Characterization of Levan Fructan Produced by a Gluconobacter japonicus Strain Isolated from a Sugarcane Processing Facility
Abstract
:1. Introduction
2. Materials and Methods
2.1. G. japonicus LASM12 Levansucrase Sequence Analysis
2.2. Culturing and EPS Isolation
2.3. Bacterial Culturing for Precipitation of Secreted Proteins
2.4. Methods for MS-Based Identification of Secreted EPS-Forming Enzyme
2.5. Glycosyl Composition Analysis
2.6. Glycosyl Linkage Analysis
2.7. Determination of Molecular Weight
2.8. H-NMR
2.9. Fourier Transform Infrared (FTIR) Spectroscopy
3. Results
3.1. Species-Level Identification of Gluconobacter Isolates from a Sugarcane Processing Facility
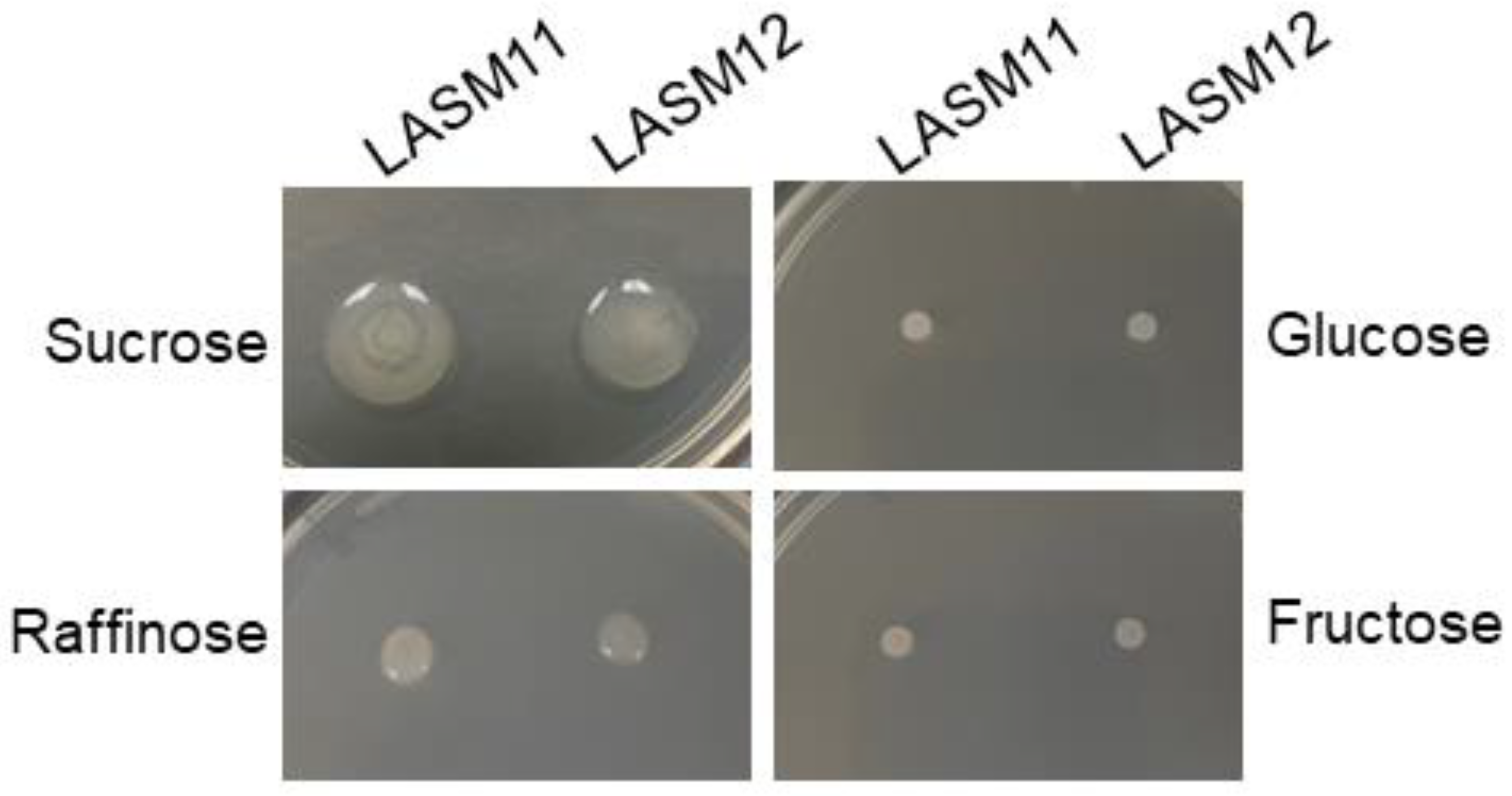
3.2. Analysis of G. japonicus EPS Production on Various Sugars
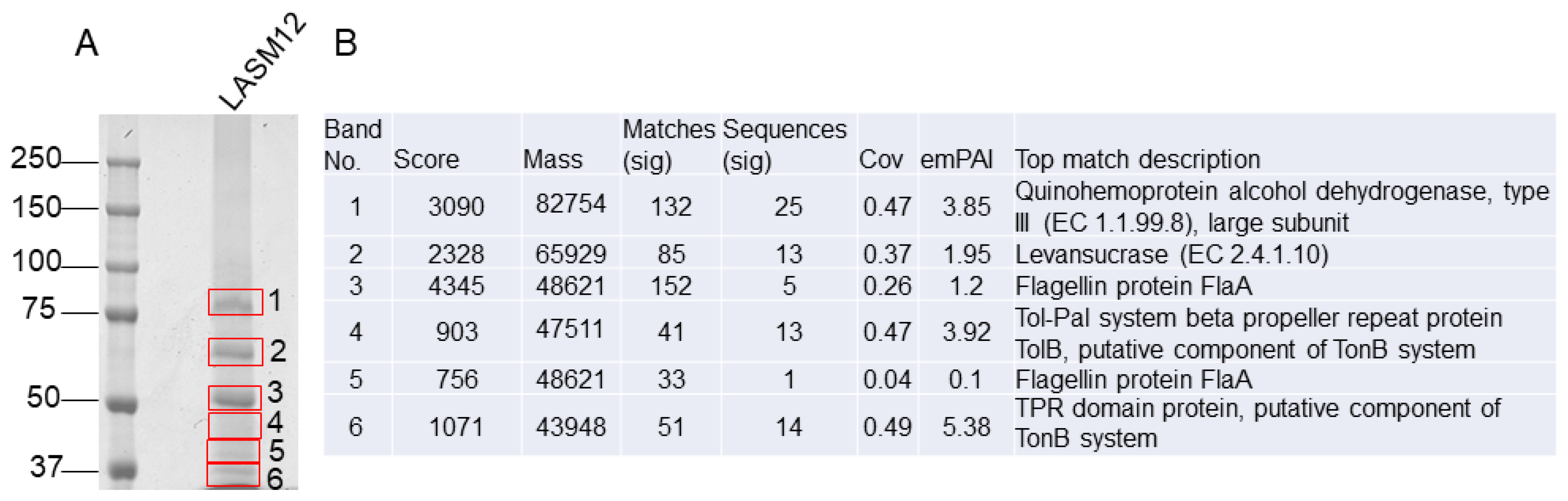
3.3. Identification of Exopolysaccharide-Forming Enzymes
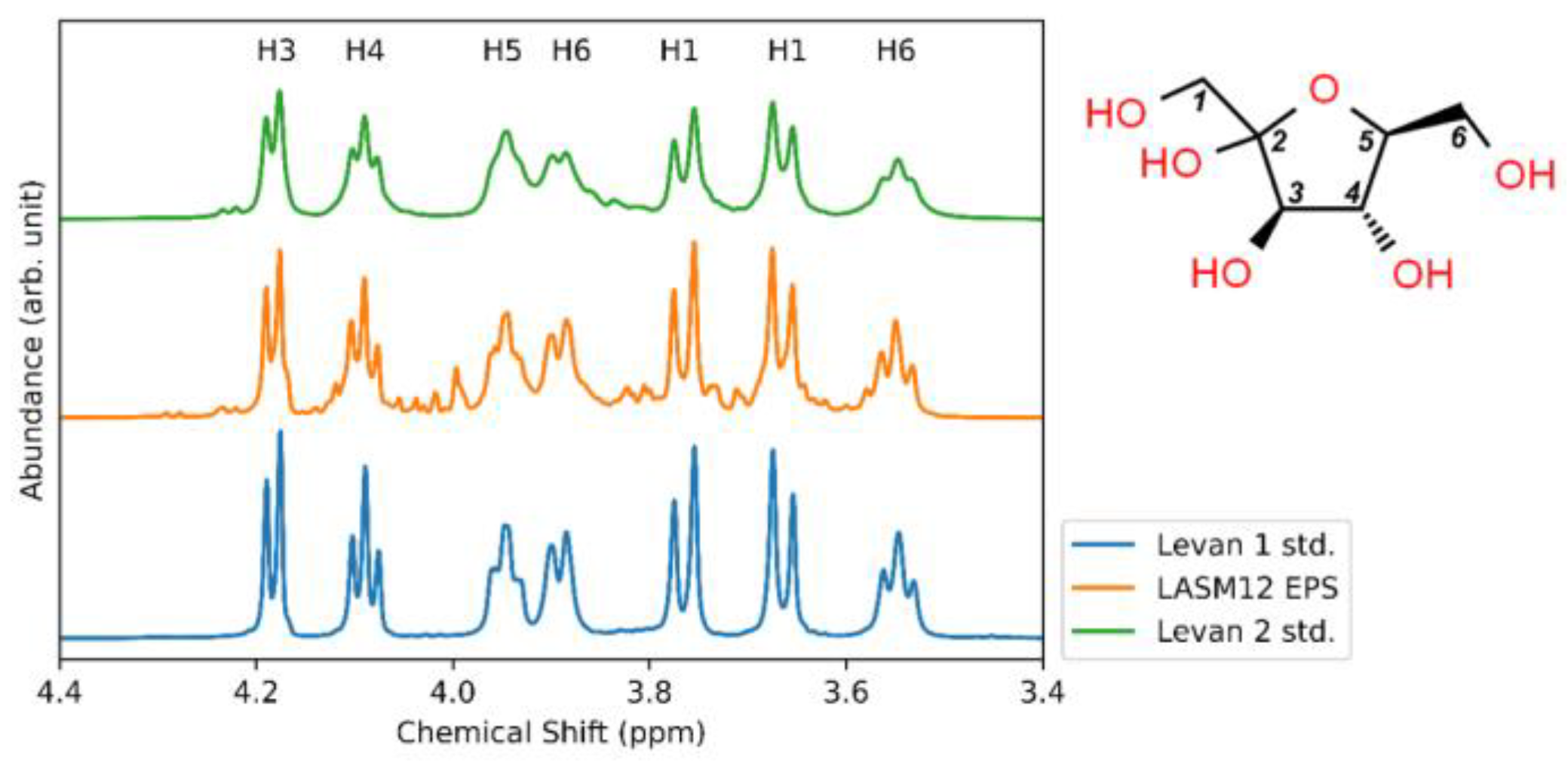
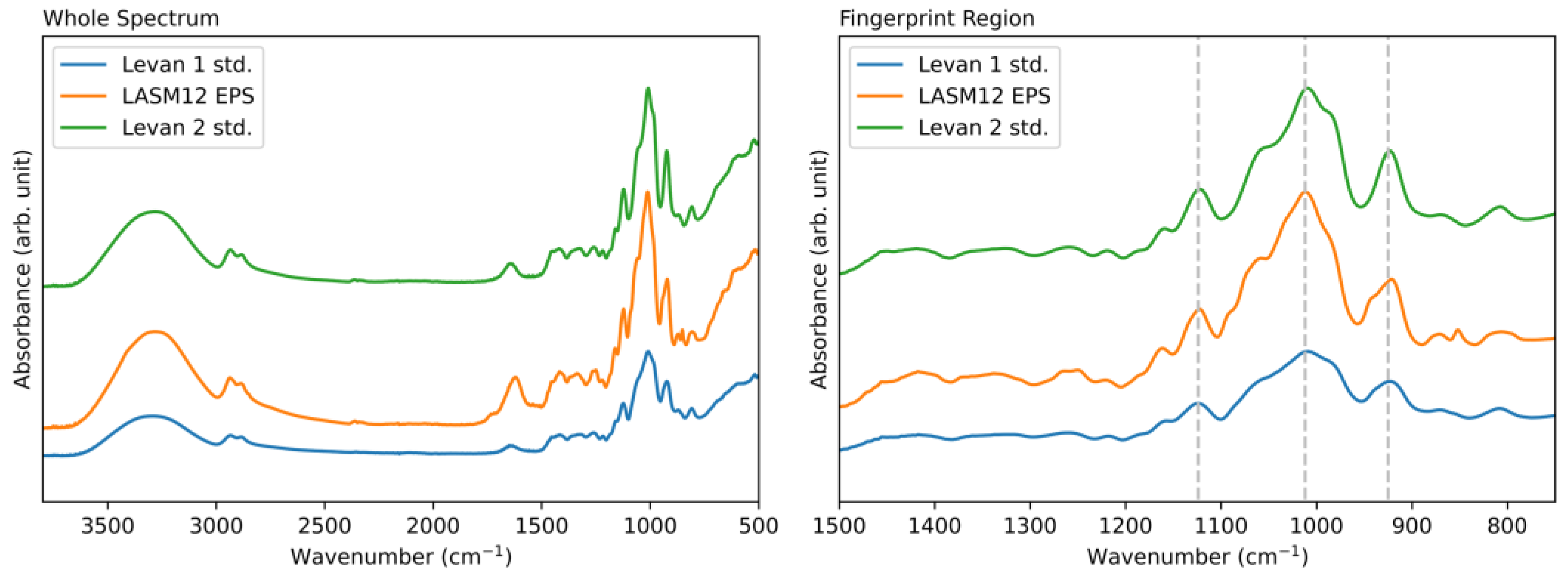
3.4. Structural Analysis of G. japonicus Exopolysaccharide
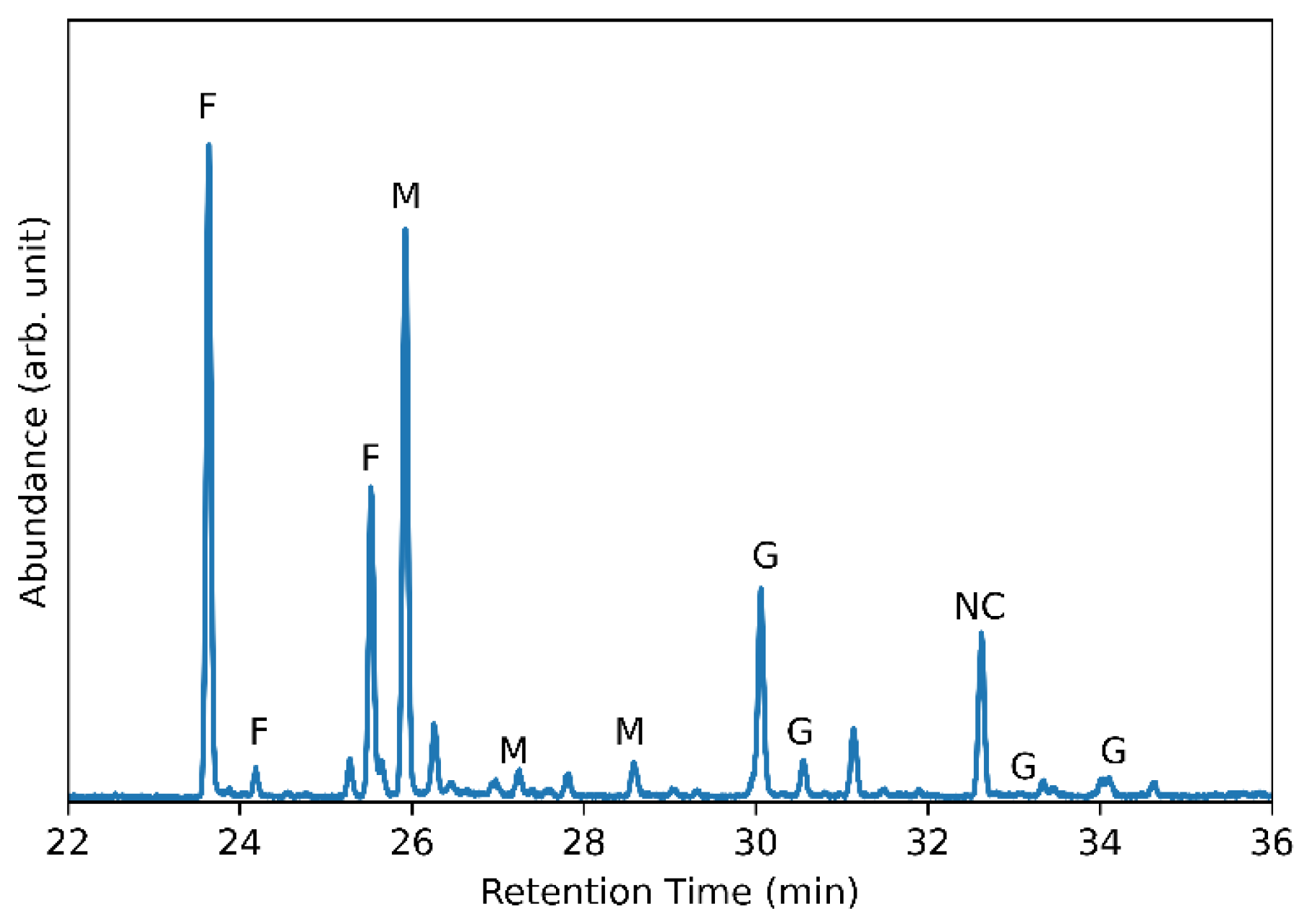
3.5. Glycosyl Monomer and Glycosyl Linkage Analysis of G. japonicus LASM12 Exopolysaccharide
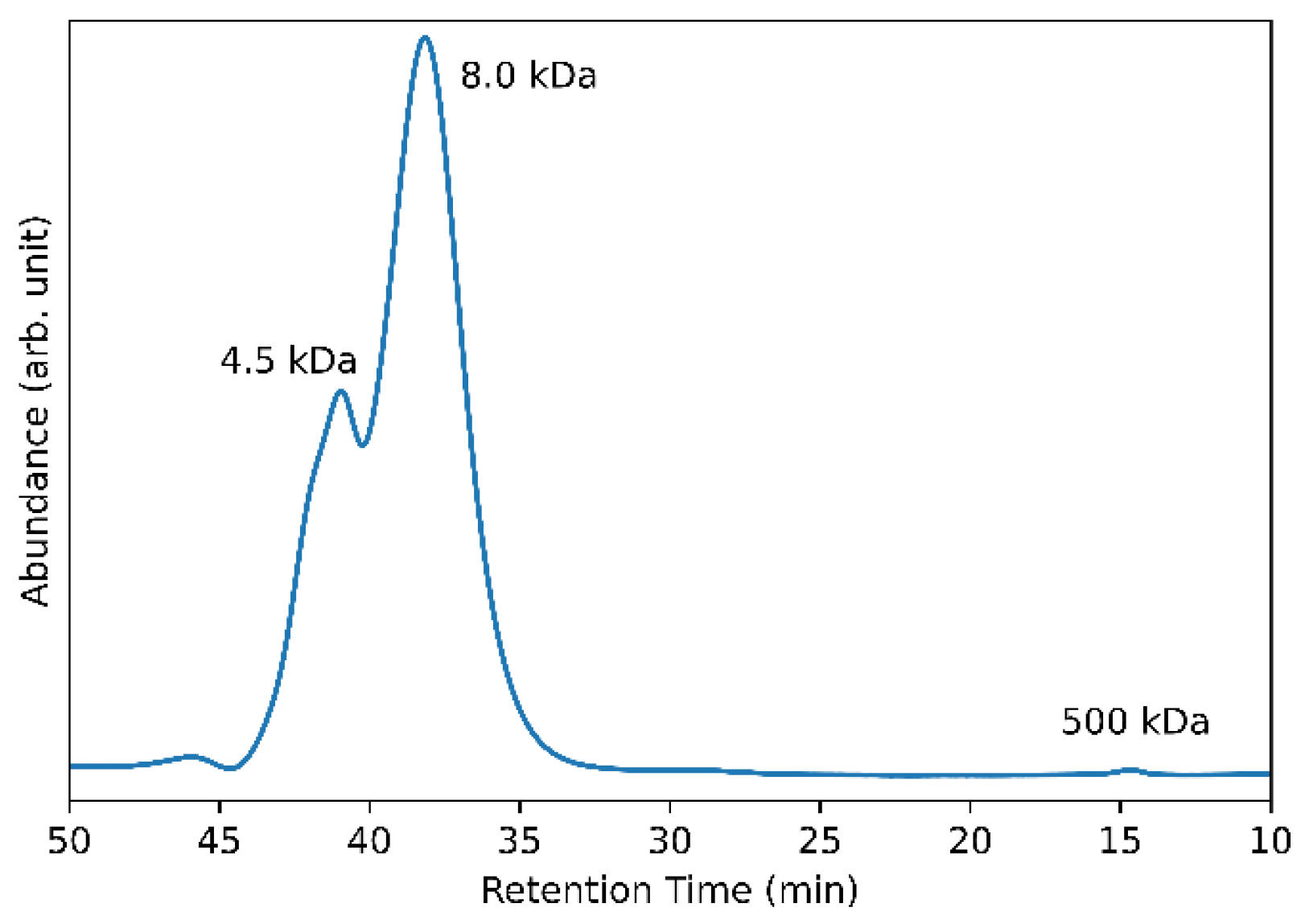
3.6. Determination of Molecular Size of Exopolysaccharide Produced by G. japonicus LASM12
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eggleston, G. Deterioration of cane juice—Sources and indicators. Food Chem. 2002, 78, 95–103. [Google Scholar] [CrossRef]
- Marques, W.L.; Raghavendran, V.; Stambuk, B.U.; Gombert, A.K. Sucrose and Saccharomyces cerevisiae: A relationship most sweet. FEMS Yeast Res. 2016, 16, fov107. [Google Scholar] [CrossRef] [PubMed]
- Bruni, G.O.; Qi, Y.; Klasson, K.T.; Lima, I.M.; Terrell, E. Isolation and analysis of microbial contaminants from Louisiana raw sugarcane factories. Int. Sugar J. 2022, 124, 530–538. [Google Scholar]
- Versluys, M.; Kirtel, O.; Toksoy Öner, E.; Van den Ende, W. The fructan syndrome: Evolutionary aspects and common themes among plants and microbes. Plant Cell Environ. 2018, 41, 16–38. [Google Scholar] [CrossRef] [PubMed]
- Öner, E.T.; Hernández, L.; Combie, J. Review of Levan polysaccharide: From a century of past experiences to future prospects. Biotechnol. Adv. 2016, 34, 827–844. [Google Scholar] [CrossRef] [PubMed]
- Jakob, F.; Gebrande, C.; Bichler, R.M.; Vogel, R.F. Insights into the pH-dependent, extracellular sucrose utilization and concomitant levan formation by Gluconobacter albidus TMW 2.1191. Antonie Van. Leeuwenhoek 2020, 113, 863–873. [Google Scholar] [CrossRef]
- Ragab, T.I.M.; Malek, R.A.; Elsehemy, I.A.; Farag, M.M.S.; Salama, B.M.; Abd El-Baseer, M.A.; Gamal-Eldeen, A.M.; El Enshasy, H.A.; Esawy, M.A. Scaling up of levan yield in Bacillus subtilis M and cytotoxicity study on levan and its derivatives. J. Biosci. Bioeng. 2019, 127, 655–662. [Google Scholar] [CrossRef]
- Ortiz-Soto, M.E.; Porras-Domínguez, J.R.; Seibel, J.; López-Munguía, A. A close look at the structural features and reaction conditions that modulate the synthesis of low and high molecular weight fructans by levansucrases. Carbohydr. Polym. 2019, 219, 130–142. [Google Scholar] [CrossRef]
- Qi, Y.; Bruni, G. Draft genomes of 17 bacterial isolates from Louisiana raw sugarcane factory juices and biofilms. Microbiol. Resour. Announc. 2023, 12, e00416-23. [Google Scholar] [CrossRef]
- Jakob, F.; Meißner, D.; Vogel, R.F. Comparison of novel GH 68 levansucrases of levan-overproducing Gluconobacter species. Acetic Acid. Bact. 2012, 1, e2. [Google Scholar] [CrossRef]
- Qi, Y.; Bruni, G.O.; Klasson, K.T. Microbiome Analysis of Sugarcane Juices and Biofilms from Louisiana Raw Sugar Factories. Microbiol. Spectr. 2023, 11, e04345-22. [Google Scholar] [CrossRef] [PubMed]
- Song, K.B.; Lee, S.K.; Joo, H.K.; Rhee, S.K. Nucleotide and derived amino acid sequences of an extracellular sucrase gene (invB) of Zymomonas mobilis ZM1 (ATCC10988). Biochim. Biophys. Acta 1994, 1219, 163–166. [Google Scholar] [CrossRef] [PubMed]
- Song, K.B.; Joo, H.K.; Rhee, S.K. Nucleotide sequence of levansucrase gene (levU) of Zymomonas mobilis ZM1 (ATCC10988). Biochim. Biophys. Acta 1993, 1173, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, M.; Serizawa, R.; Tanuma, M.; Kikuchi, A.; Sadahiro, J.; Tagami, T.; Lang, W.; Kimura, A. Molecular insight into regioselectivity of transfructosylation catalyzed by GH68 levansucrase and β-fructofuranosidase. J. Biol. Chem. 2021, 296, 100398. [Google Scholar] [CrossRef] [PubMed]
- Triplett, A.; Eggleston, G.; Gaston, P.; Stewart, D. Fructans have been underestimated in the Louisiana sugarcane industry. Int. Sugar J. 2021, 123, 546–554. [Google Scholar]
- Malang, S.K.; Maina, N.H.; Schwab, C.; Tenkanen, M.; Lacroix, C. Characterization of exopolysaccharide and ropy capsular polysaccharide formation by Weissella. Food Microbiol. 2015, 46, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Gay, P.; Le Coq, D.; Steinmetz, M.; Ferrari, E.; Hoch, J.A. Cloning structural gene sacB, which codes for exoenzyme levansucrase of Bacillus subtilis: Expression of the gene in Escherichia coli. J. Bacteriol. 1983, 153, 1424–1431. [Google Scholar] [CrossRef]
- Feldman, M.; Weiss, E.I.; Ofek, I.; Shemesh, M.; Steinberg, D. In vitro real-time interactions of cranberry constituents with immobilized fructosyltransferase. J. Med. Food 2010, 13, 1153–1160. [Google Scholar] [CrossRef]
- Mummaleti, G.; Sarma, C.; Kalakandan, S.; Sivanandham, V.; Rawson, A.; Anandharaj, A. Optimization and extraction of edible microbial polysaccharide from fresh coconut inflorescence sap: An alternative substrate. LWT 2021, 138, 110619. [Google Scholar] [CrossRef]
- Kyono, K.; Yanase, H.; Tonomura, K.; Kawasaki, H.; Sakai, T. Cloning and characterization of Zymomonas mobilis genes encoding extracellular levansucrase and invertase. Biosci. Biotechnol. Biochem. 1995, 59, 289–293. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2021, 50, D571–D577. [Google Scholar] [CrossRef] [PubMed]
- Imbachi-Ordonez, S.; Eggleston, G.; Triplett, A.; Goudeau, S.; Gaston, P. Control of fructans, dextran, and mannitol at the sugarcane factory with commercial biocides. Part I: Juice deterioration studies. Int. Sugar J. 2022, 124, 46–55. [Google Scholar]
- Solomon, S. Post-harvest deterioration of sugarcane. Sugar Tech. 2009, 11, 109–123. [Google Scholar] [CrossRef]
- Combie, J.; Öner, E.T. From healing wounds to resorbable electronics, levan can fill bioadhesive roles in scores of markets. Bioinspiration Biomim. 2019, 14, 011001. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.-X.; Zhang, L.-J.; Xu, R.; Zhang, G.; Zhou, Y.-B.; Han, X.-Q.; Zhang, Y.; Sun, Y.-X. Structural characterization and immunostimulating activity of a levan-type fructan from Curcuma kwangsiensis. Int. J. Biol. Macromol. 2015, 77, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, W.; Nasir, A.; Sattar, F.; Ashfaq, I.; Chen, M.-H.; Hayat, A.; Rehman, M.; Zhao, S.; Khaliq, S.; Ghauri, M.; et al. Production of bimodal molecular weight levan by a Lactobacillus reuteri isolate from fish gut. Folia Microbiol. 2021, 67, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Ni, D.; Xu, W.; Bai, Y.; Zhang, W.; Zhang, T.; Mu, W. Biosynthesis of levan from sucrose using a thermostable levansucrase from Lactobacillus reuteri LTH5448. Int. J. Biol. Macromol. 2018, 113, 29–37. [Google Scholar] [CrossRef]
- Runyon, J.R.; Nilsson, L.; Ulmius, M.; Castro, A.; Ionescu, R.; Andersson, C.; Schmidt, C. Characterizing changes in levan physicochemical properties in different pH environments using asymmetric flow field-flow fractionation Field-Flow Fractionation. Anal. Bioanal. Chem. 2014, 406, 1597–1605. [Google Scholar] [CrossRef]
- Srikanth, R.; Reddy, C.H.S.S.S.; Siddartha, G.; Ramaiah, M.J.; Uppuluri, K.B. Review on production, characterization and applications of microbial levan. Carbohydr. Polym. 2015, 120, 102–114. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Haynes, W.C.; Wickerham, L.J.; Hesseltine, C.W. Maintenance of Cultures of Industrially Important Microorganisms. Appl. Environ. Microbiol. 1955, 3, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass Spectrometric Sequencing of Proteins from Silver-Stained Polyacrylamide Gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Santander, J.; Martin, T.; Loh, A.; Pohlenz, C.; Gatlin, D.M.; Curtiss, R. Mechanisms of intrinsic resistance to antimicrobial peptides of Edwardsiella ictaluri and its influence on fish gut inflammation and virulence. Microbiology 2013, 159, 1471–1486. [Google Scholar] [CrossRef] [PubMed]
- Heiss, C.; Stacey Klutts, J.; Wang, Z.; Doering, T.L.; Azadi, P. The structure of Cryptococcus neoformans galactoxylomannan contains β-d-glucuronic acid. Carbohydr. Res. 2009, 344, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Anumula, K.R.; Taylor, P.B. A comprehensive procedure for preparation of partially methylated alditol acetates from glycoprotein carbohydrates. Anal. Biochem. 1992, 203, 101–108. [Google Scholar] [CrossRef]
- McCormick, C.L.; Callais, P.A.; Hutchinson, B.H. Solution studies of cellulose in lithium chloride and N,N-dimethylacetamide. Macromolecules 1985, 18, 2394–2401. [Google Scholar] [CrossRef]
- Yanagisawa, M.; Shibata, I.; Isogai, A. SEC-MALLS analysis of softwood kraft pulp using LiCl/1,3-dimethyl-2-imidazolidinone as an eluent. Cellulose 2005, 12, 151–158. [Google Scholar] [CrossRef]
- Helmus, J.J.; Jaroniec, C.P. Nmrglue: An open source Python package for the analysis of multidimensional NMR data. J. Biomol. NMR 2013, 55, 355–367. [Google Scholar] [CrossRef]
- Ciufo, S.; Kannan, S.; Sharma, S.; Badretdin, A.; Clark, K.; Turner, S.; Brover, S.; Schoch, C.L.; Kimchi, A.; DiCuccio, M. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018, 68, 2386–2392. [Google Scholar] [CrossRef] [PubMed]
- Federhen, S.; Rossello-Mora, R.; Klenk, H.-P.; Tindall, B.J.; Konstantinidis, K.T.; Whitman, W.B.; Brown, D.; Labeda, D.; Ussery, D.; Garrity, G.M. Meeting report: GenBank microbial genomic taxonomy workshop (12–13 May, 2015). Stand. Genom. Sci. 2016, 11, 15. [Google Scholar] [CrossRef]
- Malik, A.; Radji, M.; Kralj, S.; Dijkhuizen, L. Screening of lactic acid bacteria from Indonesia reveals glucansucrase and fructansucrase genes in two different Weissella confusa strains from soya. FEMS Microbiol. Lett. 2009, 300, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Bagos, P.G.; Nikolaou, E.P.; Liakopoulos, T.D.; Tsirigos, K.D. Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics 2010, 26, 2811–2817. [Google Scholar] [CrossRef] [PubMed]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Armenteros, J.J.A.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv 2022. bioRxiv:2022.2004.2008.487609. [Google Scholar] [CrossRef]
- Hövels, M.; Kosciow, K.; Kniewel, J.; Jakob, F.; Deppenmeier, U. High yield production of levan-type fructans by Gluconobacter japonicus LMG 1417. Int. J. Biol. Macromol. 2020, 164, 295–303. [Google Scholar] [CrossRef]
- Wagh, V.S.; Said, M.S.; Bennale, J.S.; Dastager, S.G. Isolation and structural characterization of exopolysaccharide from marine Bacillus sp. and its optimization by Microbioreactor. Carbohydr. Polym. 2022, 285, 119241. [Google Scholar] [CrossRef]
- Xu, M.; Pan, L.; Zhou, Z.; Han, Y. Structural characterization of levan synthesized by a recombinant levansucrase and its application as yogurt stabilizers. Carbohydr. Polym. 2022, 291, 119519. [Google Scholar] [CrossRef]
- Liu, J.; Luo, J.; Ye, H.; Sun, Y.; Lu, Z.; Zeng, X. Production, characterization and antioxidant activities in vitro of exopolysaccharides from endophytic bacterium Paenibacillus polymyxa EJS-3. Carbohydr. Polym. 2009, 78, 275–281. [Google Scholar] [CrossRef]
- Liu, J.; Luo, J.; Ye, H.; Sun, Y.; Lu, Z.; Zeng, X. Medium optimization and structural characterization of exopolysaccharides from endophytic bacterium Paenibacillus polymyxa EJS-3. Carbohydr. Polym. 2010, 79, 206–213. [Google Scholar] [CrossRef]
- Haddar, A.; Hamed, M.; Bouallegue, A.; Bastos, R.; Coelho, E.; Coimbra, M.A. Structural elucidation and interfacial properties of a levan isolated from Bacillus mojavensis. Food Chem. 2021, 343, 128456. [Google Scholar] [CrossRef] [PubMed]
- Abid, Y.; Azabou, S.; Casillo, A.; Gharsallah, H.; Jemil, N.; Lanzetta, R.; Attia, H.; Corsaro, M.M. Isolation and structural characterization of levan produced by probiotic Bacillus tequilensis-GM from Tunisian fermented goat milk. Int. J. Biol. Macromol. 2019, 133, 786–794. [Google Scholar] [CrossRef] [PubMed]
- González-Garcinuño, Á.; Tabernero, A.; Sánchez-Álvarez, J.M.; Galán, M.A.; Martin Del Valle, E.M. Effect of bacteria type and sucrose concentration on levan yield and its molecular weight. Microb. Cell Fact. 2017, 16, 91. [Google Scholar] [CrossRef] [PubMed]
- Raga-Carbajal, E.; López-Munguía, A.; Alvarez, L.; Olvera, C. Understanding the transfer reaction network behind the non-processive synthesis of low molecular weight levan catalyzed by Bacillus subtilis levansucrase. Sci. Rep. 2018, 8, 15035. [Google Scholar] [CrossRef]
- Ortiz-Soto, M.E.; Rivera, M.; Rudiño-Piñera, E.; Olvera, C.; López-Munguía, A. Selected mutations in Bacillus subtilis levansucrase semi-conserved regions affecting its biochemical properties. Protein Eng. Des. Sel. 2008, 21, 589–595. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bruni, G.O.; Qi, Y.; Terrell, E.; Dupre, R.A.; Mattison, C.P. Characterization of Levan Fructan Produced by a Gluconobacter japonicus Strain Isolated from a Sugarcane Processing Facility. Microorganisms 2024, 12, 107. https://doi.org/10.3390/microorganisms12010107
Bruni GO, Qi Y, Terrell E, Dupre RA, Mattison CP. Characterization of Levan Fructan Produced by a Gluconobacter japonicus Strain Isolated from a Sugarcane Processing Facility. Microorganisms. 2024; 12(1):107. https://doi.org/10.3390/microorganisms12010107
Chicago/Turabian StyleBruni, Gillian O., Yunci Qi, Evan Terrell, Rebecca A. Dupre, and Christopher P. Mattison. 2024. "Characterization of Levan Fructan Produced by a Gluconobacter japonicus Strain Isolated from a Sugarcane Processing Facility" Microorganisms 12, no. 1: 107. https://doi.org/10.3390/microorganisms12010107
APA StyleBruni, G. O., Qi, Y., Terrell, E., Dupre, R. A., & Mattison, C. P. (2024). Characterization of Levan Fructan Produced by a Gluconobacter japonicus Strain Isolated from a Sugarcane Processing Facility. Microorganisms, 12(1), 107. https://doi.org/10.3390/microorganisms12010107





