Phage Lytic Protein CHAPSH3b Encapsulated in Niosomes and Gelatine Films
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical Compounds, Bacterial Strains, and Proteins
2.2. Preparation and Characterization of Niosomes
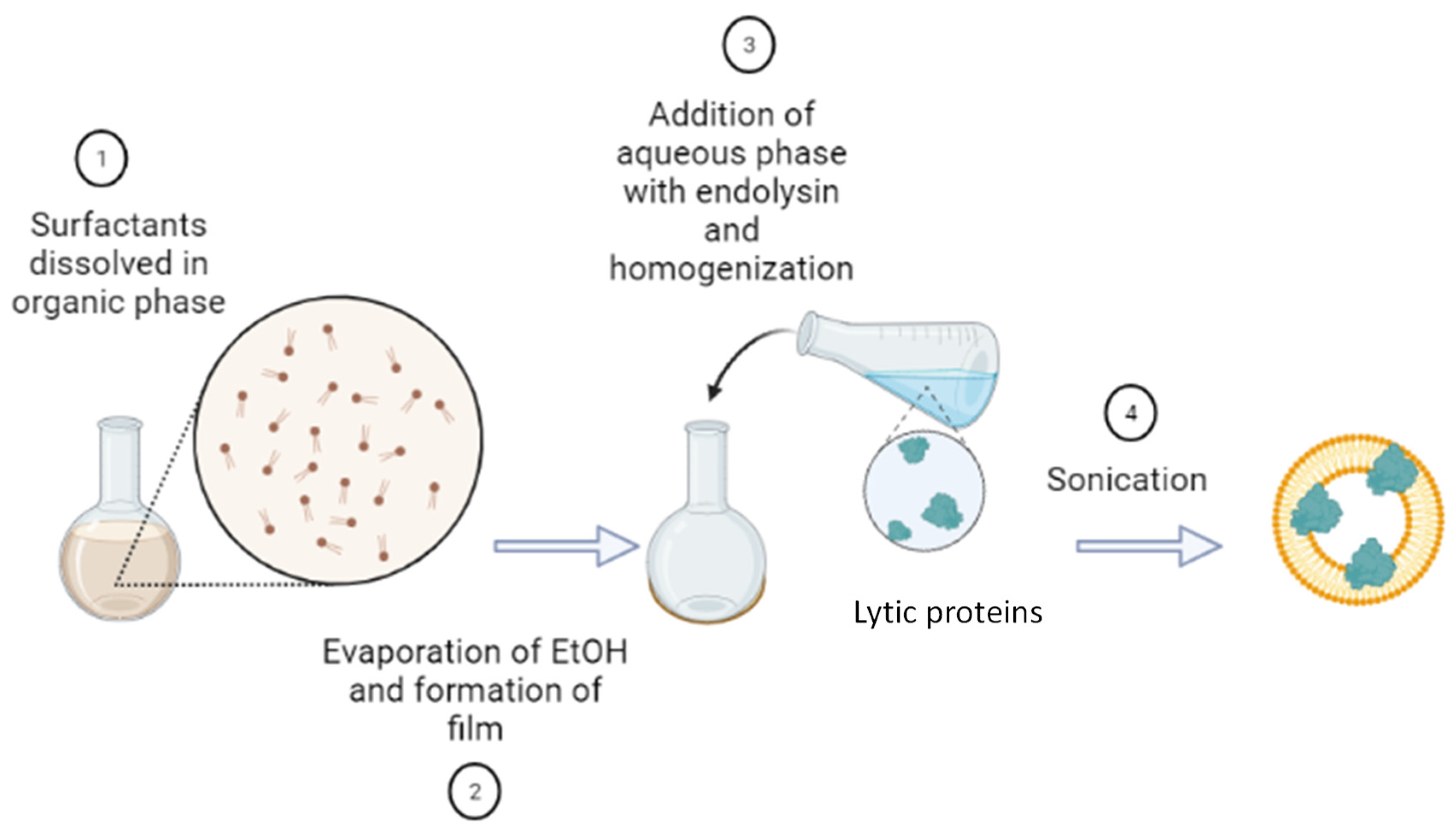
2.2.1. Preparation of Niosomes
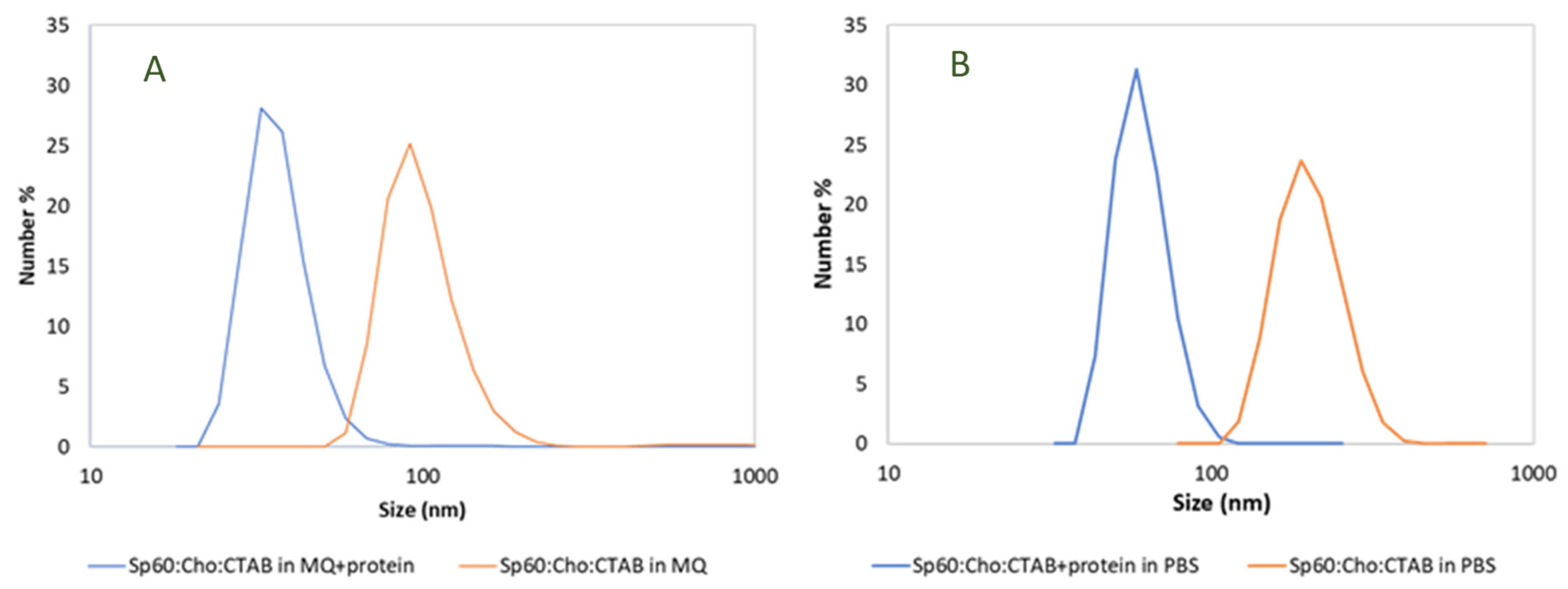
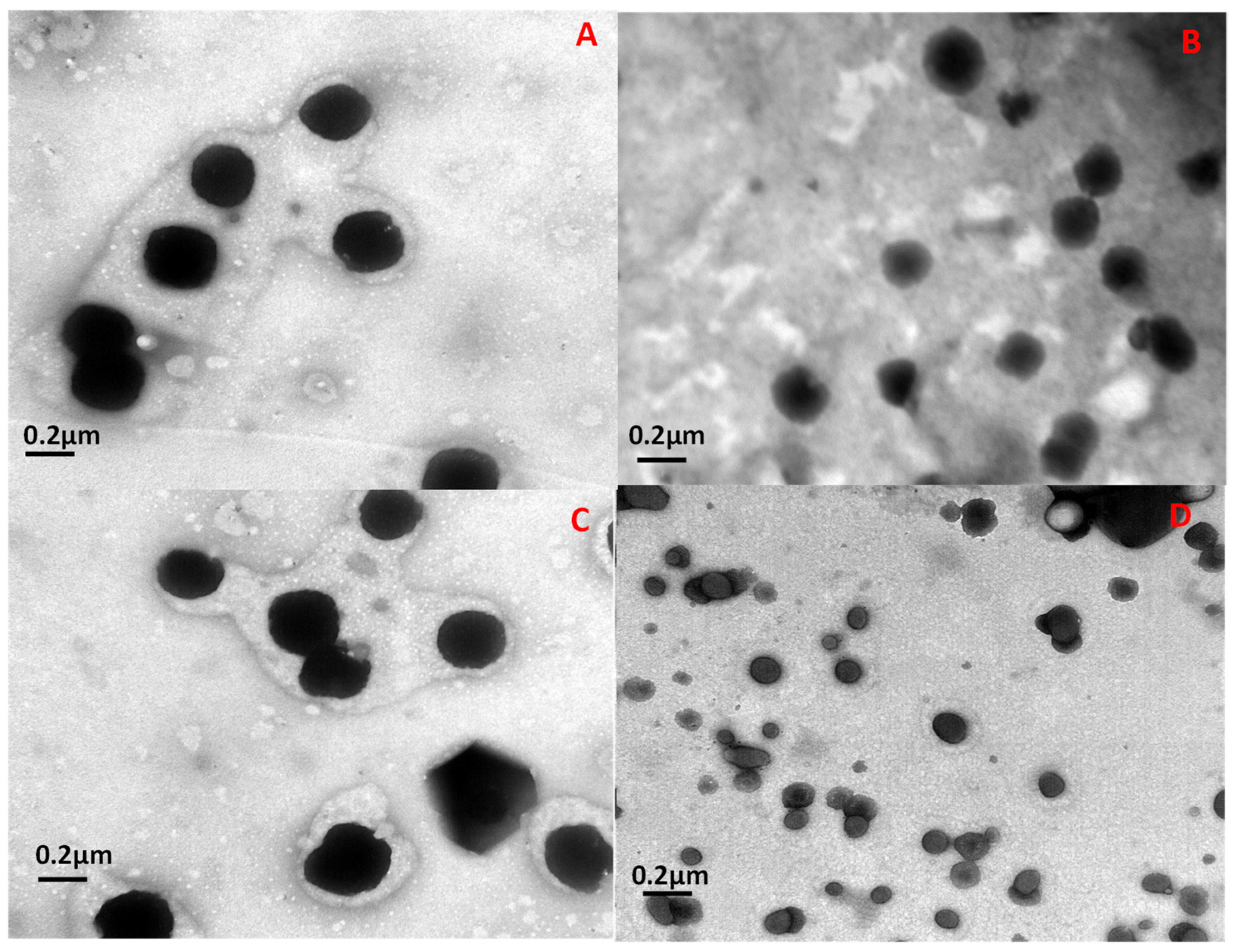
2.2.2. Size and Morphology Characterization
2.2.3. Niosome Purification
2.2.4. Determination of the Protein Encapsulation Efficiency (EE)
2.2.5. Antimicrobial Activity of Niosomes
2.3. Synthesis and Characterization of Gelatine Films
2.3.1. Films Characterization
2.3.2. Films’ Antimicrobial Activity
2.4. Statistical Analysis of Data
3. Results
3.1. Protein Encapsulation and Characterization of the Prepared Niosomes
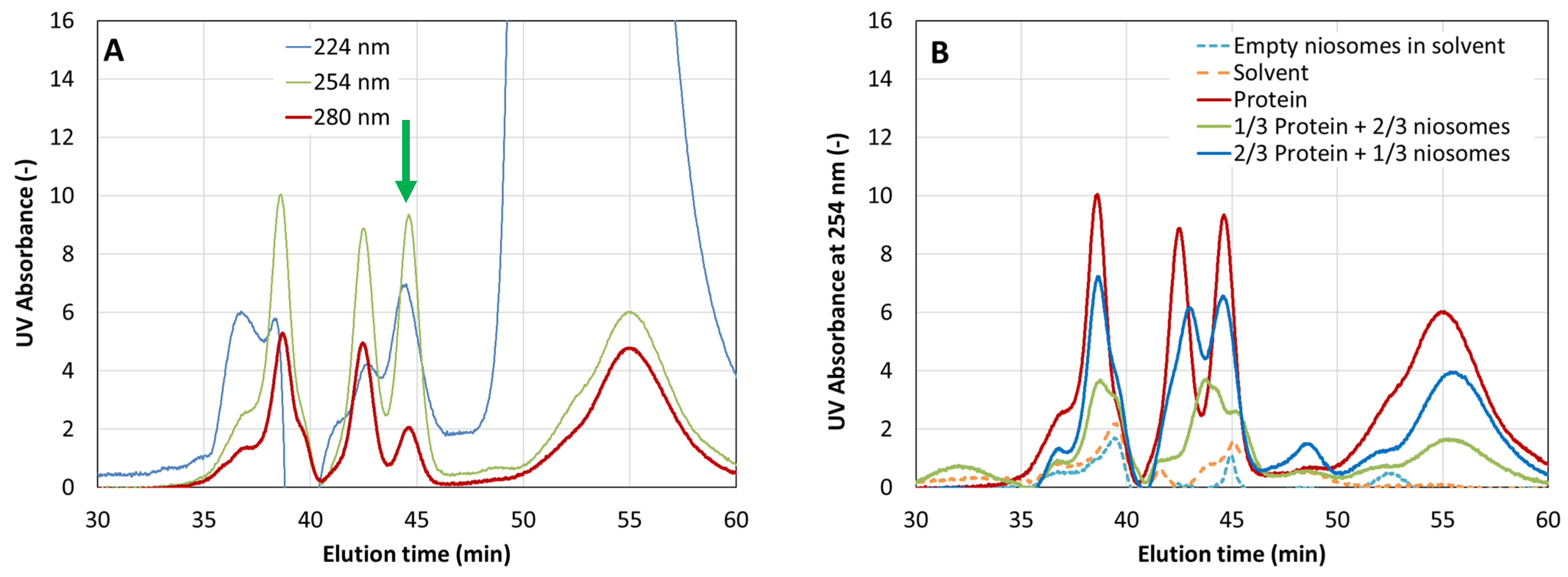
3.2. Protein Encapsulation Efficiency (EE) Determined by Size Exclusion Chromatography
3.3. Antimicrobial Activity of CHAPSH3b-Loaded Niosomes
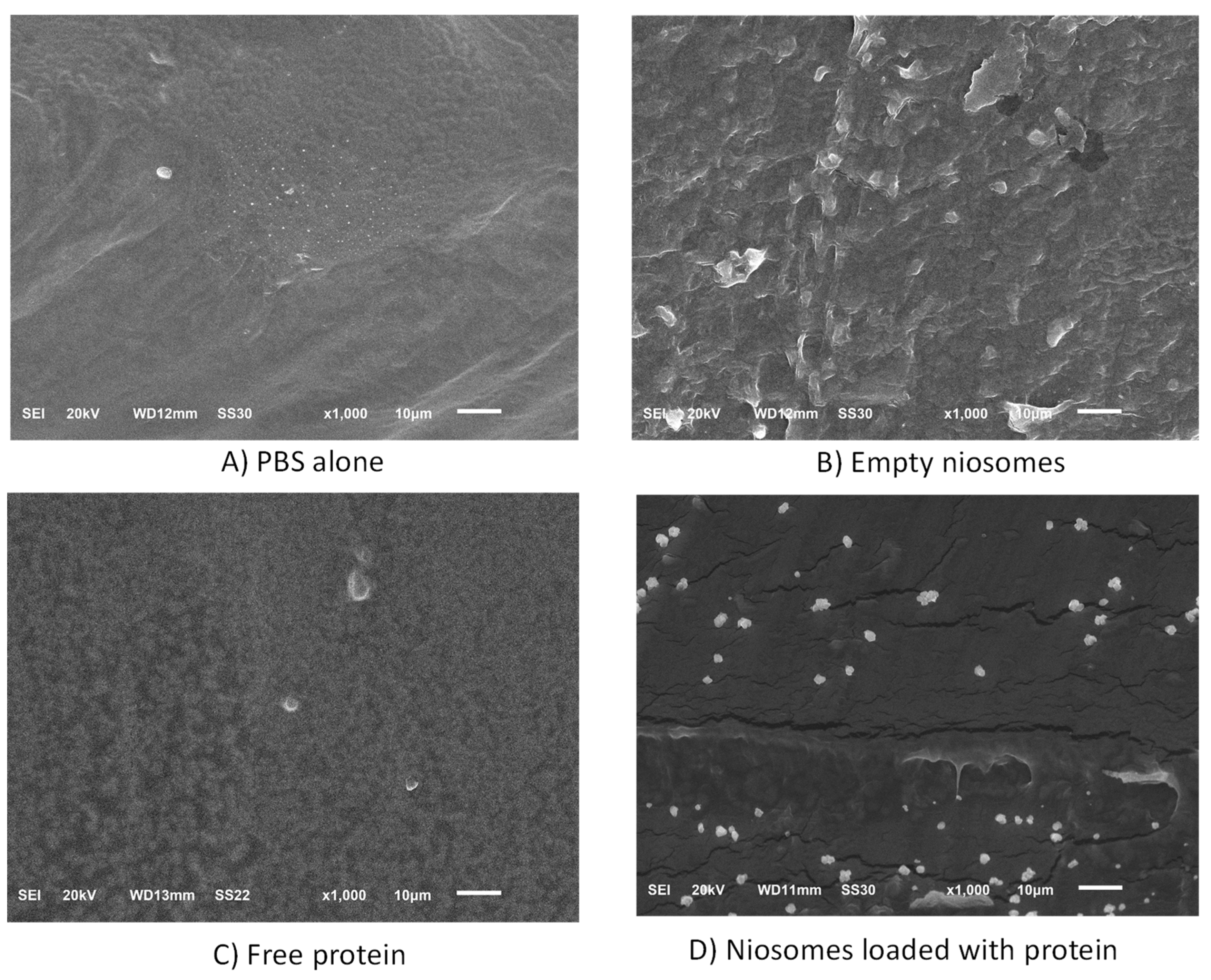
3.4. Characterization of Gelatine Films Containing Encapsulated or Free CHAPSH3b
3.5. Antimicrobial Activity of CHAPSH3b-Containing Gelatine Films
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- WHO. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- European Centre for Disease Prevention and Control. Antimicrobial Resistance Surveillance in Europe; European Centre for Disease Prevention and Control: Solna, Sweden, 2022. [Google Scholar]
- Pinto, R.M.; Lopes-De-Campos, D.; Martins, M.C.L.; Van Dijck, P.; Nunes, C.; Reis, S. Impact of Nanosystems in Staphylococcus Aureus Biofilms Treatment. FEMS Microbiol. Rev. 2019, 43, 622–641. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.W. Biofilms and Antibiotic Therapy: Is There a Role for Combating Bacterial Resistance by the Use of Novel Drug Delivery Systems? Adv. Drug Deliv. Rev. 2005, 57, 1539–1550. [Google Scholar] [CrossRef] [PubMed]
- Hatfull, G.F.; Dedrick, R.M.; Schooley, R.T. Phage Therapy for Antibiotic-Resistant Bacterial Infections. Annu. Rev. Med. 2022, 73, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.C.; Schmelcher, M.; Rodriguez-Rubio, L.; Klumpp, J.; Pritchard, D.G.; Dong, S.; Donovan, D.M. Endolysins as antimicrobials. Adv. Virus Res. 2012, 83, 299–365. [Google Scholar] [PubMed]
- Durr, H.A.; Leipzig, N.D. Advancements in bacteriophage therapies and delivery for bacterial infection. Mater Adv. 2023, 4, 1249–1257. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.M.; Silva, M.D.; Pastrana, L.M.; Bañobre-López, M.; Sillankorva, S. The clinical path to deliver encapsulated phages and lysins. FEMS Microbiol. Rev. 2021, 45, fuab019. [Google Scholar] [CrossRef]
- Forier, K.; Raemdonck, K.; De Smedt, S.C.; Demeester, J.; Coenye, T.; Braeckmans, K. Lipid and Polymer Nanoparticles for Drug Delivery to Bacterial Biofilms. J. Control. Release 2014, 190, 607–623. [Google Scholar]
- Bhardwaj, P.; Tripathi, P.; Gupta, R.; Pandey, S. Niosomes: A review on niosomal research in the last decade. J. Drug Deliv. Sci. Technol. 2020, 56, 101581. [Google Scholar] [CrossRef]
- Moghassemi, S.; Hadjizadeh, A. Nano-niosomes as nanoscale drug delivery systems: An illustrated review. J. Control. Release 2014, 185, 22–36. [Google Scholar] [CrossRef]
- Gondil, V.S.; Chhibber, S. Bacteriophage and Endolysin Encapsulation Systems: A Promising Strategy to Improve Therapeutic Outcomes. Front. Pharmacol. 2021, 12, 675440. [Google Scholar] [CrossRef]
- Bai, J.; Yang, E.; Chang, P.S.; Ryu, S. Preparation and Characterization of Endolysin-Containing Liposomes and Evaluation of Their Antimicrobial Activities against Gram-Negative Bacteria. Enzym. Microb. Technol. 2019, 128, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Quan, K.; Hou, J.; Zhang, Z.; Ren, Y.; Peterson, B.W.; Flemming, H.C.; Mayer, C.; Busscher, H.J.; van der Mei, H.C. Water in bacterial biofilms: Pores and channels, storage and transport functions. Crit. Rev. Microbiol. 2022, 48, 283–302. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Jiang, Y.Z.; Tan, L. Positively-charged microcrystalline cellulose microparticles: Rapid killing effect on bacteria, trapping behavior and excellent elimination efficiency of biofilm matrix from water environment. J. Hazard. Mater. 2022, 424, 127299. [Google Scholar] [CrossRef] [PubMed]
- Marchianò, V.; Matos, M.; López, M.; Weng, S.; Serrano-Pertierra, E.; Luque, S.; Blanco-López, M.C.; Gutiérrez, G. Nanovesicles as Vanillin Carriers for Antimicrobial Applications. Membranes 2023, 13, 95. [Google Scholar] [CrossRef] [PubMed]
- Irkin, R.; Esmer, O.K. Novel Food Packaging Systems with Natural Antimicrobial Agents. J. Food Sci. Technol. 2015, 52, 6095–6111. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; González, S.; Campelo, A.B.; Martínez, B.; Rodríguez, A.; García, P. Downregulation of Autolysin-Encoding Genes by Phage-Derived Lytic Proteins Inhibits Biofilm Formation in Staphylococcus Aureus. Antimicrob. Agents Chemother. 2017, 61, e02724-16. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; Donovan, D.M.; García, P. Enhanced Staphylolytic Activity of the Staphylococcus Aureus Bacteriophage VB_SauS-PhiiPla88 HydH5 Virion-Associated Peptidoglycan Hydrolase: Fusions, Deletions, and Synergy with LysH5. Appl. Environ. Microbiol. 2012, 78, 2241–2248. [Google Scholar] [CrossRef]
- De Maesschalck, V.; Gutiérrez, D.; Paeshuyse, J.; Lavigne, R.; Briers, Y. Advanced Engineering of Third-Generation Lysins and Formulation Strategies for Clinical Applications. Crit. Rev. Microbiol. 2020, 46, 548–564. [Google Scholar] [CrossRef]
- Tyuftin, A.A.; Kerr, J.P. Gelatin films: Study review of barrier properties and implications for future studies employing biopolymer films. Food Packag. Shelf Life 2021, 29, 100688. [Google Scholar] [CrossRef]
- Wang, Y.; Khanal, D.; Alreja, A.B.; Yang, H.; YK Chang, R.; Tai, W.; Li, M.; Nelson, D.C.; Britton, W.J.; Chan, H.K. Bacteriophage Endolysin Powders for Inhaled Delivery against Pulmonary Infections. Int. J. Pharm. 2023, 635, 122679. [Google Scholar] [CrossRef]
- Valle, J.; Toledo-Arana, A.; Berasain, C.; Ghigo, J.-M.; Amorena, B.; Penadés, J.R.; Lasa, I. SarA and Not ΣB Is Essential for Biofilm Development by Staphylococcus Aureus. Mol. Microbiol. 2003, 48, 1075–1087. [Google Scholar] [CrossRef] [PubMed]
- Obeso, J.M.; Martínez, B.; Rodríguez, A.; García, P. Lytic Activity of the Recombinant Staphylococcal Bacteriophage ΦH5 Endolysin Active against Staphylococcus Aureus in Milk. Int. J. Food Microbiol. 2008, 128, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Marchianò, V.; Matos, M.; Marcet, I.; Cabal, M.P.; Gutiérrez, G.; Blanco-López, M.C. Stability of Non-Ionic Surfactant Vesicles Loaded with Rifamycin S. Pharmaceutics 2022, 14, 2626. [Google Scholar] [CrossRef] [PubMed]
- Fathi, H.A.; Yousry, C.; Elsabahy, M.; El-Badry, M.; El Gazayerly, O.N. Effective loading of incompatible drugs into nanosized vesicles: A strategy to allow concurrent administration of furosemide and midazolam in simulated clinical settings. Int. J. Pharm. 2023, 636, 122852. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.C.; Fernández, L.; De Maesschalck, V.; Gutiérrez, D.; Campelo, A.B.; Briers, Y.; Lavigne, R.; Rodríguez, A.; García, P. Synergistic Action of Phage PhiIPLA-RODI and Lytic Protein CHAPSH3b: A Combination Strategy to Target Staphylococcus Aureus Biofilms. Npj Biofilms Microbiomes 2021, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Arana, A.; Merino, N.; Vergara-Irigaray, M.; Débarbouillé, M.; Penadés, J.R.; Lasa, I. Staphylococcus Aureus Develops an Alternative, Ica-Independent Biofilm in the Absence of the ArlRS Two-Component System. J. Bacteriol. 2005, 187, 5318–5329. [Google Scholar] [CrossRef]
- Silva, M.D.; Paris, J.L.; Gama, F.M.; Silva, B.F.B.; Sillankorva, S. Sustained Release of a Streptococcus Pneumoniae Endolysin from Liposomes for Potential Otitis Media Treatment. ACS Infect. Dis. 2021, 7, 2127–2137. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Garrido, V.; Fernández, L.; Portilla, S.; Rodríguez, A.; Grilló, M.J.; García, P. Phage Lytic Protein LysRODI Prevents Staphylococcal Mastitis in Mice. Front. Microbiol. 2020, 11, 7. [Google Scholar] [CrossRef]
- Mal, A.; Bag, S.; Ghosh, S.; Moulik, S.P. Physicochemistry of CTAB-SDS Interacted Catanionic Micelle-Vesicle Forming System: An Extended Exploration. Colloids Surf. A 2018, 553, 633–644. [Google Scholar] [CrossRef]
- Sobral, C.N.C.; Soto, M.A.; Carmona-Ribeiro, A.M. Characterization of DODAB/DPPC Vesicles. Chem. Phys. Lipids 2008, 152, 38–45. [Google Scholar] [CrossRef]
- Qian, Y.; Hu, X.; Wang, J.; Li, Y.; Liu, Y.; Xie, L. Polyzwitterionic Micelles with Antimicrobial-Conjugation for Eradication of Drug-Resistant Bacterial Biofilms. Colloids Surf. B Biointerfaces 2023, 231, 113542. [Google Scholar] [CrossRef] [PubMed]
- Grijalvo, S.; Puras, G.; Zárate, J.; Sainz-Ramos, M.; Qtaish, N.A.L.; López, T.; Mashal, M.; Attia, N.; Díaz Díaz, D.; Pons, R.; et al. Cationic Niosomes as Non-Viral Vehicles for Nucleic Acids: Challenges and Opportunities in Gene Delivery. Pharmaceutics 2019, 11, 50. [Google Scholar] [CrossRef] [PubMed]
- Dehnad, D.; Emadzadeh, B.; Ghorani, B.; Rajabzadeh, G.; Kharazmi, M.S.; Jafari, S.M. Nano-vesicular carriers for bioactive compounds and their applications in food formulations. Crit. Rev. Food Sci. Nutr. 2022, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Melis, V.; Letizia Manca, M.; Bullita, E.; Tamburini, E.; Castangia, I.; Cardia, M.C.; Valenti, D.; Fadda, A.M.; Peris, J.E.; Manconi, M. Inhalable Polymer-Glycerosomes as Safe and Effective Carriers for Rifampicin Delivery to the Lungs. Colloids Surf. B Biointerfaces 2016, 143, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Olii, A.T.; Nugroho, A.K.; Martien, R.; Riyanto, S. Effect of Ratio Span 60—Cholesterol on the Characteristic of Niosomes Vitamin D3. Res. J. Pharm. Technol. 2022, 15, 5551–5554. [Google Scholar] [CrossRef]
- Machado, N.D.; García-Manrique, P.; Fernández, M.A.; Blanco-López, M.C.; Matos, M.; Gutiérrez, G. Cholesterol free niosome production by microfluidics: Comparative with other conventional methods. Chem. Eng. Res. Des. 2020, 162, 162–171. [Google Scholar] [CrossRef]
- Bucci, A.R.; Marcelino, L.; Mendes, R.K.; Etchegaray, A. The Antimicrobial and Antiadhesion Activities of Micellar Solutions of Surfactin, CTAB and CPCl with Terpinen-4-Ol: Applications to Control Oral Pathogens. World J. Microbiol. Biotechnol. 2018, 34, 86. [Google Scholar] [CrossRef]
- Morais, D.; Tanoeiro, L.; Marques, A.T.; Gonçalves, T.; Duarte, A.; Matos, A.P.A.; Vital, J.S.; Cruz, M.E.M.; Carvalheiro, M.C.; Anes, E.; et al. Liposomal Delivery of Newly Identified Prophage Lysins in a Pseudomonas Aeruginosa Model. Int. J. Mol. Sci. 2022, 23, 10143. [Google Scholar] [CrossRef]
- Cui, S.; Qiao, J.; Xiong, M.P. Antibacterial and Biofilm-Eradicating Activities of PH-Responsive Vesicles against Pseudomonas aeruginosa. Mol. Pharm. 2022, 19, 2406–2417. [Google Scholar] [CrossRef]
- Zhao, Y.; Dai, X.; Wei, X.; Yu, Y.; Chen, X.; Zhang, X.; Li, C. Near-Infrared Light-Activated Thermosensitive Liposomes as Efficient Agents for Photothermal and Antibiotic Synergistic Therapy of Bacterial Biofilm. ACS Appl. Mater. Interfaces 2018, 10, 14426–14437. [Google Scholar] [CrossRef]
- Selvamani, V. Stability Studies on Nanomaterials Used in Drugs. Charact. Biol. Nanomater. Drug Deliv. 2019, 425–444. [Google Scholar] [CrossRef]
- Portilla, S.; Fernández, L.; Gutiérrez, D.; Rodríguez, A.; García, P. Encapsulation of the Antistaphylococcal Endolysin Lysrodi in Ph-Sensitive Liposomes. Antibiotics 2020, 9, 242. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Kour, A.; Panda, J.J.; Harjai, K.; Chhibber, S. Exploring Endolysin-Loaded Alginate-Chitosan Nanoparticles as Future Remedy for Staphylococcal Infections. AAPS PharmSciTech 2020, 21, 233. [Google Scholar] [PubMed]
- Vázquez, R.; Caro-León, F.J.; Nakal, A.; Ruiz, S.; Doñoro, C.; García-Fernández, L.; Vázquez-Lasa, B.; San Román, J.; Sanz, J.; García, P.; et al. DEAE-Chitosan Nanoparticles as a Pneumococcus-Biomimetic Material for the Development of Antipneumococcal Therapeutics. Carbohydr. Polym. 2021, 273, 118605. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Ma, Q.; Li, S.; Wang, W.; Ma, Y.; Zhao, H.; Sun, J.; Wang, J. Preparation and Characterization of Biodegradable Composited Films Based on Potato Starch/Glycerol/Gelatin. J. Food Qual. 2021, 2021, 6633711. [Google Scholar] [CrossRef]
- de Dicastillo, C.L.; Settier-Ramírez, L.; Gavara, R.; Hernández-Muñoz, P.; Carballo, G.L. Development of Biodegradable Films Loaded with Phages with Antilisterial Properties. Polymers 2021, 13, 327. [Google Scholar] [CrossRef]
- Weng, S.; López, A.; Sáez-Orviz, S.; Marcet, I.; García, P.; Rendueles, M.; Díaz, M. Effectiveness of Bacteriophages Incorporated in Gelatine Films against Staphylococcus Aureus. Food Control 2021, 121, 107666. [Google Scholar] [CrossRef]
- Kim, S.; Chang, Y. Anti-Salmonella Polyvinyl Alcohol Coating Containing a Virulent Phage PBSE191 and Its Application on Chicken Eggshell. Food Res. Int. 2022, 162, 111971. [Google Scholar] [CrossRef]


| Time (min) | Gradient (Percentage of Solvent B by Volume) |
| 0 | 5 |
| 15 | 90 |
| 17 | 90 |
| 17.1 | 5 |
| 21 | 5 |
| Formulation | Aqueous Phase | Size (nm) | Zeta Potential (mV) |
|---|---|---|---|
| Sp60: Cho: CTAB (no protein) | Milli-Q water | 100 ± 27 | 55 ± 2 |
| Sp60: Cho: CTAB + CHAPSH3b (8 µM) | Milli-Q water | 38 ± 18 | 46 ± 5 |
| Sp60: Cho: CTAB (no protein) | PBS buffer | 205 ± 46 | 28 ± 2 |
| Sp60: Cho: CTAB + CHAPSH3b (8 µM) | PBS buffer | 77 ± 21 | 30 ± 4 |
| Values Correspond to Log (CFU/cm2) | ||||
|---|---|---|---|---|
| Time (h) | PBS Buffer | CHAPSH3b (8 µM) | Empty Niosomes | CHAPSH3b (8 µM) Loaded Niosomes |
| 1 | 8.42 ± 0.06 | 7.85 ± 0.02 | 5.27 ± 0.90 | 3.74 ± 1.71 ** |
| 2 | 8.24 ± 0.13 | 7.28 ± 0.60 | 4.13 ± 3.60 * | 3.90 ± 3.40 ** |
| 4 | 7.62 ± 0.71 | 6.50 ± 0.46 | 1.33 ± 2.30 **** | 0.00 ± 0.00 **** |
| 6 | 8.72 ± 0.58 | 7.32 ± 0.06 | 1.15 ± 1.99 **** | 0.00 ± 0.00 **** |
| 24 | 6.81 ± 0.30 | 6.36 ± 0.68 | 3.22 ± 2.79 * | 3.07 ± 2.66 * |
| Values Correspond to Log (CFU/mL) | ||||
|---|---|---|---|---|
| Incubation Time (h) | PBS | Free CHAPSH3b (8 μM) | CHAPSH3b (8 µM) Loaded Niosomes | Empty Niosomes |
| After preparation | ||||
| 4 | 8.6 ± 0.30 | 5.18 ± 0.17 * | 0.00 ± 0.00 * | 0.00 ± 0.00 * |
| 24 | 9.50 ± 0.17 | 7.43 ± 0.32 * | 0.00 ± 0.00 * | 0.00 ± 0.00 * |
| After 14 days of storage | ||||
| 4 | 9.02 ± 0.12 | 8.57 ± 0.22 | 0.00 ± 0.00 * | 0.00 ± 0.00 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchianò, V.; Duarte, A.C.; Agún, S.; Luque, S.; Marcet, I.; Fernández, L.; Matos, M.; Blanco, M.d.C.; García, P.; Gutiérrez, G. Phage Lytic Protein CHAPSH3b Encapsulated in Niosomes and Gelatine Films. Microorganisms 2024, 12, 119. https://doi.org/10.3390/microorganisms12010119
Marchianò V, Duarte AC, Agún S, Luque S, Marcet I, Fernández L, Matos M, Blanco MdC, García P, Gutiérrez G. Phage Lytic Protein CHAPSH3b Encapsulated in Niosomes and Gelatine Films. Microorganisms. 2024; 12(1):119. https://doi.org/10.3390/microorganisms12010119
Chicago/Turabian StyleMarchianò, Verdiana, Ana Catarina Duarte, Seila Agún, Susana Luque, Ismael Marcet, Lucía Fernández, María Matos, Mª del Carmen Blanco, Pilar García, and Gemma Gutiérrez. 2024. "Phage Lytic Protein CHAPSH3b Encapsulated in Niosomes and Gelatine Films" Microorganisms 12, no. 1: 119. https://doi.org/10.3390/microorganisms12010119








