The Impact and Effects of Host Immunogenetics on Infectious Disease Studies Using Non-Human Primates in Biomedical Research
Abstract
1. Introduction
2. Importance of Host Immunogenetics in Biomedical Research
3. Diversity at Non-Human Primate MHC Loci
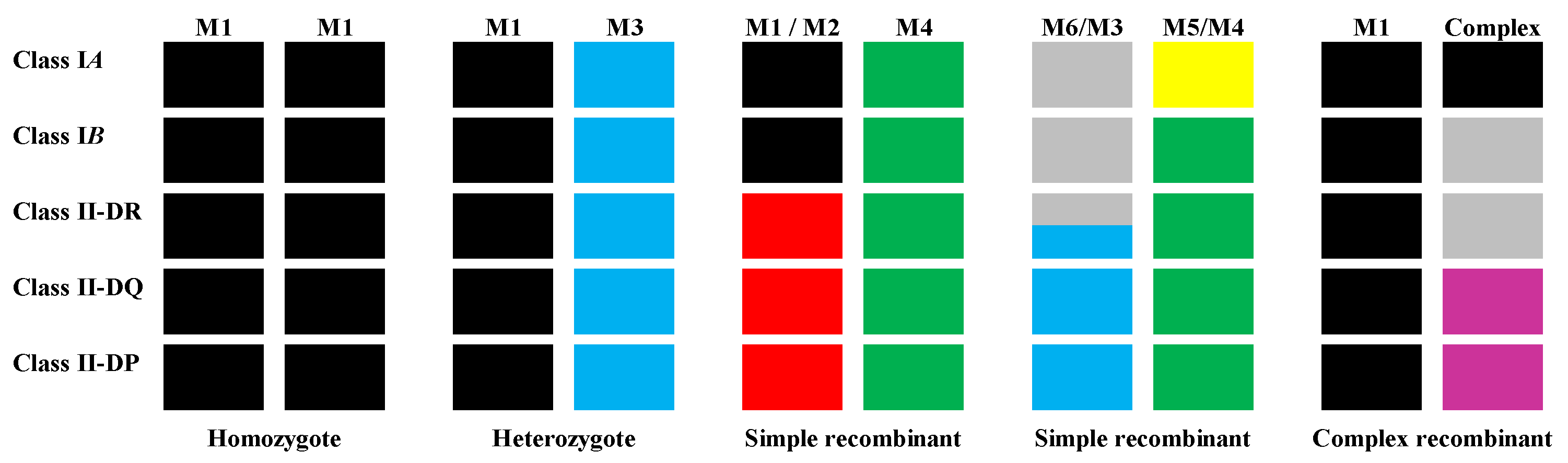
4. Geographic Origins and the Genetic Diversity of Asian Macaques
5. Specific Host MHC Differences in NHPs Used in Biomedical Research
6. New World Hosts
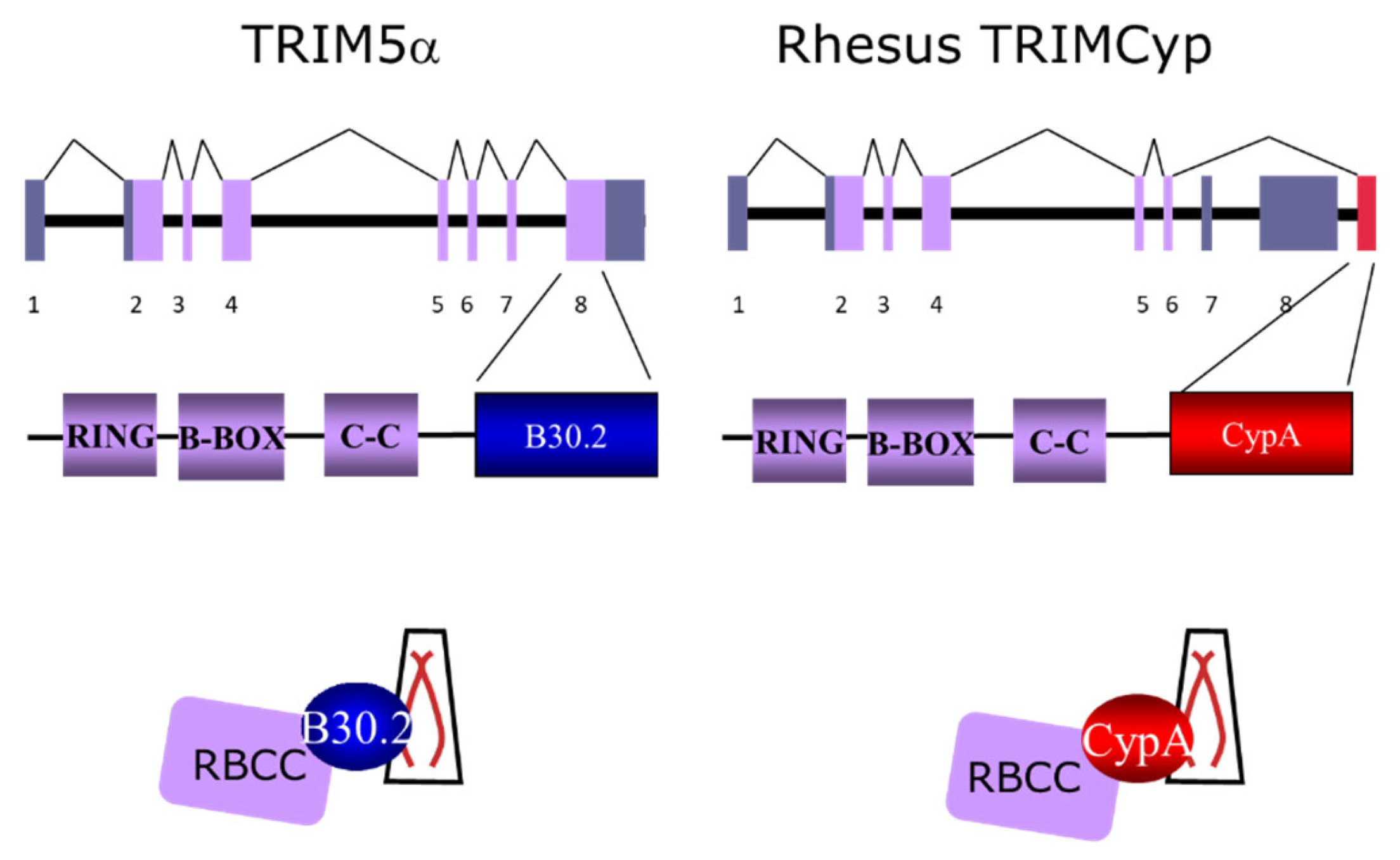
7. The TRIM5 Protein Family and Innate Immunity in NHP Models
7.1. Cyclophilin A and TRIMcyp in Cynolmolgus Macaques
7.2. Impact of TRIM5 Locus Diversity and TRIM5α Expression on Experimental Studies: An SIV Case Study
8. Transcriptomics Analyses of NHPs
9. NHPs as Model Systems for Emerging Viruses
10. Summary and Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Estes, J.; Wong, S.W.; Brenchley, J. Nonhuman primate models of human viral infections. Nat. Rev. Immunol. 2018, 18, 390–404. [Google Scholar] [CrossRef]
- Heijmans, C.M.; de Groot, N.; Bontrop, R.E. Comparative genetics of the major histocompatibility complex in humans and nonhuman primates. Int. J. Immunogenet. 2020, 47, 243–260. [Google Scholar] [CrossRef] [PubMed]
- Hood, S.P.; Mee, E.T.; Perkins, H.; Bowen, O.; Dale, J.M.; Almond, N.M.; Karayiannis, P.; Bright, H.; Berry, N.J.; Rose, N.J. Changes in immune cell populations in the periphery and liver of GBV-B-infected and convalescent tamarins (Saguinus labiatus). Virus Res. 2014, 179, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Marnata, C.; Saulnier, A.; Mompelat, D.; Krey, T.; Cohen, L.; Boukadida, C.; Warter, L.; Fresquet, J.; Vasiliauskaite, I.; Escriou, N.; et al. Determinants Involved in Hepatitis C Virus and GB Virus B Primate Host Restriction. J. Virol. 2015, 89, 12131–12144. [Google Scholar] [CrossRef]
- O’Connor, D.H.; Mothe, B.R.; Weinfurter, J.T.; Fuenger, S.; Rehrauer, W.M.; Jing, P.; Rudersdorf, R.R.; Liebl, M.E.; Krebs, K.; Vasquez, J.; et al. Major histocompatibility complex class I alleles associated with slow simian immunodeficiency virus disease progression bind epitopes recognized by dominant acute-phase cytotoxic-T-lymphocyte responses. J. Virol. 2003, 77, 9029–9040. [Google Scholar] [CrossRef] [PubMed]
- Mothe, B.R.; Weinfurter, J.; Wang, C.; Rehrauer, W.; Wilson, N.; Allen, T.M.; Allison, D.B.; Watkins, D.I. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 2003, 77, 2736–2740. [Google Scholar] [CrossRef]
- Yant, L.J.; Friedrich, T.C.; Johnson, R.C.; May, G.E.; Maness, N.J.; Enz, A.M.; Lifson, J.D.; O’Connor, D.H.; Carrington, M.; Watkins, D.I. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 2006, 80, 5074–5077. [Google Scholar] [CrossRef]
- Giraldo-Vela, J.P.; Rudersdorf, R.; Chung, C.; Qi, Y.; Wallace, L.T.; Bimber, B.; Borchardt, G.J.; Fisk, D.L.; Glidden, C.E.; Loffredo, J.T.; et al. The major histocompatibility complex class II alleles Mamu-DRB1*1003 and -DRB1*0306 are enriched in a cohort of simian immunodeficiency virus-infected rhesus macaque elite controllers. J. Virol. 2008, 82, 859–870. [Google Scholar] [CrossRef]
- Loffredo, J.T.; Friedrich, T.C.; Leon, E.J.; Stephany, J.J.; Rodrigues, D.S.; Spencer, S.P.; Bean, A.T.; Beal, D.R.; Burwitz, B.J.; Rudersdorf, R.A.; et al. CD8 T cells from SIV elite controller macaques recognize Mamu-B*08-bound epitopes and select for widespread viral variation. PLoS ONE 2007, 2, e1152. [Google Scholar] [CrossRef]
- Loffredo, J.T.; Maxwell, J.; Qi, Y.; Glidden, C.E.; Borchardt, G.J.; Soma, T.; Bean, A.T.; Beal, D.R.; Wilson, N.A.; Rehrauer, W.M.; et al. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J. Virol. 2007, 81, 8827–8832. [Google Scholar] [CrossRef]
- Loffredo, J.T.; Bean, A.T.; Beal, D.R.; Leon, E.J.; May, G.E.; Piaskowski, S.M.; Furlott, J.R.; Reed, J.; Musani, S.K.; Rakasz, E.G.; et al. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J. Virol. 2008, 82, 1723–1738. [Google Scholar] [CrossRef] [PubMed]
- Nelson, G.W.; Kaslow, R.; Mann, D.L. Frequency of HLA allele-specific peptide motifs in HIV-1 proteins correlates with the allele’s association with relative rates of disease progression after HIV-1 infection. Proc. Natl. Acad. Sci. USA 1997, 94, 9802–9807. [Google Scholar] [CrossRef] [PubMed]
- McNeil, A.J.; Yap, P.L.; Gore, S.M.; Brettle, R.P.; McColl, M.; Wyld, R.; Davidson, S.; Weightman, R.; Richardson, A.M.; Robertson, J.R. Association of HLA types A1-B8-DR3 and B27 with rapid and slow progression of HIV disease. QJM 1996, 89, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, G.M.; Kaul, R.; Dong, T.; Yang, H.B.; Rostron, T.; Bwayo, J.J.; Kiama, P.; Peto, T.; Plummer, F.A.; McMichael, A.J.; et al. Cross-reactive cytotoxic T lymphocytes against a HIV-1 p24 epitope in slow progressors with B*57. AIDS 2002, 16, 961–972. [Google Scholar] [CrossRef] [PubMed]
- Kaslow, R.A.; Carrington, M.; Apple, R.; Park, L.; Munoz, A.; Saah, A.J.; Goedert, J.J.; Winkler, C.; O’Brien, S.J.; Rinaldo, C.; et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 1996, 2, 405–411. [Google Scholar] [CrossRef]
- Kiepiela, P.; Leslie, A.J.; Honeyborne, I.; Ramduth, D.; Thobakgale, C.; Chetty, S.; Rathnavalu, P.; Moore, C.; Pfafferott, K.J.; Hilton, L.; et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 2004, 432, 769–775. [Google Scholar] [CrossRef] [PubMed]
- Lacap, P.A.; Huntington, J.D.; Luo, M.; Nagelkerke, N.J.; Bielawny, T.; Kimani, J.; Wachihi, C.; Ngugi, E.N.; Plummer, F.A. Associations of human leukocyte antigen DRB with resistance or susceptibility to HIV-1 infection in the Pumwani Sex Worker Cohort. AIDS 2008, 22, 1029–1038. [Google Scholar] [CrossRef]
- Martin, M.P.; Qi, Y.; Gao, X.; Yamada, E.; Martin, J.N.; Pereyra, F.; Colombo, S.; Brown, E.E.; Shupert, W.L.; Phair, J.; et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 2007, 39, 733–740. [Google Scholar] [CrossRef]
- Migueles, S.A.; Sabbaghian, M.S.; Shupert, W.L.; Bettinotti, M.P.; Marincola, F.M.; Martino, L.; Hallahan, C.W.; Selig, S.M.; Schwartz, D.; Sullivan, J.; et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 2000, 97, 2709–2714. [Google Scholar] [CrossRef]
- Ndung’u, T.; Gaseitsiwe, S.; Sepako, E.; Doualla-Bell, F.; Peter, T.; Kim, S.; Thior, I.; Novitsky, V.A.; Essex, M. Major histocompatibility complex class II (HLA-DRB and -DQB) allele frequencies in Botswana: Association with human immunodeficiency virus type 1 infection. Clin. Diagn. Lab. Immunol. 2005, 12, 1020–1028. [Google Scholar] [CrossRef]
- Daza-Vamenta, R.; Glusman, G.; Rowen, L.; Guthrie, B.; Geraghty, D.E. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 2004, 14, 1501–1515. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, S.L.; Blasky, A.J.; Pendley, C.J.; Becker, E.A.; Wiseman, R.W.; Karl, J.A.; Hughes, A.L.; O’Connor, D.H. Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics 2007, 59, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Wiseman, R.W.; Karl, J.A.; Bohn, P.S.; Nimityongskul, F.A.; Starrett, G.J.; O’Connor, D.H. Haplessly hoping: Macaque major histocompatibility complex made easy. ILAR J. 2013, 54, 196–210. [Google Scholar] [CrossRef] [PubMed]
- Sussman, R.; Tattersall, I. Distribution, abundance, and putative ecological strategy of Macaca fascicularis on the island of Mauritius, Southwestern Indian Ocean. Folia Primatol. 1986, 46, 28–43. [Google Scholar] [CrossRef]
- Blancher, A.; Bonhomme, M.; Crouau-Roy, B.; Terao, K.; Kitano, T.; Saitou, N. Mitochondrial DNA sequence phylogeny of 4 populations of the widely distributed cynomolgus macaque (Macaca fascicularis). J. Hered. 2008, 99, 254–264. [Google Scholar] [CrossRef]
- Kanthaswamy, S.; Smith, D.G. Effects of geographic origin on captive Macaca mulatta mitochondrial DNA variation. Comp. Med. 2004, 54, 193–201. [Google Scholar]
- Kawamoto, Y.; Kawamoto, S.; Matsubayashi, K.; Nozawa, K.; Watanabe, T.; Stanley, M.A.; Perwitasari-Farajallah, D. Genetic diversity of longtail macaques (Macaca fascicularis) on the island of Mauritius: An assessment of nuclear and mitochondrial DNA polymorphisms. J. Med. Primatol. 2008, 37, 45–54. [Google Scholar] [CrossRef]
- Lawler, S.H.; Sussman, R.W.; Taylor, L.L. Mitochondrial DNA of the Mauritian macaques (Macaca fascicularis): An example of the founder effect. Am. J. Phys. Anthropol. 1995, 96, 133–141. [Google Scholar] [CrossRef]
- Smith, D.G.; McDonough, J. Mitochondrial DNA variation in Chinese and Indian rhesus macaques (Macaca mulatta). Am. J. Primatol. 2005, 65, 1–25. [Google Scholar] [CrossRef]
- Stevison, L.S.; Kohn, M.H. Determining genetic background in captive stocks of cynomolgus macaques (Macaca fascicularis). J. Med. Primatol. 2008, 37, 311–317. [Google Scholar] [CrossRef]
- Leuchte, N.; Berry, N.; Kohler, B.; Almond, N.; LeGrand, R.; Thorstensson, R.; Titti, F.; Sauermann, U. MhcDRB-sequences from cynomolgus macaques (Macaca fascicularis) of different origin. Tissue Antigens 2004, 63, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Mee, E.T.; Badhan, A.; Karl, J.A.; Wiseman, R.W.; Cutler, K.; Knapp, L.A.; Almond, N.; O’Connor, D.H.; Rose, N.J. MHC haplotype frequencies in a UK breeding colony of Mauritian cynomolgus macaques mirror those found in a distinct population from the same geographic origin. J. Med. Primatol. 2009, 38, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Otting, N.; de Vos-Rouweler, A.J.; Heijmans, C.M.; de Groot, N.G.; Doxiadis, G.G.; Bontrop, R.E. MHC class I A region diversity and polymorphism in macaque species. Immunogenetics 2007, 59, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Pendley, C.J.; Becker, E.A.; Karl, J.A.; Blasky, A.J.; Wiseman, R.W.; Hughes, A.L.; O’Connor, S.L.; O’Connor, D.H. MHC class I characterization of Indonesian cynomolgus macaques. Immunogenetics 2008, 60, 339–351. [Google Scholar] [CrossRef]
- Penedo, M.C.; Bontrop, R.E.; Heijmans, C.M.; Otting, N.; Noort, R.; Rouweler, A.J.; de Groot, N.; de Groot, N.G.; Ward, T.; Doxiadis, G.G.M. Microsatellite typing of the rhesus macaque MHC region. Immunogenetics 2005, 57, 198–209. [Google Scholar] [CrossRef]
- Kondo, M.; Kawamoto, Y.; Nozawa, K.; Matsubayashi, K.; Watanabe, T.; Griffiths, O.; Stanley, M.-A. Population genetics of crab eating macaques (Macaca fascicularis) on the island of Mauritius. Am. J. Primatol. 1993, 29, 167–182. [Google Scholar] [CrossRef]
- Krebs, K.C.; Jin, Z.; Rudersdorf, R.; Hughes, A.L.; O’Connor, D.H. Unusually high frequency MHC class I alleles in Mauritian origin cynomolgus macaques. J. Immunol. 2005, 175, 5230–5239. [Google Scholar] [CrossRef]
- Tosi, A.J.; Coke, C.S. Comparative phylogenetics offer new insights into the biogeographic history of Macaca fascicularis and the origin of the Mauritian macaques. Mol. Phylogenet. Evol. 2007, 42, 498–504. [Google Scholar] [CrossRef]
- Sano, K.; Shiina, T.; Kohara, S.; Yanagiya, K.; Hosomichi, K.; Shimizu, S.; Anzai, T.; Watanabe, A.; Ogasawara, K.; Torii, R.; et al. Novel cynomolgus macaque MHC-DPB1 polymorphisms in three South-East Asian populations. Tissue Antigens 2006, 67, 297–306. [Google Scholar] [CrossRef]
- Shiina, T.; Yamada, Y.; Aarnink, A.; Suzuki, S.; Masuya, A.; Ito, S.; Ido, D.; Yamanaka, H.; Iwatani, C.; Tsuchiya, H.; et al. Discovery of novel MHC-class I alleles and haplotypes in Filipino cynomolgus macaques (Macaca fascicularis) by pyrosequencing and Sanger sequencing: Mafa-class I polymorphism. Immunogenetics 2015, 67, 563–578. [Google Scholar] [CrossRef]
- Doxiadis, G.G.; Rouweler, A.J.; de Groot, N.G.; Louwerse, A.; Otting, N.; Verschoor, E.J.; Bontrop, R.E. Extensive sharing of MHC class II alleles between rhesus and cynomolgus macaques. Immunogenetics 2006, 58, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Reimann, K.A.; Parker, R.A.; Seaman, M.S.; Beaudry, K.; Beddall, M.; Peterson, L.; Williams, K.C.; Veazey, R.S.; Montefiori, D.C.; Mascola, J.R.; et al. Pathogenicity of simian-human immunodeficiency virus SHIV-89.6P and SIVmac is attenuated in cynomolgus macaques and associated with early T-lymphocyte responses. J. Virol. 2005, 79, 8878–8885. [Google Scholar] [CrossRef] [PubMed]
- Berry, N.; Ham, C.; Mee, E.T.; Rose, N.J.; Mattiuzzo, G.; Jenkins, A.; Page, M.; Elsley, W.; Robinson, M.; Smith, D.; et al. Early potent protection against heterologous SIVsmE660 challenge following live attenuated SIV vaccination in Mauritian cynomolgus macaques. PLoS ONE 2011, 6, e23092. [Google Scholar] [CrossRef] [PubMed]
- De Groot, N.G.; Blokhuis, J.H.; Otting, N.; Doxiadis, G.G.; Bontrop, R.E. Co-evolution of the MHC class I and KIR gene families in rhesus macaques: Ancestry and plasticity. Immunol. Rev. 2015, 267, 228–245. [Google Scholar] [CrossRef] [PubMed]
- Ellis-Connell, A.L.; Kannal, N.M.; Balgeman, A.J.; O’Connor, S.L. Characterization of major histocompatibility complex-related molecule 1 sequence variants in non-human primates. Immunogenetics 2019, 71, 109–121. [Google Scholar] [CrossRef]
- Walter, L.; Ansari, A.A. MHC and KIR Polymorphisms in Rhesus Macaque SIV Infection. Front. Immunol. 2015, 6, 540. [Google Scholar] [CrossRef] [PubMed]
- Silver, Z.A.; Watkins, D.I. The role of MHC class I gene products in SIV infection of macaques. Immunogenetics 2017, 69, 511–519. [Google Scholar] [CrossRef]
- Caskey, J.R.; Wiseman, R.W.; Karl, J.A.; Baker, D.A.; Lee, T.; Maddox, R.J.; Raveendran, M.; Harris, R.A.; Hu, J.; Muzny, D.M.; et al. MHC genotyping from rhesus macaque exome sequences. Immunogenetics 2019, 71, 531–544. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, Q.; Jin, Y.; Liu, B.; Zhuo, M.; Ling, F. First identification of the MHC-DPB2 alleles in the rhesus macaques (Macaca mulatta). Immunogenet 2014, 66, 575–580. [Google Scholar] [CrossRef]
- Karl, J.A.; Prall, T.M.; Bussan, H.E.; Varghese, J.M.; Pal, A.; Wiseman, R.W.; O’Connor, D.H. Complete sequencing of a cynomolgus macaque major histocompatibility complex haplotype. Genome Res. 2023, 33, 448–462. [Google Scholar] [CrossRef]
- Naruse, T.K.; Chen, Z.; Yanagida, R.; Yamashita, T.; Saito, Y.; Mori, K.; Akari, H.; Yasutomi, Y.; Miyazawa, M.; Matano, T.; et al. Diversity of MHC class I genes in Burmese-origin rhesus macaques. Immunogenetics 2010, 62, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Karl, J.A.; Wiseman, R.W.; Campbell, K.J.; Blasky, A.J.; Hughes, A.L.; Ferguson, B.; Read, D.S.; O’Connor, D.H. Identification of MHC class I sequences in Chinese-origin rhesus macaques. Immunogenetics 2008, 60, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.F.; Zhou, Y.Y.; Xu, H.L. Comprehensive identification of MHC class II alleles in a cohort of Chinese rhesus macaques. HLA 2018, 92, 188–190. [Google Scholar] [CrossRef]
- Morgan, R.A.; Karl, J.A.; Bussan, H.E.; Heimbruch, K.E.; O’Connor, D.H.; Dudley, D.M. Restricted MHC class I A locus diversity in olive and hybrid olive/yellow baboons from the Southwest National Primate Research Center. Immunogenetics 2018, 70, 449–458. [Google Scholar] [CrossRef]
- Heimbruch, K.E.; Karl, J.A.; Wiseman, R.W.; Dudley, D.M.; Johnson, Z.; Kaur, A.; O’Connor, D.H. Novel MHC class I full-length allele and haplotype characterization in sooty mangabeys. Immunogenetics 2015, 67, 437–445. [Google Scholar] [CrossRef]
- De Groot, N.G.; Otting, N.; Robinson, J.; Blancher, A.; Lafont, B.A.; Marsh, S.G.; O’Connor, D.H.; Shiina, T.; Walter, L.; Watkins, D.I.; et al. Nomenclature report on the major histocompatibility complex genes and alleles of Great Ape, Old and New World monkey species. Immunogenetics 2012, 64, 615–631. [Google Scholar] [CrossRef] [PubMed]
- De Groot, N.G.; Otting, N.; Maccari, G.; Robinson, J.; Hammond, J.A.; Blancher, A.; Lafont, B.A.P.; Guethlein, L.A.; Wroblewski, E.E.; Marsh, S.G.E.; et al. Nomenclature report 2019: Major histocompatibility complex genes and alleles of Great and Small Ape and Old and New World monkey species. Immunogenetics 2020, 72, 25–36. [Google Scholar] [CrossRef]
- McBrien, J.B.; Kumar, N.A.; Silvestri, G. Mechanisms of CD8+ T cell-mediated suppression of HIV/SIV replication. Eur. J. Immunol. 2018, 48, 898–914. [Google Scholar] [CrossRef]
- Lim, S.Y.; Chan, T.; Gelman, R.S.; Whitney, J.B.; O’Brien, K.L.; Barouch, D.H.; Goldstein, D.B.; Haynes, B.F.; Letvin, N.L. Contributions of Mamu-A*01 status and TRIM5 allele expression, but not CCL3L copy number variation, to the control of SIVmac251 replication in Indian-origin rhesus monkeys. PLoS Genet. 2010, 6, e1000997. [Google Scholar] [CrossRef]
- Muhl, T.; Krawczak, M.; Ten Haaft, P.; Hunsmann, G.; Sauermann, U. MHC class I alleles influence set-point viral load and survival time in simian immunodeficiency virus-infected rhesus monkeys. J. Immunol. 2002, 169, 3438–3446. [Google Scholar] [CrossRef]
- Sauermann, U.; Siddiqui, R.; Suh, Y.S.; Platzer, M.; Leuchte, N.; Meyer, H.; Matz-Rensing, K.; Stoiber, H.; Nürnberg, H.; Hunsmann, G.; et al. Mhc class I haplotypes associated with survival time in simian immunodeficiency virus (SIV)-infected rhesus macaques. Genes. Immun. 2008, 9, 69–80. [Google Scholar] [CrossRef]
- Zhang, Z.Q.; Fu, T.M.; Casimiro, D.R.; Davies, M.E.; Liang, X.; Schleif, W.A.; Handt, L.; Tussey, L.; Chen, M.; Tang, A.; et al. Mamu-A*01 allele-mediated attenuation of disease progression in simian-human immunodeficiency virus infection. J. Virol. 2002, 76, 12845–12854. [Google Scholar] [CrossRef]
- Wojcechowskyj, J.A.; Yant, L.J.; Wiseman, R.W.; O’Connor, S.L.; O’Connor, D.H. Control of simian immunodeficiency virus SIVmac239 is not predicted by inheritance of Mamu-B*17-containing haplotypes. J. Virol. 2007, 81, 406–410. [Google Scholar] [CrossRef]
- Harris, M.; Burns, C.M.; Becker, E.A.; Braasch, A.T.; Gostick, E.; Johnson, R.C.; Broman, K.W.; Price, D.A.; Friedrich, T.C.; O’Connor, S.L. Acute-phase CD8 T cell responses that select for escape variants are needed to control live attenuated simian immunodeficiency virus. J. Virol. 2013, 87, 9353–9364. [Google Scholar] [CrossRef] [PubMed]
- Adnan, S.; Colantonio, A.D.; Yu, Y.; Gillis, J.; Wong, F.E.; Becker, E.A.; Piatak, M., Jr.; Reeves, R.K.; Lifson, J.D.; O’Connor, S.L.; et al. CD8 T cell response maturation defined by anentropic specificity and repertoire depth correlates with SIVΔnef-induced protection. PLoS Pathog. 2015, 17, 11. [Google Scholar] [CrossRef] [PubMed]
- Stebbings, R.; Berry, N.; Waldmann, H.; Bird, P.; Hale, G.; Stott, J.; North, D.; Hull, R.; Hall, J.; Lines, J.; et al. CD8+ lymphocytes do not mediate protection against acute superinfection 20 days after vaccination with a live attenuated simian immunodeficiency virus. J. Virol. 2005, 79, 12264–12272. [Google Scholar] [CrossRef] [PubMed]
- Fukazawa, Y.; Park, H.; Cameron, M.J.; Lefebvre, F.; Lum, R.; Coombes, N.; Mahyari, E.; Hagen, S.I.; Bae, J.Y.; Reyes, M.D., 3rd; et al. Lymph node T cell responses predict the efficacy of live attenuated SIV vaccines. Nat. Med. 2012, 18, 1673–1681. [Google Scholar] [CrossRef]
- Jia, B.; Ng, S.K.; DeGottardi, M.Q.; Piatak, M.; Yuste, E.; Carville, A.; Mansfield, K.G.; Li, W.; Richardson, B.A.; Lifson, J.D.; et al. Immunization with single-cycle SIV significantly reduces viral loads after an intravenous challenge with SIV(mac)239. PLoS Pathog. 2009, 5, e1000272. [Google Scholar] [CrossRef]
- Berry, N.; Manoussaka, M.; Ham, C.; Ferguson, D.; Tudor, H.; Mattiuzzo, M.; Klaver, B.; Page, M.; Stebbings, R.; Das, A.T.; et al. Role of Occult and Post-acute Phase Replication in Protective Immunity Induced with a Novel Live Attenuated SIV Vaccine. PLoS Pathog. 2016, 12, e1006083. [Google Scholar] [CrossRef]
- Hansen, S.G.; Marshall, E.E.; Malouli, D.; Ventura, A.B.; Hughes, C.M.; Ainslie, E.; Ford, J.C.; Morrow, D.; Gilbride, R.M.; Bae, J.Y.; et al. A live-attenuated RhCMV/SIV vaccine shows long-term efficacy against heterologous SIV challenge. Sci. Transl. Med. 2019, 11, eaaw2607. [Google Scholar] [CrossRef]
- Hansen, S.G.; Hancock, M.H.; Malouli, D.; Marshall, E.E.; Hughes, C.M.; Randall, K.T.; Morrow, D.; Ford, J.C.; Gilbride, R.M.; Selseth, A.N.; et al. Myeloid cell tropism enables MHC-E-restricted CD8+ T cell priming and vaccine efficacy by the RhCMV/SIV vaccine. Sci. Immunol. 2022, 7, eabn9301. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.L.; Wiseman, R.W.; Hughes, C.M.; Webb, G.M.; Abdulhaqq, S.A.; Bimber, B.N.; Hammond, K.B.; Reed, J.S.; Gao, L.; Burwitz, B.J.; et al. The Role of MHC-E in T Cell Immunity Is Conserved among Humans, Rhesus Macaques, and Cynomolgus Macaques. J. Immunol. 2018, 200, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, H.R.; Bowyer, G.; Brackenridge, S.; Lambe, T. HLA-E: Exploiting pathogen-host interactions for vaccine development. Clin. Exp. Immunol. 2019, 196, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Picker, L.J.; Lifson, J.D.; Gale, M.; Hansen, S.G.; Früh, K. Programming cytomegalovirus as an HIV vaccine. Trends Immunol. 2023, 44, 287–304. [Google Scholar] [CrossRef]
- Campbell, K.J.; Detmer, A.M.; Karl, J.A.; Wiseman, R.W.; Blasky, A.J.; Hughes, A.L.; Bimber, B.N.; O’Connor, S.L.; O’Connor, D.H. Characterization of 47 MHC class I sequences in Filipino cynomolgus macaques. Immunogenetics 2009, 61, 177–187. [Google Scholar] [CrossRef]
- Creager, H.M.; Becker, E.A.; Sandman, K.K.; Karl, J.A.; Lank, S.M.; Bimber, B.N.; Wiseman, R.W.; Hughes, A.L.; O’Connor, S.L.; O’Connor, D.H. Characterization of full-length MHC class II sequences in Indonesian and Vietnamese cynomolgus macaques. Immunogenetics 2011, 63, 611–618. [Google Scholar] [CrossRef]
- Karl, J.A.; Graham, M.E.; Wiseman, R.W.; Heimbruch, K.E.; Gieger, S.M.; Doxiadis, G.G.; Bontrop, R.E.; O’Connor, D.H. Major histocompatibility complex haplotyping and long-amplicon allele discovery in cynomolgus macaques from Chinese breeding facilities. Immunogenetics 2017, 69, 211–229. [Google Scholar] [CrossRef]
- Shortreed, C.G.; Wiseman, R.W.; Karl, J.A.; Bussan, H.E.; Baker, D.A.; Prall, T.M.; Haj, A.K.; Moreno, G.K.; Penedo, M.C.; O’Connor, D.H. Characterization of 100 extended major histocompatibility complex haplotypes in Indonesian cynomolgus macaques. Immunogenetics 2020, 72, 225–239. [Google Scholar] [CrossRef]
- Mee, E.T.; Berry, N.; Ham, C.; Sauermann, U.; Maggiorella, M.T.; Martinon, F.; Verschoor, E.J.; Heeney, J.L.; Grand, R.L.; Titti, F.; et al. Mhc haplotype H6 is associated with sustained control of SIVmac251 infection in Mauritian cynomolgus macaques. Immunogenetics 2009, 1, 327–339. [Google Scholar] [CrossRef]
- Mee, E.T.; Berry, N.; Ham, C.; Aubertin, A.; Lines, J.; Hall, J.; Stebbings, R.; Page, M.; Almond, N.; Rose, N.J. Mhc haplotype M3 is associated with early control of SHIVsbg infection in Mauritian cynomolgus macaques. Tissue Antigens 2010, 76, 223–229. [Google Scholar] [CrossRef]
- O’Connor, S.L.; Lhost, J.J.; Becker, E.A.; Detmer, A.M.; Johnson, R.C.; Macnair, C.E.; Wiseman, R.W.; Karl, J.A.; Greene, J.M.; Burwitz, B.J.; et al. MHC heterozygote advantage in simian immunodeficiency virus-infected Mauritian cynomolgus macaques. Sci. Transl. Med. 2010, 2, 22ra18. [Google Scholar] [CrossRef]
- Burwitz, B.J.; Pendley, C.J.; Greene, J.M.; Detmer, A.M.; Lhost, J.J.; Karl, J.A.; Piaskowski, S.M.; Rudersdorf, R.A.; Wallace, L.T.; Bimber, B.N.; et al. Mauritian cynomolgus macaques share two exceptionally common major histocompatibility complex class I alleles that restrict simian immunodeficiency virus-specific CD8+ T cells. J. Virol. 2009, 83, 6011–6019. [Google Scholar] [CrossRef] [PubMed]
- Wiseman, R.W.; Wojcechowskyj, J.A.; Greene, J.M.; Blasky, A.J.; Gopon, T.; Soma, T.; Friedrich, T.C.; O’Connor, S.L.; O’Connor, D.H. Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J. Virol. 2007, 81, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Budde, M.L.; Wiseman, R.W.; Karl, J.A.; Hanczaruk, B.; Simen, B.B.; O’Connor, D.H. Characterization of Mauritian cynomolgus macaque major histocompatibility complex class I haplotypes by high-resolution pyrosequencing. Immunogenetics 2010, 62, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Schrago, C.G.; Russo, C.A. Timing the Origin of New World Monkeys. Mol. Biol. Evol. 2003, 20, 1620–1625. [Google Scholar] [CrossRef] [PubMed]
- Mee, E.T.; Greenhow, J.; Rose, N.J. Characterisation of Mhc class I and class II DRB polymorphism in red-bellied tamarins (Saguinus labiatus). Immunogenetics 2011, 63, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Van der Wiel, M.; Otting, N.; de Groot, N.; Doxiadis, G.; Bontrop, R. The repertoire of MHC class I genes in the common marmoset: Evidence for functional plasticity. Immunogenetics 2013, 65, 841–849. [Google Scholar] [CrossRef]
- Gyllensten, U.; Bergström, T.; Josefsson, A.; Sundvall, M.; Savage, A.; Blumer, E.S.; Giraldo, L.H.; Soto, L.H.; Watkins, D.I. The cotton-top tamarin revisited: Mhc class I polymorphism of wild tamarins, and polymorphism and allelic diversity of the class II DQA1, DQB1, and DRB loci. Immunogenetics 1994, 40, 167–176. [Google Scholar] [CrossRef]
- Watkins, D.I.; Garber, T.L.; Chen, Z.W.; Toukatly, G.; Hughes, A.L.; Letvin, N.L. Unusually limited nucleotide sequence variation of the expressed major histocompatibility complex class I genes of a New World primate species (Saguinus oedipus). Immunogenetics 1991, 33, 79–89. [Google Scholar] [CrossRef]
- Stremlau, M.; Owens, C.M.; Perron, M.J.; Kiessling, M.; Autissier, P.; Sodroski, J. The cytoplasmic body component TRIM5 alpha restricts HIV-1 infection in Old World monkeys. Nature 2004, 427, 848–853. [Google Scholar] [CrossRef]
- Yap, M.W.; Nisole, S.; Lynch, C.; Stoye, J.P. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 2004, 101, 10786–10791. [Google Scholar] [CrossRef] [PubMed]
- Pertel, T.; Hausmann, S.; Morger, D.; Züger, S.; Guerra, J.; Lascano, J.; Reinhard, C.; Santoni, F.A.; Uchil, P.D.; Chatel, L.; et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 2011, 472, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Stremlau, M.; Perron, M.; Welikala, S.; Sodroski, J. Species-specific variation in the B30.2(SPRY) domain of TRIM5 alpha determines the potency of human immunodeficiency virus restriction. J. Virol. 2005, 79, 3139–3145. [Google Scholar] [CrossRef] [PubMed]
- Sawyer, S.L.; Wu, L.I.; Emerman, M.; Malik, H.S. Positive selection of primate TRIM5 alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 2005, 102, 2832–2837. [Google Scholar] [CrossRef] [PubMed]
- Stremlau, M.; Perron, M.; Lee, M.; Li, Y.; Song, B.; Javanbakht, H.; Diaz-Griffero, F.; Anderson, D.J.; Sundquist, W.I.; Sodroski, J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5 alpha restriction factor. Proc. Natl. Acad. Sci. USA 2006, 103, 5514–5519. [Google Scholar] [CrossRef]
- Nisole, S.; Lynch, C.; Stoye, J.P.; Yap, M.W. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. USA 2004, 101, 13324–13328. [Google Scholar] [CrossRef]
- Sayah, D.M.; Sokolskaja, E.; Berthoux, L.; Luban, J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 2004, 430, 569–573. [Google Scholar] [CrossRef]
- Brennan, G.; Kozyrev, Y.; Hu, S.L. TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc. Natl. Acad. Sci. USA 2008, 105, 3569–3574. [Google Scholar] [CrossRef]
- Liao, C.H.; Kuang, Y.Q.; Liu, H.L.; Zheng, Y.T.; Su, B. A novel fusion gene, TRIM5-Cyclophilin A in the pig-tailed macaque determines its susceptibility to HIV-1 infection. AIDS 2007, 21, S19–S26. [Google Scholar] [CrossRef]
- Newman, R.M.; Hall, L.; Kirmaier, A.; Pozzi, L.-A.; Pery, E.; Farzan, M.; O’Neil, S.P.; Johnson, W. Evolution of a TRIM5-CypA splice isoform in Old World monkeys. PLoS Pathog. 2008, 4, e1000003. [Google Scholar] [CrossRef]
- Virgen, C.A.; Kratovac, Z.; Bieniasz, P.D.; Hatziioannou, T. Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc. Natl. Acad. Sci. USA 2008, 105, 3563–3568. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.J.; Webb, B.L.; Ylinen, L.M.; Verschoor, E.; Heeney, J.L.; Towers, G.J. Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc. Natl. Acad. Sci. USA 2008, 105, 3557–3562. [Google Scholar] [CrossRef] [PubMed]
- De Groot, N.G.; Heijmans, C.M.C.; Koopman, G.; Verschoor, E.J.; Bogers, W.M.; Bontrop, R. TRIM5 allelic polymorphism in macaque species/populations of different geographic origins: Its impact on SIV vaccine studies. Tissue Antigens 2011, 78, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Ylinen, L.M.J.; Price, A.J.; Rasaiyaah, J.; Hué, S.; Rose, N.J.; Marzetta, F.; James, L.C.; Towers, G.J. Conformational adaptation of Asian macaque TRIMCyp directs lineage specific antiviral activity. PLoS Pathog. 2010, 6, e1001062. [Google Scholar] [CrossRef]
- Liu, H.F.; Wang, Y.Q.; Liao, C.H.; Kuang, Y.Q.; Zheng, Y.T.; Su, B. Adaptive evolution of primate TRIM5 alpha, a gene restricting HIV-1 infection. Gene 2005, 362, 109–116. [Google Scholar] [CrossRef]
- Wilson, S.J.; Webb, B.L.; Maplanka, C.; Newman, R.M.; Verschoor, E.J.; Heeney, J.L.; Towers, G.J. Rhesus macaque TRIM5 alleles have divergent antiretroviral specificities. J. Virol. 2008, 82, 7243–7247. [Google Scholar] [CrossRef]
- Nakayama, E.E.; Shioda, T. Anti-retroviral activity of TRIM5 alpha. Rev. Med. Virol. 2010, 20, 77–92. [Google Scholar] [CrossRef]
- Lim, S.Y.; Rogers, T.; Chan, T.; Whitney, J.B.; Kim, J.; Sodroski, J.; Letvin, N.L. TRIM5alpha modulates immunodeficiency virus control in rhesus monkeys. PLoS Pathog. 2010, 6, e1000738. [Google Scholar] [CrossRef]
- Reynolds, M.R.; Sacha, J.B.; Weiler, A.M.; Borchardt, G.J.; Glidden, C.E.; Sheppard, N.C.; Norante, F.A.; Castrovinci, P.A.; Harris, J.J.; Robertson, H.T.; et al. TRIM5α genotype of Rhesus macaques affects acquisition of SIVsmE660 infection after repeated limiting-dose intrarectal challenge. J. Virol. 2011, 85, 9637–9640. [Google Scholar] [CrossRef]
- Kirmaier, A.; Wu, F.; Newman, R.M.; Hall, L.R.; Morgan, J.S.; O’Connor, S.; Marx, P.A.; Meythaler, M.; Goldstein, S.; Buckler-White, A.; et al. TRIM5 suppresses crossspecies transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 2010, 8, e1000462. [Google Scholar] [CrossRef]
- Berry, N.J.; Marzetta, F.; Towers, G.J.; Rose, N.J. Diversity of TRIM5α and TRIMCyp sequences in cynomolgus macaques from different geographical origins. Immunogenetics 2012, 64, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, E.A.; Brennan, G.; Ferguson, B.; Wiseman, R.W.; O’Connor, D.; Hu, S.L. Variable prevalence and functional diversity of the antiretroviral restriction factor TRIMCyp in Macaca fascicularis. J. Virol. 2011, 85, 9956–9963. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Kawamoto, Y.; Higashino, A.; Yoshida, T.; Ikoma, T.; Suzaki, Y.; Ami, Y.; Shioda, T.; Nakayama, E.E.; Akari, H. Allele frequency of antiretroviral host factor TRIMcyp in wild-caught cynomolgus macaques (Macaca fascicularis). Front. Microbiol. 2012, 3, 314. [Google Scholar] [CrossRef]
- Mattiuzzo, G.; Rose, N.J.; Almond, N.; Towers, G.J.; Berry, N. Upregulation of TRIM5α gene expression after live-attenuated simian immunodeficiency virus vaccination in Mauritian cynomolgus macaques, but TRIM5α genotype has no impact on virus acquisition or vaccination outcome. J. Gen. Virol. 2013, 94, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Warren, W.C.; Harris, R.A.; Haukness, M.; Fiddes, I.T.; Murali, S.C.; Fernandes, J.; Dishuck, P.C.; Storer, J.M.; Raveendran, M.; Hillier, L.W.; et al. Sequence diversity analyses of an improved rhesus macaque genome enhance its biomedical utility. Science 2020, 370, eabc661. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Wei, X.; Liu, C.; Volpe, G.; Zhuang, Z.; Zou, X.; Wang, Z.; Pan, T.; Yuan, Y.; Zhang, X.; et al. Cell transcriptomic atlas of the non-human primate. Macaca fascicularis. Nature 2022, 604, 723–731. [Google Scholar] [PubMed]
- Hu, Q.; Huang, X.; Jin, Y.; Zhang, R.; Zhao, A.; Wang, Y.; Zhou, C.; Liu, W.; Liu, X.; Li, C.; et al. Long-read assembly of major histocompatibility complex and killer cell immunoglobulin-like receptor genome regions in cynomolgus macaque. Biol. Direct 2022, 17, 36. [Google Scholar] [CrossRef]
- Rogers, J.; Gibbs, R.A. Comparative primate genomics: Emerging patterns of genome content and dynamics. Nat. Rev. Genet. 2014, 15, 347–359. [Google Scholar] [CrossRef]
- Maudhoo, M.D.; Ren, D.; Gradnigo, J.S.; Gibbs, R.M.; Lubker, A.C.; Moriyama, E.N.; French, J.A.; Norgren, R.B., Jr. De novo assembly of the common marmoset transcriptome from NextGen mRNA sequences. Gigascience 2014, 3, 14. [Google Scholar] [CrossRef]
- Rockx, B.; Kuiken, T.; Herfst, S.; Bestebroer, T.; Lamers, M.M.; Oude Munnink, B.B.; de Meulder, D.; van Amerongen, G.; van den Brand, J.; Okba, N.M.A.; et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 2020, 368, 1012–1015. [Google Scholar] [CrossRef]
- Koo, B.S.; Oh, H.; Kim, G.; Hwang, E.H.; Jung, H.; Lee, Y.; Kang, P.; Park, J.H.; Ryu, C.M.; Hong, J.J. Transient Lymphopenia and Interstitial Pneumonia With Endotheliitis in SARS-CoV-2-Infected Macaques. J. Infect. Dis. 2020, 222, 1596–1600. [Google Scholar] [CrossRef] [PubMed]
- Salguero, F.J.; White, A.D.; Slack, G.S.; Fotheringham, S.A.; Bewley, K.R.; Gooch, K.E.; Longet, S.; Humphries, H.E.; Watson, R.J.; Hunter, L.; et al. Comparison of rhesus and cynomolgus macaques as an infection model for COVID-19. Nat. Commun. 2021, 12, 1260. [Google Scholar] [CrossRef] [PubMed]
- Munster, V.J.; Feldmann, F.; Williamson, B.N.; van Doremalen, N.; Pérez-Pérez, L.; Schulz, J.; Meade-White, K.; Okumura, A.; Callison, J.; Brumbaugh, B.; et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 2020, 585, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.K.; Singh, B.; Ganatra, S.R.; Gazi, M.; Cole, J.; Thippeshappa, R.; Alfson, K.J.; Clemmons, E.; Gonzalez, O.; Escobedo, R.; et al. Responses to acute infection with SARS-CoV-2 in the lungs of rhesus macaques, baboons and marmosets. Nat. Microbiol. 2021, 6, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Hartman, A.L.; Nambulli, S.; McMillen, C.M.; White, A.G.; Tilston-Lunel, N.L.; Albe, J.R.; Cottle, E.; Dunn, M.D.; Frye, L.J.; Gilliland, T.H.; et al. SARS-CoV-2 infection of African green monkeys results in mild respiratory disease discernible by PET/CT imaging and shedding of infectious virus from both respiratory and gastrointestinal tracts. PLoS Pathog. 2020, 16, e1008903. [Google Scholar] [CrossRef]
- Berry, N.; Ferguson, D.; Kempster, S.; Hall, J.; Ham, C.; Jenkins, A.; Rannow, V.; Giles, E.; Leahy, R.; Goulding, S.; et al. Intrinsic host susceptibility among multiple species to intranasal SARS-CoV-2 identifies diverse virological, biodistribution and pathological outcomes. Sci. Rep. 2022, 12, 18694. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, G.; Wang, Y.; Ren, W.; Zhao, X.; Ji, F.; Zhu, Y.; Feng, F.; Gong, M.; Ju, X.; et al. Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2021, 118, e2025373118. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Lambe, T.; Spencer, A.; Belij-Rammerstorfer, S.; Purushotham, J.N.; Port, J.R.; Avanzato, V.A.; Bushmaker, T.; Flaxman, A.; Ulaszewska, M.; et al. ChAdOx1nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 2020, 586, 578–582. [Google Scholar] [CrossRef]
- Corbett, K.S.; Nason, M.C.; Flach, B.; Gagne, M.; O’Connell, S.; Johnston, T.S.; Shah, S.N.; Edara, V.V.; Floyd, K.; Lai, L.; et al. Immune correlates of protection by mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. Science 2021, 373, eabj0299. [Google Scholar] [CrossRef]
- Neil, J.A.; Griffith, M.; Godfrey, D.I.; Purcell, D.F.J.; Deliyannis, G.; Jackson, D.; Rockman, S.; Subbarao, K.; Nolan, T. Nonhuman primate models for evaluation of SARS-CoV-2 vaccines. Expert Rev. Vaccines 2022, 21, 1055–1070. [Google Scholar] [CrossRef]
- Muñoz-Fontela, C.; Widerspick, L.; Albrecht, R.A.; Beer, M.; Carroll, M.W.; de Wit, E.; Diamond, M.S.; Dowling, W.E.; Funnell, S.G.P.; García-Sastre, A.; et al. Advances and gaps in SARS-CoV-2 infection models. PLoS Pathog. 2022, 13, 18. [Google Scholar] [CrossRef] [PubMed]
- Solforosi, L.; Kuipers, H.; Jongeneelen, M.; Huber, S.K.R.; van der Lubbe, J.E.M.; Dekking, L.; Czapska-Casey, D.N.; Gil, A.I.; Baert, M.R.M.; Drijver, J.; et al. Immunogenicity and efficacy of one and two doses of Ad26.COV2.S COVID vaccine in adult and aged NHP. J. Exp. Med. 2021, 218, e20202756. [Google Scholar] [CrossRef]
- Roozendaal, R.; Solforosi, L.; Stieh, D.J.; Serroyen, J.; Straetemans, R.; Dari, A.; Boulton, M.; Wegmann, F.; Rosendahl-Huber, S.K.; van der Lubbe, J.E.; et al. SARS-CoV-2 binding and neutralizing antibody levels after Ad26.COV2.S vaccination predict durable protection in rhesus macaques. Nat. Commun. 2021, 12, 5877. [Google Scholar] [CrossRef] [PubMed]
- McMahan, K.; Yu, J.; Mercado, N.B.; Loos, C.; Tostanoski, L.H.; Chandrashekar, A.; Liu, J.; Peter, L.; Atyeo, C.; Zhu, A.; et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 2021, 590, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yu, J.; McMahan, K.; Jacob-Dolan, C.; He, X.; Giffin, V.; Wu, C.; Sciacca, M.; Powers, O.; Nampanya, F.; et al. CD8 T cells contribute to vaccine protection against SARS-CoV-2 in macaques. Sci. Immunol. 2022, 7, eabq7647. [Google Scholar] [CrossRef] [PubMed]
- Berry, N.; Ferguson, D.; Ham, C.; Hall, J.; Jenkins, A.; Giles, E.; Devshi, D.; Kempster, S.; Rose, N.; Dowall, S.; et al. High susceptibility, viral dynamics and persistence of South American Zika virus in New World monkey species. Sci. Rep. 2019, 9, 14495. [Google Scholar] [CrossRef]
- Jiang, X.; Fan, Z.; Li, S.; Yin, H. A review on zoonotic pathogens associated with non-human primates: Understanding the potential threats to humans. Microorganisms 2023, 11, 246. [Google Scholar] [CrossRef]
- Broeckel, R.M.; Feldmann, F.; McNally, K.L.; Chiramel, A.I.; Sturdevant, G.L.; Leung, J.M.; Hanley, P.W.; Lovaglio, J.; Rosenke, R.; Scott, D.P.; et al. A pigtailed macaque model of Kyasanur Forest disease virus and Alkhurma hemorrhagic disease virus pathogenesis. PLoS Pathog. 2021, 17, e1009678. [Google Scholar] [CrossRef]
- Tiemessen, M.M.; Solforosi, L.; Dekking, L.; Czapska-Casey, D.; Serroyen, J.; Sullivan, N.J.; Volkmann, A.; Pau, M.G.; Callendret, B.; Schuitemaker, H.; et al. Protection against Marburg Virus and Sudan Virus in NHP by an Adenovector-Based Trivalent Vaccine Regimen Is Correlated to Humoral Immune Response Levels. Vaccines 2022, 10, 1263. [Google Scholar] [CrossRef]
- Bockstal, V.; Leyssen, M.; Heerwegh, D.; Spiessens, B.; Robinson, C.; Stoop, J.N.; Roozendaal, R.; Van Effelterre, T.; Gaddah, A.; Van Roey, G.A.; et al. Non-human primate to human immunobridging demonstrates a protective effect of Ad26.ZEBOV, MVA-BN-Filo vaccine against Ebola. NPJ Vaccines 2022, 7, 156. [Google Scholar] [CrossRef]
- Roozendaal, R.; Hendriks, J.; van Effelterre, T.; Spiessens, B.; Dekking, L.; Solforosi, L.; Czapska-Casey, D.; Bockstal, V.; Stoop, J.; Splinter, D.; et al. Nonhuman primate to human immunobridging to infer the protective effect of an Ebola virus vaccine candidate. NPJ Vaccines 2020, 5, 112. [Google Scholar] [CrossRef] [PubMed]
- Berry, N.; Kempster, S.; Ham, C.; Jenkins, A.; Hall, J.; Page, M.; Mattiuzzo, G.; Adedeji, Y.; Hewson, R.; Giles, E.; et al. Passive immunisation of convalescent human anti-Zika plasma protects against challenge with New World Zika virus in cynomolgus macaques. NPJ Vaccines 2020, 15, 86. [Google Scholar] [CrossRef] [PubMed]
- Finch, C.L.; Dowling, W.E.; King, T.H.; Martinez, C.; Nguyen, B.V.; Roozendaal, R.; Rustomjee, R.; Skiadopoulos, M.H.; Vert-Wong, E.; Yellowlees, A.; et al. Bridging Animal and Human Data in Pursuit of Vaccine Licensure. Vaccines 2022, 10, 1384. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berry, N.; Mee, E.T.; Almond, N.; Rose, N.J. The Impact and Effects of Host Immunogenetics on Infectious Disease Studies Using Non-Human Primates in Biomedical Research. Microorganisms 2024, 12, 155. https://doi.org/10.3390/microorganisms12010155
Berry N, Mee ET, Almond N, Rose NJ. The Impact and Effects of Host Immunogenetics on Infectious Disease Studies Using Non-Human Primates in Biomedical Research. Microorganisms. 2024; 12(1):155. https://doi.org/10.3390/microorganisms12010155
Chicago/Turabian StyleBerry, Neil, Edward T. Mee, Neil Almond, and Nicola J. Rose. 2024. "The Impact and Effects of Host Immunogenetics on Infectious Disease Studies Using Non-Human Primates in Biomedical Research" Microorganisms 12, no. 1: 155. https://doi.org/10.3390/microorganisms12010155
APA StyleBerry, N., Mee, E. T., Almond, N., & Rose, N. J. (2024). The Impact and Effects of Host Immunogenetics on Infectious Disease Studies Using Non-Human Primates in Biomedical Research. Microorganisms, 12(1), 155. https://doi.org/10.3390/microorganisms12010155



