS ONE 2019, 14, e0216085. [Google Scholar] [CrossRef] [PubMed]
- Surpeta, B.; Grulich, M.; Brezovsky, J. Quorum quenching enzymes—Understanding molecular determinants responsible for activity of N-terminal serine hydrolases to increase their strong antibacterial potency. Biophys. J. 2021, 120, 305a. [Google Scholar] [CrossRef]
- Surpeta, B.; Grulich, M.; Palyzová, A.; Marešová, H.; Brezovsky, J. Common dynamic determinants govern quorum quenching activity in N-terminal serine hydrolases. ACS Catal. 2022, 12, 6359–6374. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, Y.; Ge, Y.; Zhu, X.; Pan, J. Regulatory mechanisms and promising applications of quorum sensing-inhibiting agents in control of bacterial biofilm formation. Front. Microbiol. 2020, 11, 589640. [Google Scholar] [CrossRef]
- Dulcey, C.E.; Dekimpe, V.; Fauvelle, D.-A.; Milot, S.; Groleau, M.-C.; Doucet, N.; Rahme, L.G.; Lepine, F.; Deziel, E. The end of an old hypothesis: The Pseudomonas signaling molecules 4-hydroxy-2-alkylquinolines derive from fatty acids, not 3-ketofatty acids. Chem. Biol. 2013, 20, 1481–1491. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef]
- O’Loughlin, C.T.; Miller, L.C.; Siryaporn, A.; Drescher, K.; Semmelhack, M.F.; Semmelhack, M.F.; Bassler, B.L. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. USA 2013, 110, 17981–17986. [Google Scholar] [CrossRef]
- Liu, Y.C.; Chan, K.G.; Chang, C.Y. Modulation of host biology by Pseudomonas aeruginosa quorum sensing signal molecules: Messengers or traitors. Front. Microbiol. 2016, 6, 1226. [Google Scholar] [CrossRef]
- Ramos, A.N.; Peral, M.C.; Valdez, J.C. Differences between Pseudomonas aeruginosa in a clinical sample and in a colony isolated fromit: Comparison of virulence capacity and susceptibility of biofilm to inhibitors. Comp. Immunol. Microbiol. Infect. Dis. 2010, 33, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Li, Y.; Zhou, Q.; Xu, Y. Regulation of gene expression in Pseudomonas aeruginosa M18 by phenazine-1-carboxylic acid. Appl. Microbiol. Biotechnol. 2015, 99, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Vilaplana, L.; Marco, M.P. Phenazines as potential biomarkers of Pseudomonas aeruginosa infections: Synthesis regulation, pathogenesis and analytical methods for their detection. Anal. Bioanal. Chem. 2020, 412, 5897–5912. [Google Scholar] [CrossRef]
- Sass, G.; Nazik, H.; Chatterjee, P.; Stevens, D.A. Under nonlimiting iron conditions pyocyanin is a major antifungal molecule, and differences between prototypic Pseudomonas aeruginosa strains. Med. Mycol. 2021, 59, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Saunders, S.H.; Tse, E.C.M.; Yates, M.D.; Otero, F.J.; Trammell, S.A.; Stemp, E.D.A.; Barton, J.K.; Tender, L.M.; Newman, D.K. Extracellular DNA promotes efficient extracellular electron transfer by pyocyanin in Pseudomonas aeruginosa biofilms. Cell 2020, 182, 919–932.e19. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, P.N.; Koch, G.; Thompson, J.A.; Xavier, K.B.; Cool, R.H.; Quax, W.J. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2012, 76, 46–65. [Google Scholar] [CrossRef]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Luptáková, D.; Patil, R.H.; Dobiáš, R.; Stevens, D.A.; Pluháček, T.; Palyzová, A.; Káňová, M.; Navrátil, M.; Vrba, Z.; Hubáček, P.; et al. Siderophore-based noninvasive differentiation of Aspergillus fumigatus colonization and invasion in pulmonary aspergillosis. Microbiol. Spectr. 2023, 11, e0406822. [Google Scholar] [CrossRef]
- Walsh, T.J.; Wissel, M.C.; Grantham, K.J.; Petraitiene, R.; Petraitis, V.; Kasai, M.; Francesconi, A.; Cotton, M.P.; Hughes, J.E.; Greene, L.; et al. Molecular detection and species-specific identification of medically important Aspergillus species by real-time PCR in experimental invasive pulmonary aspergillosis. J. Clin. Microbiol. 2011, 49, 4150–4157. [Google Scholar] [CrossRef]
- Joly, B.; Pierre, M.; Auvin, S.; Colin, F.; Gottrand, F.; Guery, B.; Husson, M.O. Relative expression of Pseudomonas aeruginosa virulence genes analyzed by a real time RT-PCR method during lung infection in rats. FEMS Microbiol. Lett. 2005, 243, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Patil, R.H.; Kotta-Loizou, I.; Palyzová, A.; Pluháček, T.; Coutts, R.H.A.; Stevens, D.A.; Havlíček, V. Freeing Aspergillus fumigatus of polymycovirus infection renders it more resistant to competition with Pseudomonas aeruginosa due to altered iron-acquiring tactics. J. Fungi 2021, 7, 497. [Google Scholar] [CrossRef] [PubMed]
- Winson, M.K.; Camara, M.; Latifi, A.; Foglino, M.; Chhabra, S.R.; Daykin, M.; Bally, M.; Chapon, V.; Salmond, G.P.C.; Bycroft, B.W.; et al. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1995, 92, 9427–9431. [Google Scholar] [CrossRef]
- Chuang, S.K.; Vrla, G.D.; Fröhlich, K.S.; Gitai, Z. Surface association sensitizes Pseudomonas aeruginosa to quorum sensing. Nat. Commun. 2019, 10, 4118. [Google Scholar] [CrossRef] [PubMed]
- Pearson, J.P.; Gray, K.M.; Passador, L.; Tucker, K.D.; Eberhard, A.; Iglewski, B.H.; Greenberg, E.P. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 1994, 91, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Schaefer, A.L.; Parsek, M.R.; Moninger, T.O.; Welsh, M.J.; Greenberg, E.P. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 2000, 407, 762–764. [Google Scholar] [CrossRef] [PubMed]
- Reimmann, C.; Beyeler, M.; Latifi, A.; Winteler, H.; Foglino, M.; Lazdunski, A.; Haas, D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide and lipase. Mol. Microbiol. 1997, 24, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Favre-Bonté, S.; Kohler, T.; Van Delden, C. Biofilm formation by Pseudomonas aerugnosa: Role of the C4-HSL cell-to-cell signal and inhibition by azithromycin. J. Antimicrobiol. Chem. 2003, 52, 598–604. [Google Scholar] [CrossRef]
- Mellbye, B.; Schuster, M. Physiological framework for the regulation of quorum sensing-dependent public goods in Pseudomonas aeruginosa. J. Bacteriol. 2014, 196, 1155–1164. [Google Scholar] [CrossRef]
- Zain, N.M.M.; Webb, K.; Stewart, I.; Halliday, N.; Barrett, D.A.; Nash, E.F.; Whitehouse, J.L.; Honeybourne, D.; Smyth, A.R.; Forrester, D.L.; et al. 2-Alkyl-4-quinolone quorum sensing molecules are biomarkers for culture-independent Pseudomonas aeruginosa burden in adults with cystic fibrosis. J. Med. Microbiol. 2021, 70, 001420. [Google Scholar] [CrossRef]
- Dobiáš, R.; Škríba, A.; Pluháček, T.; Petřík, M.; Palyzová, A.; Káňová, M.; Čubová, E.; Houšť, J.; Novák, J.; Stevens, D.A.; et al. Noninvasive combined diagnosis and monitoring of Aspergillus and Pseudomonas infections: Proof of concept. J. Fungi 2021, 7, 730. [Google Scholar] [CrossRef] [PubMed]
- Dandela, R.; Mantin, D.; Cravatt, B.F.; Rayo, J.; Meijler, M.M. Proteome-wide mapping of PQS-interacting proteins in Pseudomonas aeruginosa. Chem. Sci. 2018, 9, 2290–2294. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.C.; Loughlin, C.T.O.; Zhang, Z.; Siryaporn, A.; Slipe, J.E.; Bassler, B.L.; Semmelhack, M.F. Development of potential inhibitors of pyocyanin production in Pseudomonas aeruginosa. J. Med. Chem. 2015, 58, 1298–1306. [Google Scholar] [CrossRef]
- Kumar, L.; Patel, S.K.S.; Kharga, K.; Kumar, R.; Kumar, P.; Pandohee, J.; Kulshresha, S.; Harjai, K.; Chhiber, S. Molecular mechanism and apllications of N-acyl homoserine lactone-mediated quorum sensing in bacteria. Molecules 2022, 27, 7584. [Google Scholar] [CrossRef]
- Kerr, J.R.; Taylor, G.W.; Rutman, A.; Høiby, N.; Cole, P.J.; Wilson, R. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J. Clin. Pathol. 1999, 52, 385–387. [Google Scholar] [CrossRef]
- Ostapska, H.; Le Mauff, F.; Gravelat, F.N.; Snarr, B.D.; Bamford, N.C.; Van Loon, J.C.; McKay, G.; Nguyen, D.; Howell, P.L.; Sheppard, D.C. Co-operative biofilm interactions between Aspergillus fumigatus and Pseudomonas aeruginosa through secreted galactosaminogalactan exopolysaccharide. J. Fungi 2022, 8, 336. [Google Scholar] [CrossRef]
- McKnight, S.L.; Iglewski, B.H.; Pesci, E.C. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 2702–2708. [Google Scholar] [CrossRef] [PubMed]
- Saraswathy, A.; Hallberg, R. Mycelial pellet formation by Penicillium ochrochloron species due to exposure to pyrene. Microbiol Res. 2005, 160, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liao, W.; Chen, S. Study of pellet formation of filamentous fungi Rhizopus oryzae using a multiple logistic regression model. Biotechnol. Bioeng. 2008, 99, 117–128. [Google Scholar] [CrossRef]
- Papagianni, M. Fungal morphology and metabolite production in submerged mycelial processes. Biotechnol. Adv. 2004, 22, 189–259. [Google Scholar] [CrossRef]
- Nair, R.B.; Lennartsson, P.R.; Taherzadeh, M.J. Mycelial pellet formation by edible ascomycete filamentous fungi, Neurospora intermedia. AMB Express. 2016, 6, 31. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, J. The filamentous fungal pellet and forces driving its formation. Crit. Rev. Biotechnol. 2016, 36, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Anyan, M.E.; Amiri, A.; Harvey, C.W.; Tierra, G.; Morales-Soto, N.; Driscoll, C.M.; Alber, M.S.; Shrout, J.D. Type IV pili interactions promote intercellular association and moderate swarming of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2014, 16, 18013–18018. [Google Scholar] [CrossRef]
- Durand, E.; Michel, G.; Voulhoux, R.; Kürner, J.; Bernadac, A.; Filloux, A. XcpX controls biogenesis of the Pseudomonas aeruginosa XcpT-containing pseudopilus. J. Biol. Chem. 2005, 9, 31378–31389. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liang, T.; Zhao, M.; Lv, Y.; Song, Z.; Sheng, T.; Ma, F. A review on mycelial pellets as biological carriers: Wastewater treatment and recovery for resource and energy. Bioresour. Technol. 2022, 355, 127200. [Google Scholar] [CrossRef] [PubMed]
- Veiter, L.; Rajamanickam, V.; Herwig, C. The filamentous fungal pellet-relationship between morphology and productivity. Appl. Microbiol. Biotechnol. 2018, 102, 2997–3006. [Google Scholar] [CrossRef]
- Schleheck, D.; Barraud, N.; Klebensberger, J.; Webb, J.S.; McDougald, D.; Rice, S.A.; Kjelleberg, S. Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PLoS ONE 2009, 4, e5513. [Google Scholar] [CrossRef]
- GBD 2019 Antimicrobial Resistance Collaborators. Global mortality associated with 33 bacterial pathogens in 2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2022, 17, 2221–2248. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juříková, T.; Mácha, H.; Lupjanová, V.; Pluháček, T.; Marešová, H.; Papoušková, B.; Luptáková, D.; Patil, R.H.; Benada, O.; Grulich, M.; et al. The Deciphering of Growth-Dependent Strategies for Quorum-Sensing Networks in Pseudomonas aeruginosa. Microorganisms 2023, 11, 2329. https://doi.org/10.3390/microorganisms11092329
Juříková T, Mácha H, Lupjanová V, Pluháček T, Marešová H, Papoušková B, Luptáková D, Patil RH, Benada O, Grulich M, et al. The Deciphering of Growth-Dependent Strategies for Quorum-Sensing Networks in Pseudomonas aeruginosa. Microorganisms. 2023; 11(9):2329. https://doi.org/10.3390/microorganisms11092329
Chicago/Turabian StyleJuříková, Tereza, Hynek Mácha, Vanda Lupjanová, Tomáš Pluháček, Helena Marešová, Barbora Papoušková, Dominika Luptáková, Rutuja H. Patil, Oldřich Benada, Michal Grulich, and et al. 2023. "The Deciphering of Growth-Dependent Strategies for Quorum-Sensing Networks in Pseudomonas aeruginosa" Microorganisms 11, no. 9: 2329. https://doi.org/10.3390/microorganisms11092329
APA StyleJuříková, T., Mácha, H., Lupjanová, V., Pluháček, T., Marešová, H., Papoušková, B., Luptáková, D., Patil, R. H., Benada, O., Grulich, M., & Palyzová, A. (2023). The Deciphering of Growth-Dependent Strategies for Quorum-Sensing Networks in Pseudomonas aeruginosa. Microorganisms, 11(9), 2329. https://doi.org/10.3390/microorganisms11092329





