Microbiome Diversity of Anaerobic Digesters Is Enhanced by Microaeration and Low Frequency Sound
Abstract
:1. Introduction
2. Materials and Methods
2.1. Digester Description
2.2. Chemical Analysis
2.3. Microbial Population Analysis
2.4. Bioinformatics Analysis
3. Results and Discussion
3.1. Digester Environment and Biogas Production
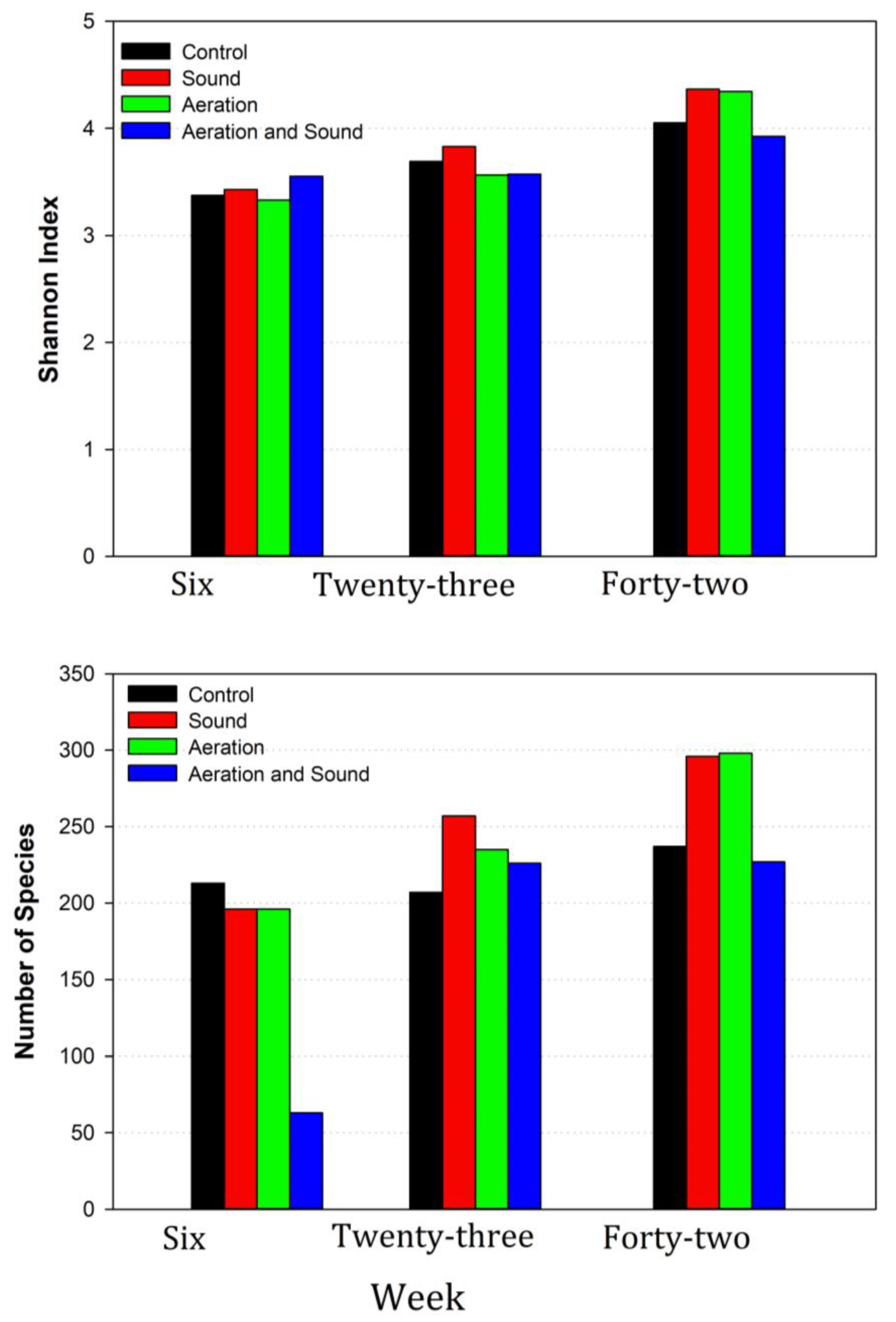
3.2. Description of Sequencing Outputs
3.3. Microbiome of Poultry Litter and Digestate Seed
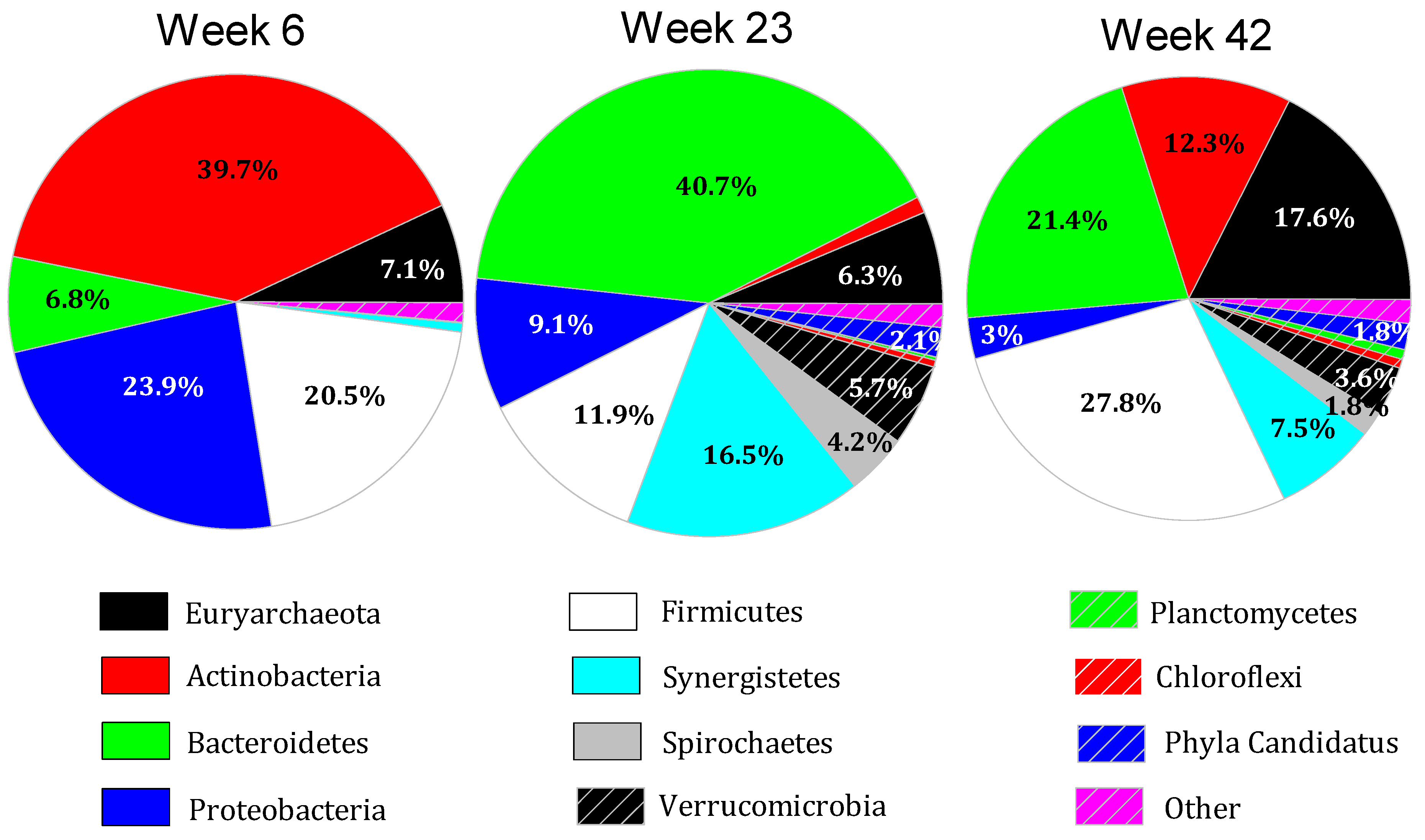
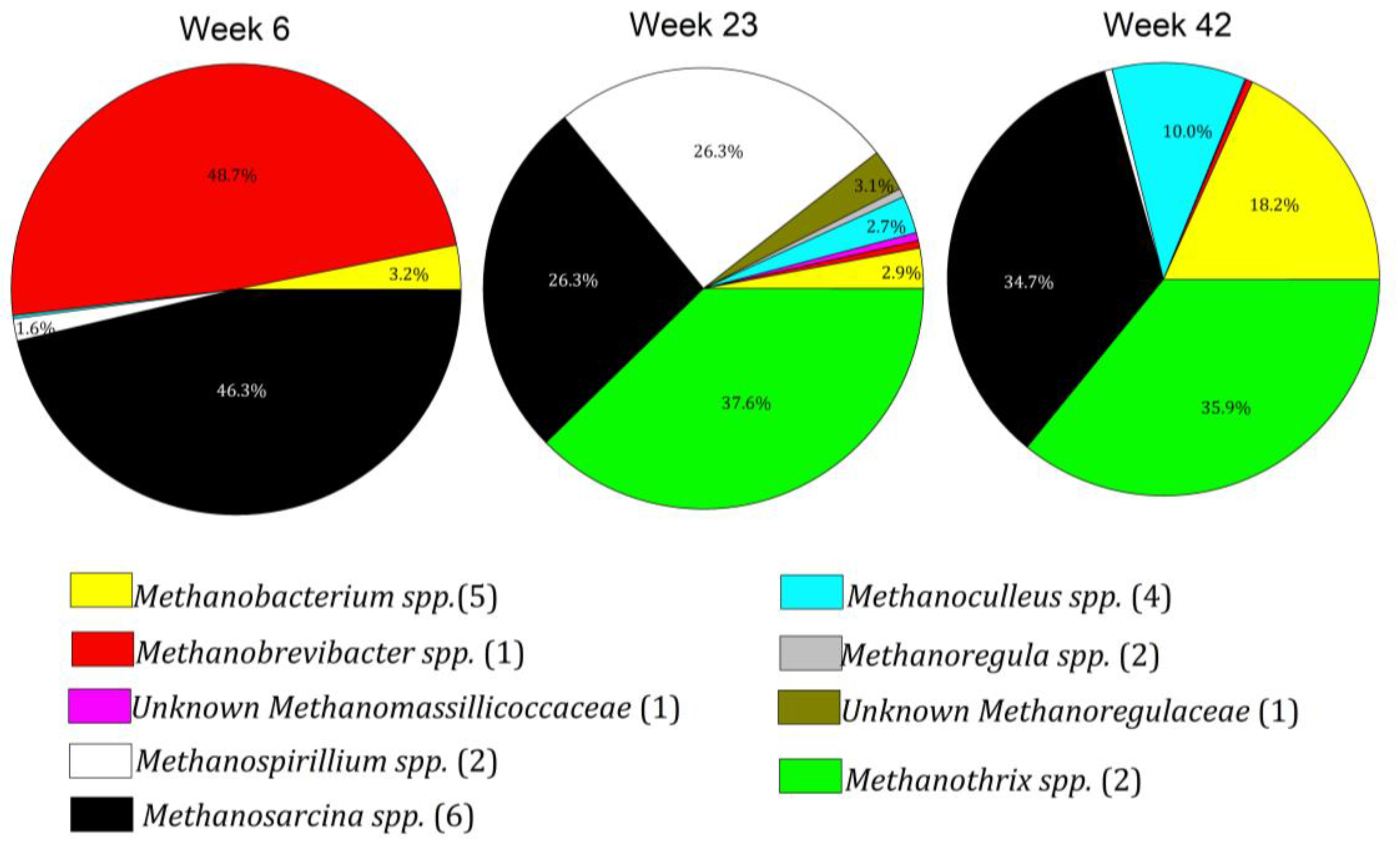
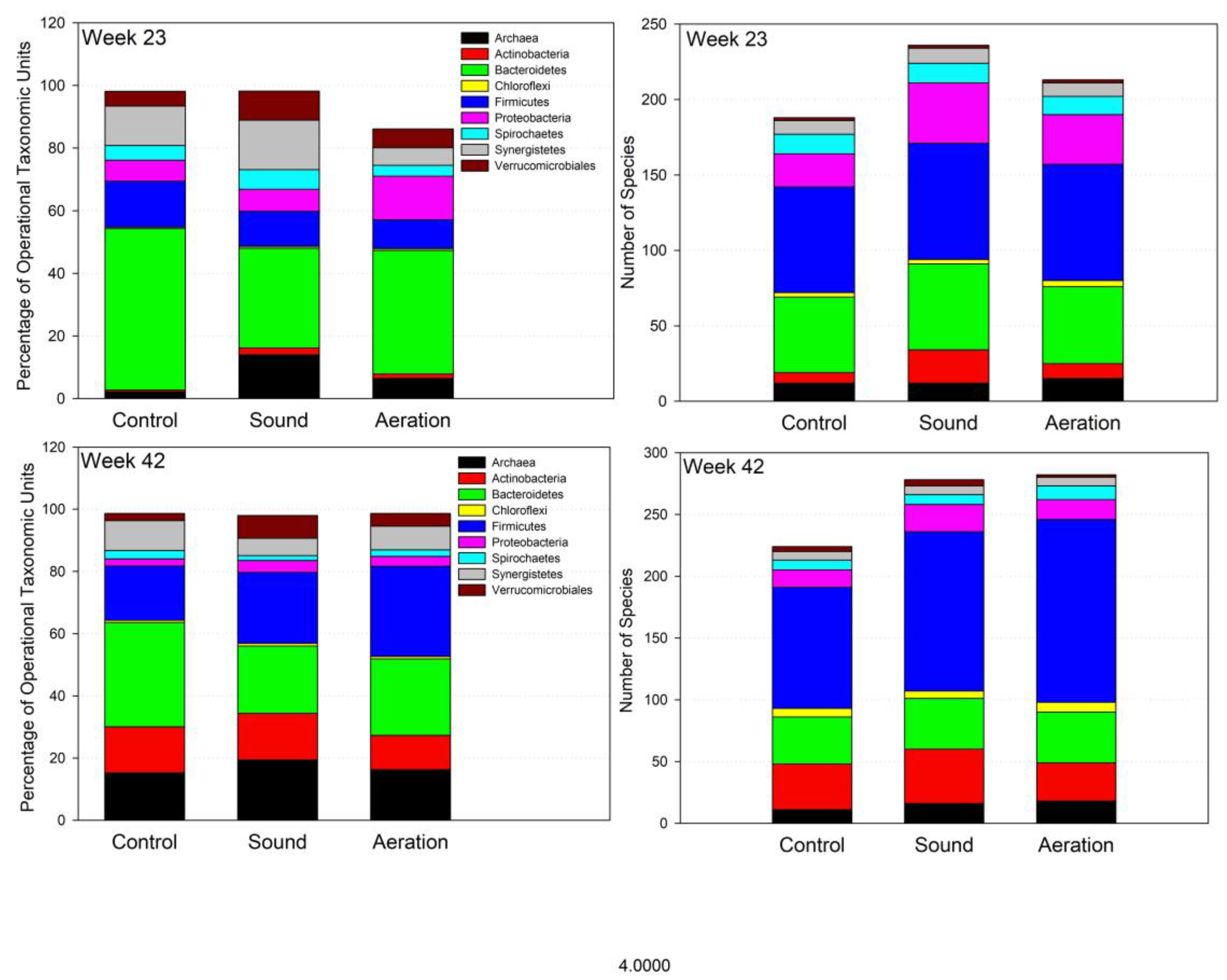
3.4. Microbiome Dynamics over Time
3.5. Interpretation of Results in Reference to Previous Research
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Auer, A.; Vande Burgt, N.H.; Abram, F.; Barry, G.; Fenton, O.; Markey, B.K.; Nolan, S.; Richards, K.; Bolton, D.; De Waal, T.; et al. Agricultural anaerobic digestion power plants in Ireland and Germany: Policy and practice. J. Sci. Food. Agric. 2017, 97, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Malekiha, M.; Isfahani, M.N.; Isfahani, N.M. A Modified Biogas Generator in Cow Farm and Rural Area of Developing Countries. In Proceedings of the Eurasia 2014 Waste Management Symposium, Istanbul, Turkey, 28–30 April 2014; pp. 1–5. Available online: https://www.ctc-n.org/resources/modified-biogas-generator-cow-farm-and-rural-area-developing-countries. (accessed on 24 July 2023).
- Negi, H.; Agrawal, R.; Verma, A.; Goel, R. Municipal solid waste to bioenergy: Current status, opportunities, and challenges in Indian context. In Microbial Biotechnology in Agro-Environmental Sustainability; Singh, S.J., Singh, D.P., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 191–203. [Google Scholar] [CrossRef]
- Klinker, B. Anaerobic digestion as a renewable energy source and waste management technology: What must be done for this technology to realize success in the United States? UMass. Law Rev. 2014, 9, 68–95. Available online: https://scholarship.law.umassd.edu/umlr/vol9/iss1/3 (accessed on 24 July 2023).
- Center, T.J. Governmental oversight of discharges from concentrated animal feeding operations. Environ. Manag. 2006, 37, 745–752. [Google Scholar] [CrossRef]
- Quist, A.J.L.; Helcomb, D.A.; Fliss, M.D.; Delamater, P.L.; Richardson, D.B.; Engel, L.S. Exposure to industrial hog operations and gastrointestinal illness in North Carolina, USA. Sci Total Environ. 2022, 830, 154823. [Google Scholar] [CrossRef] [PubMed]
- Carbella, M.l.; Omil, F.; Lema, J.M. Influence of different pretreatments on anaerobically digested sludge characteristics: Suitability for final disposal. Water Air Soil. Pollut. 2008, 199, 311–321. [Google Scholar] [CrossRef]
- Sridhar, P.; Tyagi, R.D.; Bhunia, P.; Rour, P.R.; Zhang, T.C.; Surampalli, R.Y. Greenhouse gas emissions in sludge ultrasonication followed by anaerobic digestion processes. Bioresour. Technol. 2021, 341, 125754. [Google Scholar] [CrossRef]
- Postawa, K.; Szczygiel, J.; Kulażyński, M. Innovations in anaerobic digestion: A model-based study. Biotechnol. Biofuels. 2021, 14, 19. [Google Scholar] [CrossRef]
- Yungin, L.; Dehan, W.; Shaoquan, W.; Chunmin, W. Alkali pretreatment enhances biogas production in the anaerobic digestion of pulp and paper sludge. J. Hazard. Mat. 2009, 170, 366–373. [Google Scholar] [CrossRef]
- Arreola-Vargas, J.; Ojeda-Castillo, V.; Snell-Castro, R.; Corona-González, R.I.C.; Alatriste-Mondragón, F.; Méndez-Acosta, H.O. 2015. Methane production from acid hydrolysates of Agave tequilana bagasse: Evaluation of hydrolysis conditions and methane yield. Bioresour. Technol. 2015, 181, 191–199. [Google Scholar] [CrossRef]
- Li, W.; Fang, A.; Liu, B.; Xie, G.; Lou, Y.; Xing, D. 2019. Effect of different co-treatments of waste activated sludge on biogas production and shaping microbial community in subsequent anaerobic digestion. Chem. Eng. J. 2019, 379, 122098. [Google Scholar] [CrossRef]
- Xia, Y.; Yang, C.; Zhang, T. Microbial effects of part-stream low-frequency ultrasonic pretreatment on sludge anaerobic digestion as revealed by high-throughput sequencing-based metagenomics and metatranscriptomics. Biotechnol. Biofuels. 2018, 11, 47. [Google Scholar] [CrossRef]
- Yue, L.; Cheng, J.; Tang, S.; An, X.; Hua, J.; Dong, H.; Zhou, J. Ultrasound and microwave pretreatments promote methane production potential and energy conversion during anaerobic digestion of lipid and food wastes. Energy 2021, 228, 120525. [Google Scholar] [CrossRef]
- Yu, L.; Chen, Z.-X.; Tong, X.; Li, K.; Li, W.-W. Anaerobic degradation of microcrystalline cellulose: Kinetics and micro-scale structure evolution. Chemosphere 2012, 86, 348. [Google Scholar] [CrossRef] [PubMed]
- Ahring, B.K.; Biswas, R.; Ahamed, A.; Teller, P.J.; Uellendahl, H. Making lignin accessible for anaerobic digestion by wet-explosion pretreatment. Bioresour. Technol. 2015, 175, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Muller, C.; Guevarra, K.; Summers, A.; Pierce, L.; Shahbaz, P.; Zemke, P.E.; Woodland, K.; Hollingsworth, V.; Nakhia, G.; Bell, K.; et al. A review of the practical application of micro-aeration and oxygenation for hydrogen sulfide management in anaerobic digesters. Process Saf. Environ. Prot. 2022, 165, 126–137. [Google Scholar] [CrossRef]
- Loughrin, J.H.; Lovanh, N.; Antle, S.W.; Bryant, M.D.; Berry, Z.P. Sound enhances wastewater degradation and improves anaerobic digester performance. SN Appl. Sci. 2019, 1, 533. [Google Scholar] [CrossRef]
- Loughrin, J.H.; Antle, S.W.; Sistani, K.R.; Lovanh, N. In situ acoustic treatment of anaerobic digesters to improve biogas yields. Environments 2020, 7, 11. [Google Scholar] [CrossRef]
- Loughrin, J.; Antle, S.; Bryant, M.; Berry, Z.; Lovanh, N. Evaluation of microaeration and sound to increase biogas production from poultry litter. Environments 2020, 7, 62. [Google Scholar] [CrossRef]
- Loughrin, J.; Lovanh, N. Aeration to improve biogas production by recalcitrant feedstock. Environments 2019, 6, 44. [Google Scholar] [CrossRef]
- Riesz, P.; Berdahl, D.; Christman, C.L. Free radical generation by ultrasound in aqueous and nonaqueous solutions. Environ. Health Perspect. 1985, 64, 233–252. [Google Scholar] [CrossRef]
- Merouani, S.; Hamdaoui, O.; Rezgui, Y.; Guemini, M. Computer simulation of chemical reactions occurring in collapsing acoustical bubble: Dependence of free radicals production on operational conditions. Res. Chem. Intermed. 2015, 41, 881–897. [Google Scholar] [CrossRef]
- Eaton, A.; Clesceri, L.; Greenberg, A.; Franson, M. Standard Methods for the Examination of Water and Wastewater, 20th ed.; American Public Health Association: Washington, DC, USA, 1998. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 114–120. [Google Scholar] [CrossRef]
- Kim, D.; Song, L.; Breitwieser, F.P.; Saizberg, S.L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016, 26, 1721–1729. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Clarke, K.R.; Somerfield, P.J.; Chapman, M.G. On resemblance measures for ecological studies, including taxonomic dissimilarities and a zero-adjusted Bray–Curtis coefficient for denuded assemblages. J. Exp. Marine Biol. Ecol. 2006, 330, 55–80. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Franzosa, E.A.; McIver, L.J.; Rahnavard, G.; Thompson, L.R.; Schirmer, M.; Weingart, G.; Lipson, K.S.; Knight, R.; Caporaso, J.G.; Segata, N.; et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 2018, 15, 962–968. [Google Scholar] [CrossRef]
- Suzek, B.E.; Wang, Y.; Huang, H.; McGarvey, P.B.; Wu, C.H.; UnitProt Consortium. UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformanics 2015, 31, 926–932. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummernacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc database of metabolic pathways and enzymes—A 2019 update. Nucleic Acids Res. 2020, 8, D445–D453. [Google Scholar] [CrossRef]
- Yuan, T.; Cheng, Y.; Wang, X.; Yu, Y.; Zhang, Z.; Lei, Z.; Shimizu, K.; Utsumi, Y.; Lee, D.-J. A novel anaerobic digestion system coupling biogas recirculation with MgCl2 addition for multipurpose sewage sludge treatment. J. Clean. Prod. 2019, 230, 499–507. [Google Scholar] [CrossRef]
- Lewin, G.R.; Carlos, C.; Chevrette, M.G.; Horn, H.A.; McDonald, B.R.; Stankey, R.J.; Fox, B.G.; Currie, C.R. Evolution and ecology of Actinobacteria and their bioenergy applications. Annu. Rev. Microbiol. 2016, 70, 235–254. [Google Scholar] [CrossRef]
- O’Donnell, M.M.; Harris, H.M.B.; Ross, R.P.; O’Tolle, P.W. Core fecal microbiota of domesticated herbivorous ruminant, hindgut fermenters, and monogastric animals. Microbiologyopen 2017, 6, e00509. [Google Scholar] [CrossRef]
- Poulsen, J.S.; de Jonge, N.; Macêdo, W.V.; Dalby, F.R.; Feilberg, A.; Nielsen, J.L. Characterisation of cellulose-degrading organisms in an anaerobic digester. Bioresour. Technol. 2022, 352, 126933. [Google Scholar] [CrossRef]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotecnol. 2015, 33, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Peng, Y.; Ni, B.-J.; Han, X.; Fan, L.; Yuan, Z. Dissecting microbial community structure and methane-producing pathways of a full-scale anaerobic reactor digesting activated sludge from wastewater treatment by metagenomic sequencing. Microb. Cell Factories 2015, 14, 33. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, H.; Zhang, W.; Liu, S.J. Rhodopseudomonas faecalis sp. nov., a phototrophic bacterium isolated from an anaerobic reactor that digests chicken faeces. Int. J. Syst. Evol. Microbiol. 2002, 52, 2055–2060. [Google Scholar] [CrossRef]
- Waite, D.W.; Chuvovhina, M.; Pelikan, C.; Parks, D.H.; Yilmaz, P.; Wagner, M.; Loy, A.; Naganuma, T.; Nakai, R.; Whitman, W.B.; et al. Proposal to reclassify proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int. J. Syst. Evol. Microbiol. 2020, 70, 5972–6016. [Google Scholar] [CrossRef]
- Liu, Y.; Whitman, W.B. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N.Y. Acad. Sci. 2008, 1125, 171–189. [Google Scholar] [CrossRef]
- McKee, L.S.; La Rosa, S.L.; Westereng, B.; Eijsink, V.G.; Pope, P.B.; Larsbrink, J. Polysaccharide degradation by the Bacteroidetes: Mechanisms and nomenclature. Environ. Microbiol. Rep. 2021, 13, 559–581. [Google Scholar] [CrossRef]
- Pudlo, N.A.; Urs, K.; Crawfors, R.; Pirani, A.; Atherly, T.; Jimenez, R.; Terrapon, N.; Henrissat, B.; Peterson, D.; Ziemer, C.; et al. Phenotypic and genomic diversification in complex carbohydrate-degrading human gut bacteria. mSystems 2022, 7, e00947-21. [Google Scholar] [CrossRef]
- Ivanova, N.; Sorokin, A.; Anderson, I.; Galleron, N.; Candelon, B.; Kapatral, V.; Bhattacharyya, A.; Reznik, G.; Mikhailova, N.; Lapidus, A.; et al. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 2003, 423, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Blouzard, J.-C.; Coutinho, P.M.; Fierobe, H.-P.; Henrissat, B.; Lignon, S.; Tardif, C.; Pagés, S.; de Philip, P. Modulation of cellulosome composition in Clostridium cellulolyticum: Adaptation to the polysaccharide environment revealed by proteomic and carbohydrate-active enzyme analyses. Proteomics 2010, 10, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Müller, V. Energy conservation in acetogenic bacteria. Appl. Environ. Microbiol. 2003, 69, 6345–6353. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Park, J.-H.; Kang, H.-J.; Lee, Y.H.; Lee, T.J.; Park, H.-D. Distribution and abundance of Spirochaetes in full-scale anaerobic digesters. Bioresource Technol. 2013, 145, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Tokuda, G.; Mikaelyan, A.; Fukui, C.; Brune, A. Fiber-associated spirochetes are major agents of hemicellulose degradation in the hindgut of wood-feeding higher termites. PNAS 2018, 115, E11996–E12004. [Google Scholar] [CrossRef]
- Ransom-Jones, E.; McCarthy, A.J.; Haldenby, S.; Doonan, J.; McDonald, J.E. Lignocellulose-degrading microbial communities in landfill sites represent a repository of unexplored biomass-degrading diversity. mSphere 2017, 2, e00300-17. [Google Scholar] [CrossRef]
- Montgomery, L.; Flesher, B.; Stahl, D. Transfer of Bacteroides succinogenes (Hungate) to Fibrobacter gen. nov. as Fibrobacter succinogenes comb. nov. and description of Fibrobacter intestinalis sp. nov. Int. J. Syst. Evol. Microbiol. 1988, 38, 430–435. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Shinkai, T.; Koike, S. Ecological and physiological characterization shows that Fibrobacter succinogenes is important in rumen fiber digestion. Folia Microbiol. 2008, 53, 195–200. [Google Scholar] [CrossRef]
- Siegert, M.; Li, X.-F.; Yates, M.D.; Logan, B.E. The presence of hydrogenotrophic methanogens in the inoculum improves methane gas production in microbial electrolysis cells. Front. Microbiol. 2015, 5, 778. [Google Scholar] [CrossRef]
- Yamamto, K.; Tamaki, H.; Cadillo-Quiroz, H.; Imachi, H.; Kyrpides, N.; Woyke, T.; Goodwin, L.; Zinder, S.H.; Kamagata, Y.; Liu, W.-T. Complete genome sequence of Methanoregula formicica SMSPT, a mesophilic hydrogenotrophic methanogen isolated from a methanogenic upflow anaerobic sludge blanket reactor. Genome Announc. 2014, 2, e00870-14. [Google Scholar] [CrossRef]
- Saha, S.; Basak, B.; Hwang, J.-H.; Salama, E.-S.; Chaterjee, P.K.; Jeon, B.-H. Microbial Symbiosis: A Network towards Biomethanation. Trends Microbiol. 2020, 12, 968–984. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, T.J.; Duddleston, K.N.; Buck, C.L. Effects of season and host physiological state on the diversity, density, and activity of the arctic ground squirrel cecal microbiota. Appl. Environ. Microbiol. 2014, 80, 5611–5622. [Google Scholar] [CrossRef] [PubMed]
- Kirchman, D.L. Processes in Microbial Ecology; Oxford University Press: Oxford, UK, 2018; p. 131. [Google Scholar]
- Tartakovsky, B.; Mehta, P.; Bourque, J.-S.; Guiot, S.R. Electrolysis-enhanced anaerobic digestion of wastewater. Bioresour. Technol. 2011, 102, 5685–5691. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wu, W.; Qi, D.; Ding, Y.; Zhao, Z. Review on microaeration-based anaerobic digestion: State of the art, challenges, and prospectives. Sci. Total Environ. 2020, 710, 136388. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.; Khanbal, S.K. A little breath of fresh air into an anaerobic system: How microaeration facilitates anaerobic digestion process. Biotechnol. Adv. 2018, 36, 1971–1983. [Google Scholar] [CrossRef]
- Fu, S.; Lian, S.; Angelidaki, I.; Guo, R. Micro-aeration: An attractive strategy to facilitate anaerobic digestion. Trends Biotechnol. 2023, 41, 714–726. [Google Scholar] [CrossRef]
- Lim, J.W.; Wang, J.-Y. Enhanced hydrolysis and methane yield by applying microaeration pretreatment to the anaerobic co-digestion of brown water and food waste. Waste Manag. 2013, 33, 813–819. [Google Scholar] [CrossRef]
- Guo, B.; Yu, N.; Weissbrodt, D.G.; Liu, Y. Effects of micro-aeration on microbial niches and antimicrobial resistances in blackwater anaerobic digesters. Water Res. 2021, 196, 117035. [Google Scholar] [CrossRef]
- Galagan, J.E.; Nusbaum, C.; Roy, A.; Endrizzi, M.G.; MacDonald, P.; FitzHugh, W.; Calvo, S.; Engels, R.; Smirnov, S.; Atnoor, D.; et al. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 2002, 12, 532–542. [Google Scholar] [CrossRef]
- Sun, L.; Liu, T.; Müller, B.; Schnürer, A. The microbial community structure in industrial biogas plants influences the degradation rate of straw and cellulose in batch tests. Biotechnol. Biofuels 2016, 9, 128. [Google Scholar] [CrossRef]
- Lim, J.W.; Park, T.; Tong, Y.W.; Yu, Z. The microbiome driving anaerobic digestion and microbial analysis. Adv. Bioenergy 2020, 5, 1–61. [Google Scholar] [CrossRef]
- Bao, Y.; Dolfing, J.; Guo, Z.; Chen, R.; Wu, M.; Li, Z.; Lin, X.; Feng, Y. Important ecophysiological roles of nondominant Actinobacteria in plant residue decomposition, especially in less fertile soils. Microbiome 2021, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.-B.; Yang, B.; Wu, Y.; Li, S.-C.; Liu, W.; Duan, X.-F.; Li, M.-W. Growth and physiological characteristics of E. coli in response to the exposure of sound field. Pak. J. Biol. Sci. 2013, 16, 969–975. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Yongzhu, Z.; Wu, Y. Effects of sound exposure on the growth and intracellular macromolecular synthesis of E. coli k-12. PeerJ 2016, 4, e1920. [Google Scholar] [CrossRef] [PubMed]
- Harris, A.; Lindsay, M.A.; Ganley, A.R.D.; Jeffs, A.; Villas-Boas, S.G. Sound stimulation can affect Saccharomyces cerevisiae growth and production of volatile metabolites in liquid medium. Metabolites 2021, 11, 605. [Google Scholar] [CrossRef] [PubMed]
- Everitt, C.T.; Redwood, W.R.; Haydon, D.A. Problem of boundary layers in the exchange diffusion of water across biomolecular lipid membranes. J. Theoret. Biol. 1969, 22, 20–32. [Google Scholar] [CrossRef]
- Brennen, C.E. Cavitation and Bubble Dynamics, 1st ed.; Cambridge University Press: New York, NY, USA, 2014; p. 268. [Google Scholar]
- De Freita, C.F.; Calori, I.R.; Tessaro, A.L.; Caetano, W.; Hioka, N. Rapid formation of Small Unilamellar Vesicles (SUV) through low-frequency sonication: An innovative approach. Colloids Surf. B. 2019, 181, 837–844. [Google Scholar] [CrossRef]
- Deguchi, S.; Tsujii, K.; Horikoshi, K. Cooking cellulose in hot and compressed water. Chem. Commun. 2006, 31, 3293–3295. [Google Scholar] [CrossRef]
- Leschine, S.B. Cellulose degradation in anaerobic environments. Ann. Rev. Microbiol. 1995, 49, 399–426. [Google Scholar] [CrossRef]
- Schwarz, W.H. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 2001, 56, 634–649. [Google Scholar] [CrossRef] [PubMed]
- Redlinger-Pohn, J.S.; Petkovšek, M.K.; Gordeyeva, K.; Zupanc, M.; Gordeeva, A.; Zhang, Q.; Dular, M.; Söderberg, L.D. Cavitation fibrillation of cellulose fiber. Biomacromolecules 2022, 23, 847–862. [Google Scholar] [CrossRef] [PubMed]
| Treatment | ||||
| Control | Sound | Aeration | Sound with Aeration | |
| Week 6 | ||||
| Biogas Characteristics | ||||
| CO2 (µg L−1) | 638,000 | 629,000 | 660,000 | 638,000 |
| CH4 (µg L−1) | 274,000 | 435,000 | 451,000 | 407,000 |
| Biogas Volume (L wk−1) | 50.0 | 44.3 | 49.2 | 53.1 |
| Millimoles CH4 wk−1 | 855 | 1200 | 1380 | 1350 |
| Digestate Characteristics | ||||
| pH | 6.45 | 6.48 | 6.54 | 6.50 |
| HCO3− (mM) | 15.3 | 15.4 | 19.5 | 17.2 |
| NH4+ (mM) | 1.79 | 1.74 | 1.51 | 1.74 |
| SO4− (µM) | 29.5 | 28.0 | 31.6 | 35.1 |
| Chemical oxygen demand (mg L−1) | 1140 | 1290 | 1200 | 1160 |
| Total suspended solids (mg L−1) | 59 | 50 | 61 | 50 |
| Week 23 | ||||
| Biogas Characteristics | ||||
| CO2 (µg L−1) | 594,000 | 629,000 | 660,000 | 638,000 |
| CH4 (µg L−1) | 409,000 | 415,000 | 435,000 | 441,000 |
| Biogas Volume (L wk−1) | 144 | 166 | 188 | 172 |
| Millimoles CH4 wk−1 | 3600 | 4300 | 5090 | 4730 |
| Digestate Characteristics | ||||
| pH | 7.41 | 7.41 | 7.39 | 7.38 |
| HCO3− (mM) | 87.3 | 84.8 | 84.3 | 83.6 |
| NH4+ (mM) | 2.26 | 2.17 | 3.04 | 2.13 |
| SO4− (µM) | 31.4 | 26.3 | 68.3 | 27.9 |
| Chemical oxygen demand (mg L−1) | 3420 | 3820 | 3130 | 3170 |
| Total suspended solids (mg L−1) | 160 | 568 | 211 | 183 |
| Week 42 | ||||
| Biogas Characteristics | ||||
| CO2 (µg L−1) | 691,000 | 603,000 | 651,000 | 533,000 |
| CH4 (µg L−1) | 472,000 | 460,000 | 467,000 | 427,000 |
| Biogas Volume (L wk−1) | 248 | 286 | 322 | 309 |
| Millimoles CH4 wk−1 | 6950 | 7120 | 5280 | 5660 |
| Digestate Characteristics | ||||
| pH | 7.32 | 7.32 | 7.26 | 7.31 |
| HCO3− (mM) | 108 | 118 | 146 | 130 |
| NH4+ (mM) | 5.52 | 2.17 | 3.05 | 2.13 |
| SO4− (µM) | 95.9 | 156 | 66.3 | 201 |
| Chemical oxygen demand (mg L−1) | 6950 | 7120 | 5280 | 5660 |
| Total suspended solids (mg L−1) | 877 | 1530 | 510 | 737 |
| Control | Sound | Microaeration | Microaeration and Sound | |||||||||
| Week | 6 | 23 | 42 | 6 | 23 | 42 | 6 | 23 | 42 | 6 | 23 | 42 |
| Archaea | ||||||||||||
| Orders | 3 | 4 | 3 | 3 | 4 | 4 | 3 | 3 | 5 | 3 | 4 | 3 |
| Families | 3 | 7 | 5 | 3 | 6 | 6 | 4 | 7 | 8 | 3 | 6 | 5 |
| Genera | 3 | 8 | 6 | 4 | 7 | 7 | 4 | 6 | 6 | 5 | 8 | 9 |
| Species | 4 | 12 | 11 | 7 | 12 | 16 | 10 | 15 | 18 | 5 | 10 | 10 |
| Bacteria | ||||||||||||
| Orders | 37 | 42 | 44 | 37 | 50 | 43 | 32 | 43 | 39 | 18 | 47 | 34 |
| Families | 77 | 70 | 79 | 75 | 94 | 97 | 67 | 77 | 81 | 34 | 80 | 74 |
| Genera | 131 | 107 | 123 | 115 | 136 | 157 | 114 | 120 | 147 | 38 | 117 | 121 |
| Species | 209 | 195 | 226 | 189 | 245 | 280 | 186 | 220 | 280 | 58 | 217 | 217 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loughrin, J.H.; Parekh, R.R.; Agga, G.E.; Silva, P.J.; Sistani, K.R. Microbiome Diversity of Anaerobic Digesters Is Enhanced by Microaeration and Low Frequency Sound. Microorganisms 2023, 11, 2349. https://doi.org/10.3390/microorganisms11092349
Loughrin JH, Parekh RR, Agga GE, Silva PJ, Sistani KR. Microbiome Diversity of Anaerobic Digesters Is Enhanced by Microaeration and Low Frequency Sound. Microorganisms. 2023; 11(9):2349. https://doi.org/10.3390/microorganisms11092349
Chicago/Turabian StyleLoughrin, John H., Rohan R. Parekh, Getahun E. Agga, Philip J. Silva, and Karamat R. Sistani. 2023. "Microbiome Diversity of Anaerobic Digesters Is Enhanced by Microaeration and Low Frequency Sound" Microorganisms 11, no. 9: 2349. https://doi.org/10.3390/microorganisms11092349
APA StyleLoughrin, J. H., Parekh, R. R., Agga, G. E., Silva, P. J., & Sistani, K. R. (2023). Microbiome Diversity of Anaerobic Digesters Is Enhanced by Microaeration and Low Frequency Sound. Microorganisms, 11(9), 2349. https://doi.org/10.3390/microorganisms11092349







