Gut Microbiota Ecological and Functional Modulation in Post-Stroke Recovery Patients: An Italian Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants and Clinical Data
2.2. Ethics Statement
2.3. Fecal Sample Collection
2.4. 16S rRNA Targeted Metataxonomy Analysis
2.5. Gas Chromatography Coupled to Mass Spectrometry (GC-MS) Metabolomics Analysis
- An aqueous stock standard solution was prepared from chemical reagents of acetic acid (99%), propanoic acid (99%) and butanoic acid (99%) purchased by Merck (Darmstadt, Germany) with a concentration of 200 mm for each acid. All the stock standard solutions were stored at −20 °C until used.
- For SCFAs analysis, fecal samples (250 mg) were added to 1.25 mL of H2O and homogenized in a shaker for 3 min. The samples were then acidified with HCl (3 M) to a pH of 2–3. Finally, they were centrifuged at 5000× g at 4 °C for 10 min. After centrifugation 1 mL of supernatant was centrifuged at 15,000× g at 4 °C for 15 min. The supernatant was transferred and basified at pH 6 with NaOH (5 M), after which hexanoic acid (final concentration of 0.4 mg/mL) was added as an internal standard before sample screening. The samples were then filtered through 0.45 µm and 0.22 µm filters. Finally, 1 µL of each sample was injected into GC-MS. The temperature program was: 50 °C for 2 min, a temperature increase of 10 °C per min at 70 °C, 3 °C per min at 85 °C, 5 °C per min at 110°C, 15°C per min at 200 °C, 20 °C per min at 230 °C, 10 °C per min at 240 °C, then 240 °C for 5 min, according to the adapted in house procedures performed by Zhao et al. [25]. The method was validated by in house procedures according to European Medicines Agency (EMEA) and FDA guidelines [26,27] on bioanalytical method validation. The linearity of the calibration curve for each SCFA was assessed by the calculation of the coefficient of determination (R2). Validation was based on coefficient of variability (CV) of inter- and intraday reproducibility (%), limit of detection (LOD), limit of quantification (LOQ) and % of recovery.
- For indole and skatole determinations, fecal samples (100–500 mg) were managed and analyzed as previously described by Vernocchi et al. [28] using GC-MS coupled to solid-phase microextraction (SPME).
2.6. Fecal Markers of Intestinal Inflammation and Permeability (Zonulin)
2.7. Statistical Analyses Processing
3. Results
3.1. Subject Characteristics, Anamnestic and Clinical Features
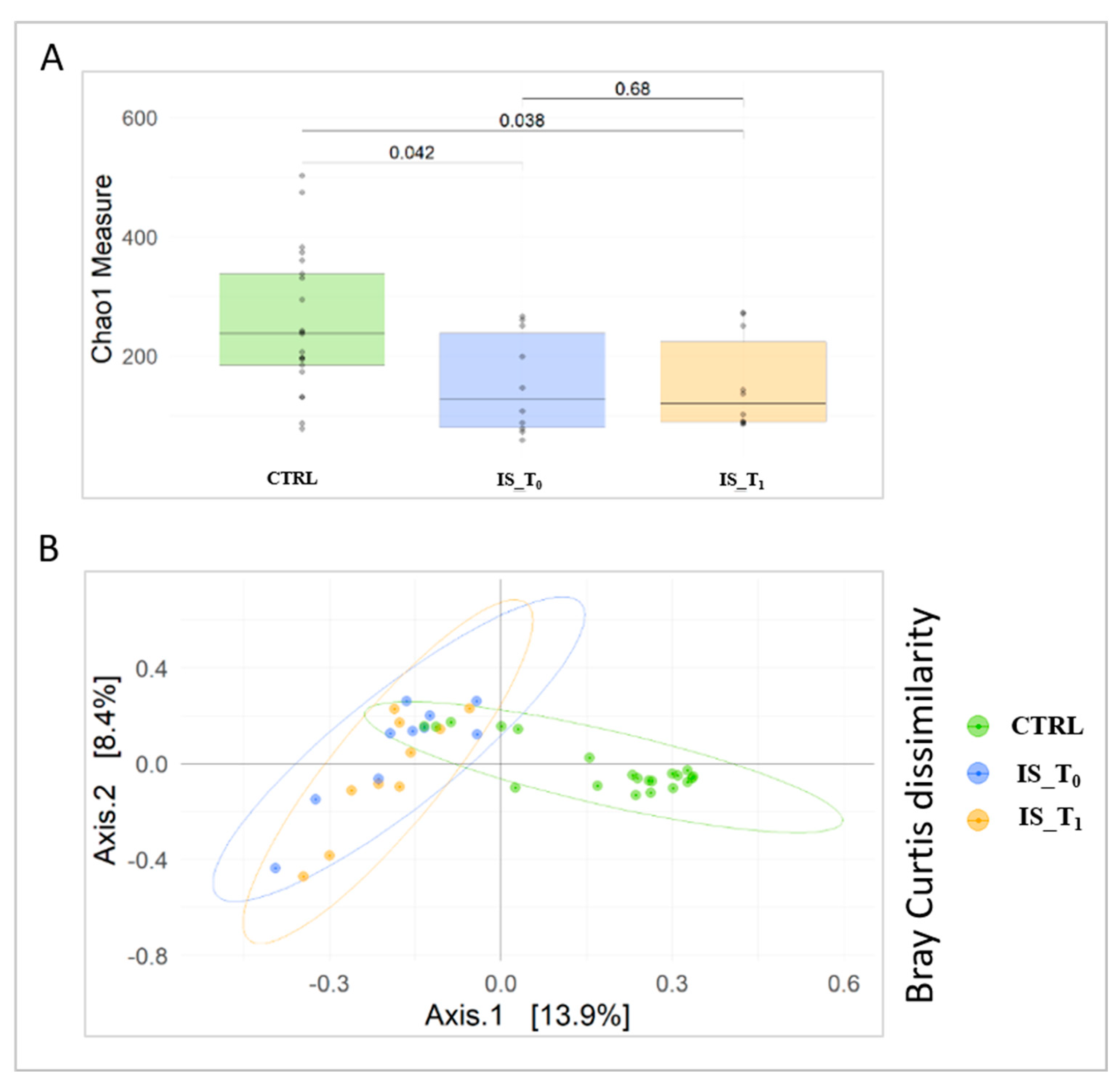
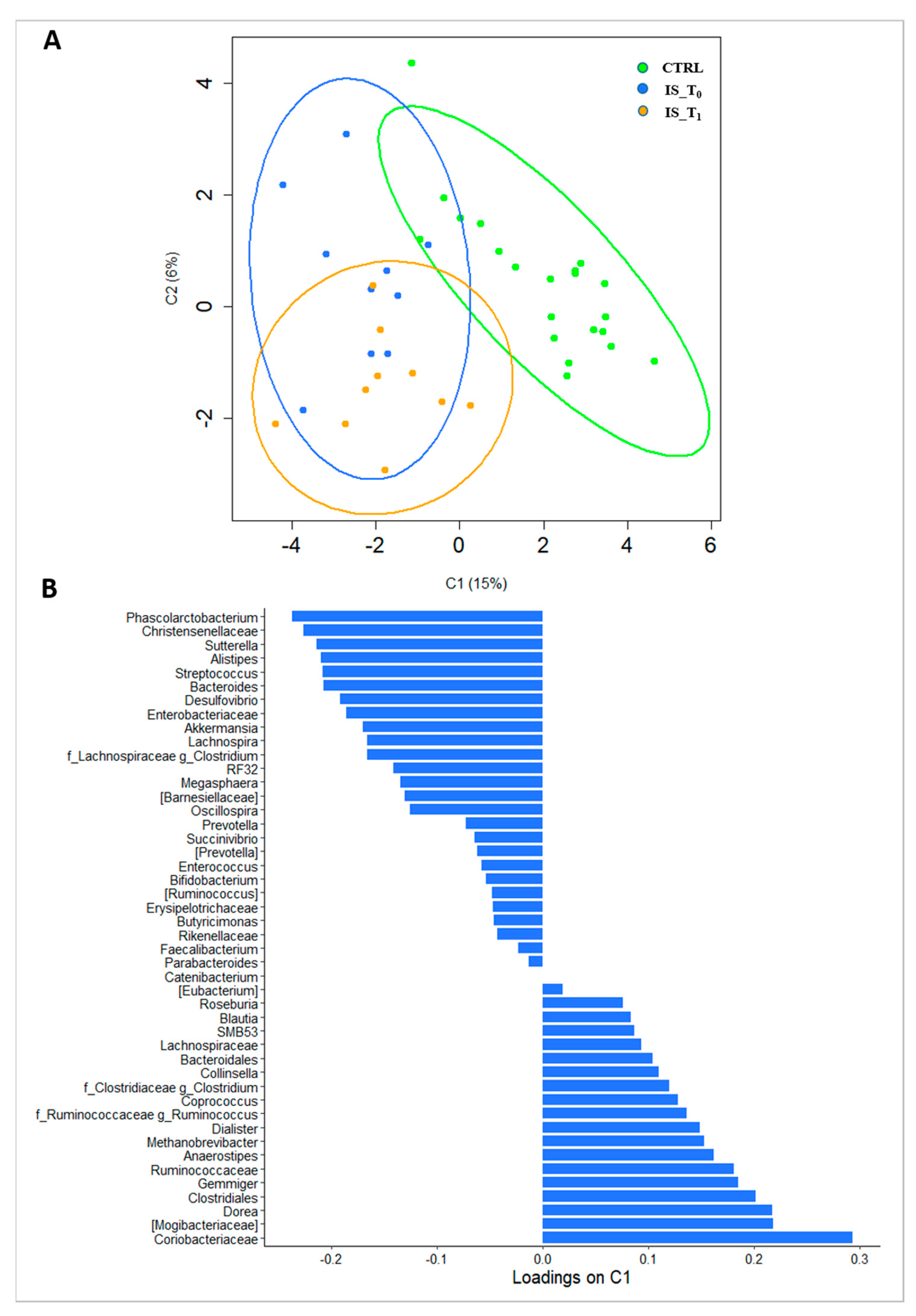
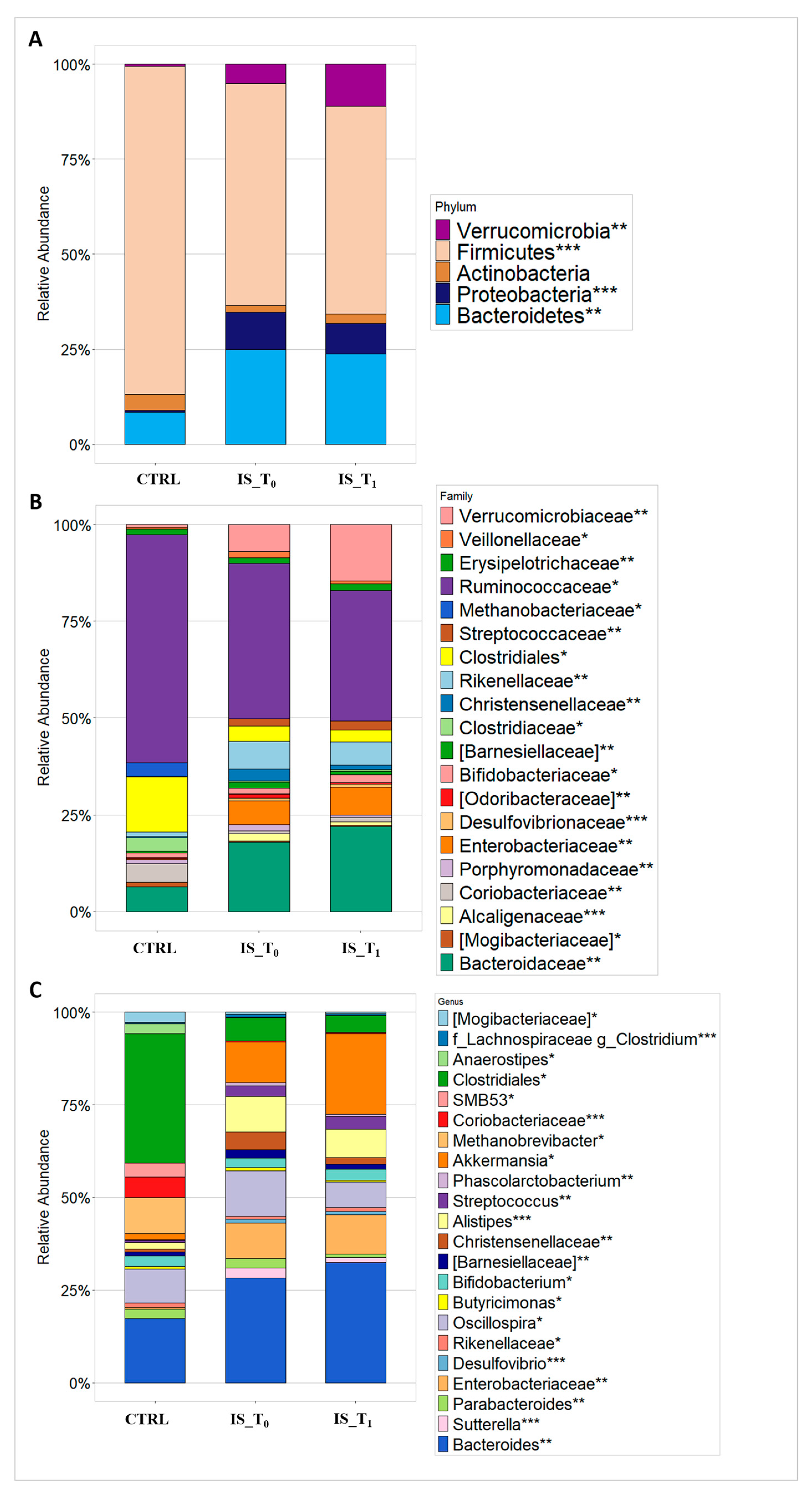
3.2. Gut Microbiota Metaxonomy
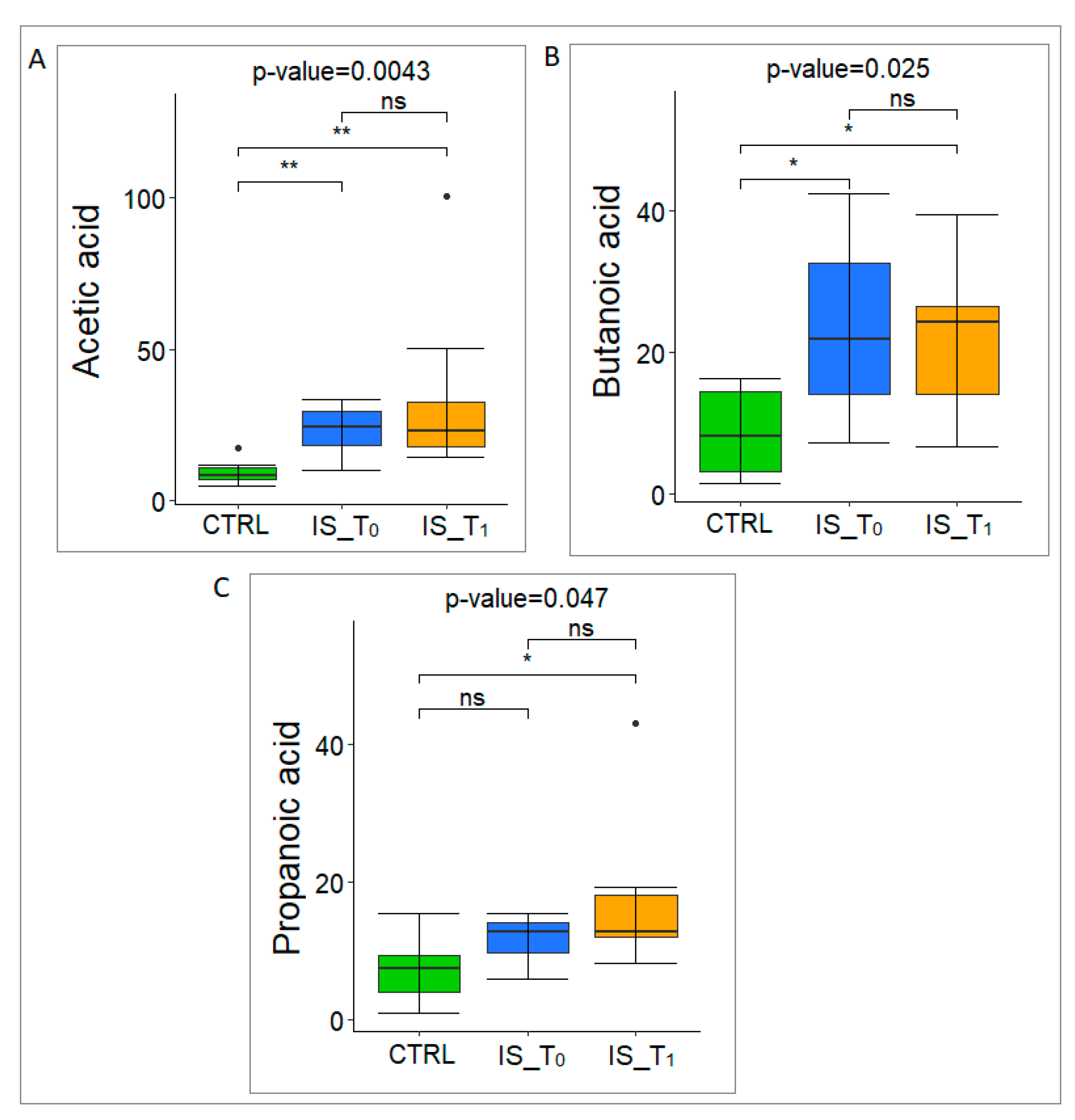
3.3. Gut Microbiota Metabolome Characterization
3.4. Fecal Markers of Intestinal Permeability (Zonulin)
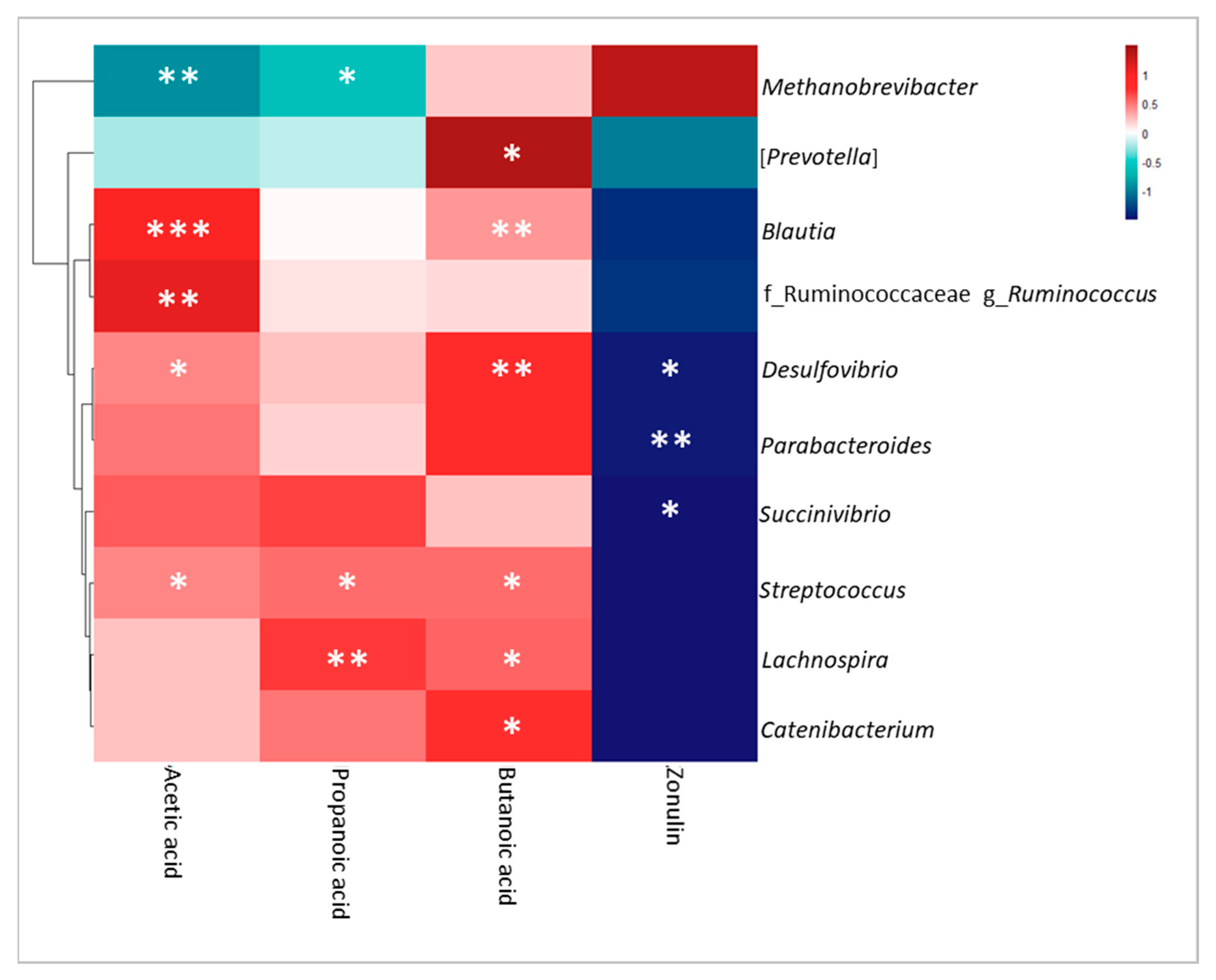
3.5. Correlation Analysis of Omics Data, Clinical Features and BBP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CNS | Central nervous system |
| ANS | Autonomous nervous system |
| ENS | Enteric nervous system |
| GM | Gut microbiota |
| GI | Calibri |
| SCFAs | Short-chain fatty acids |
| BBP | Blood biochemical parameters |
| ALT | Alanine aminotransferase |
| AST | Aspartate aminotransferase |
| GGT | Gamma-glutamyl transpeptidase |
| CTRLs | Control subjects |
| IS | Ischemic stroke patients |
| BBT | Box and Block test |
| ARAT | Action Research Arm test |
| MI_TOT | Motricity Index test |
| 10 MWT | 10 Meter Walking test |
| OPBG | Bambino Gesù Children’s Hospital |
| ASV | Amplicon Sequence Variant |
| GC | Gas chromatograph |
| MS | Mass spectrometer |
| NIST | National Institute of Standards and Technology |
| PCoA | Principal coordinate analyses |
| LEfSe | Linear discriminant analysis Effect Size |
| PICRUSt | Phylogenetic Investigation of Communities by Reconstruction of Unobserved States |
| PLS-DA | Partial least-squares-discriminant analysis |
| LOQ | Limit of Quantification |
| IBS | Inflammatory bowel disease |
| CD | Coeliac disease |
| ASD | Autism spectrum disorder |
| AHR | Aryl hydrocarbon receptor |
| SPME | Solid-phase microextraction |
References
- Sudo, N.; Chida, Y.; Aiba, Y.; Sonoda, J.; Oyama, N.; Yu, X.; Kubo, C.; Koga, Y. Postnatal Microbial Colonization Programs the Hypothalamic–Pituitary–Adrenal System for Stress Response in Mice. J. Physiol. 2004, 558, 263–275. [Google Scholar] [CrossRef]
- Zhang, Y.; Sharma, S.; Tom, L.; Liao, Y.-T.; Wu, V.C.H. Gut Phageome—An Insight into the Role and Impact of Gut Microbiome and Their Correlation with Mammal Health and Diseases. Microorganisms 2023, 11, 2454. [Google Scholar] [CrossRef]
- Bonaz, B.; Bazin, T.; Pellissier, S. The Vagus Nerve at the Interface of the Microbiota-Gut-Brain Axis. Front. Neurosci. 2018, 12, 49. [Google Scholar] [CrossRef]
- Bercik, P.; Collins, S.M.; Verdu, E.F. Microbes and the Gut-brain Axis. Neurogastroenterol. Motil. 2012, 24, 405–413. [Google Scholar] [CrossRef]
- Li, X.; You, X.; Wang, C.; Li, X.; Sheng, Y.; Zhuang, P.; Zhang, Y. Bidirectional Brain-gut-microbiota Axis in Increased Intestinal Permeability Induced by Central Nervous System Injury. CNS Neurosci. Ther. 2020, 26, 783–790. [Google Scholar] [CrossRef]
- Yarandi, S.S.; Peterson, D.A.; Treisman, G.J.; Moran, T.H.; Pasricha, P.J. Modulatory Effects of Gut Microbiota on the Central Nervous System: How Gut Could Play a Role in Neuropsychiatric Health and Diseases. J. Neurogastroenterol. Motil. 2016, 22, 201–212. [Google Scholar] [CrossRef]
- Heiss, C.N.; Olofsson, L.E. The Role of the Gut Microbiota in Development, Function and Disorders of the Central Nervous System and the Enteric Nervous System. J. Neuroendocrinol. 2019, 31, e12684. [Google Scholar] [CrossRef]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the Human Intestinal Microbial Flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Natividad, J.M.M.; Verdu, E.F. Modulation of Intestinal Barrier by Intestinal Microbiota: Pathological and Therapeutic Implications. Pharmacol. Res. 2013, 69, 42–51. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut Microbiota in Human Metabolic Health and Disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the Human Gut Microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Yamashiro, K.; Tanaka, R.; Urabe, T.; Ueno, Y.; Yamashiro, Y.; Nomoto, K.; Takahashi, T.; Tsuji, H.; Asahara, T.; Hattori, N. Gut Dysbiosis Is Associated with Metabolism and Systemic Inflammation in Patients with Ischemic Stroke. PLoS ONE 2017, 12, e0171521. [Google Scholar] [CrossRef]
- Liu, M.; Wu, B.; Wang, W.-Z.; Lee, L.-M.; Zhang, S.-H.; Kong, L.-Z. Stroke in China: Epidemiology, Prevention, and Management Strategies. Lancet Neurol. 2007, 6, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Xia, G.-H.; You, C.; Gao, X.-X.; Zeng, X.-L.; Zhu, J.-J.; Xu, K.-Y.; Tan, C.-H.; Xu, R.-T.; Wu, Q.-H.; Zhou, H.-W.; et al. Stroke Dysbiosis Index (SDI) in Gut Microbiome Are Associated With Brain Injury and Prognosis of Stroke. Front. Neurol. 2019, 10, 397. [Google Scholar] [CrossRef] [PubMed]
- Durgan, D.J.; Lee, J.; McCullough, L.D.; Bryan, R.M. Examining the Role of the Microbiota-Gut-Brain Axis in Stroke. Stroke 2019, 50, 2270–2277. [Google Scholar] [CrossRef]
- Spychala, M.S.; Venna, V.R.; Jandzinski, M.; Doran, S.J.; Durgan, D.J.; Ganesh, B.P.; Ajami, N.J.; Putluri, N.; Graf, J.; Bryan, R.M.; et al. Age-related Changes in the Gut Microbiota Influence Systemic Inflammation and Stroke Outcome. Ann. Neurol. 2018, 84, 23–36. [Google Scholar] [CrossRef]
- Statler, K.D.; Dong, L.; Nielsen, D.M.; Bratton, S.L. Pediatric Stroke: Clinical Characteristics, Acute Care Utilization Patterns, and Mortality. Childs Nerv. Syst. 2011, 27, 565–573. [Google Scholar] [CrossRef]
- Gardner, M.A.; Hills, N.K.; Sidney, S.; Johnston, S.C.; Fullerton, H.J. The 5-Year Direct Medical Cost of Neonatal and Childhood Stroke in a Population-Based Cohort. Neurology 2010, 74, 372–378. [Google Scholar] [CrossRef]
- Geroin, C.; Mazzoleni, S.; Smania, N.; Gandolfi, M.; Bonaiuti, D.; Gasperini, G.; Sale, P.; Munari, D.; Waldner, A.; Spidalieri, R.; et al. Systematic Review of Outcome Measures of Walking Training Using Electromechanical and Robotic Devices in Patients with Stroke. J. Rehabil. Med. 2013, 45, 987–996. [Google Scholar] [CrossRef]
- Mathiowetz, V.; Volland, G.; Kashman, N.; Weber, K. Adult Norms for the Box and Block Test of Manual Dexterity. Am. J. Occup. Ther. 1985, 39, 386–391. [Google Scholar] [CrossRef]
- Lyle, R.C. A Performance Test for Assessment of Upper Limb Function in Physical Rehabilitation Treatment and Research. Int. J. Rehabil. Res. 1981, 4, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Wade, D.T. Measuring Arm Impairment and Disability after Stroke. Int. Disabil. Stud. 1989, 11, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Finch, E. Physical Rehabilitation Outcome Measures: A Guide to Enhanced Clinical Decision Making; Canadian Physiotherapy Association: Hamilton, ON, Canada, 2002; ISBN 978-1-55009-220-2. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Nyman, M.; Åke Jönsson, J. Rapid Determination of Short-Chain Fatty Acids in Colonic Contents and Faeces of Humans and Rats by Acidified Water-Extraction and Direct-Injection Gas Chromatography. Biomed. Chromatogr. 2006, 20, 674–682. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency. ICH Guideline M10 on Bioanalytical Method Valida-Tion Step 2b. 2019. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/draft-ich-guideline-m10-bioanalytical-method-validation-step-2b_en.pdf (accessed on 3 April 2022).
- U.S. Department of Health and Human Services; Food and Drug Administration; Center for Drug Evaluation and Research (CDER); Center for Veterinary Medicine (CVM). Bioanalytical Method Validation—Guidance for Industry. 2018. Available online: http://www.fda.gov/downloads/drugs/guidances/ucm070107.pdf (accessed on 3 April 2022).
- Vernocchi, P.; Marini, F.; Capuani, G.; Tomassini, A.; Conta, G.; Del Chierico, F.; Malattia, C.; De Benedetti, F.; Martini, A.; Dallapiccola, B.; et al. Fused Omics Data Models Reveal Gut Microbiome Signatures Specific of Inactive Stage of Juvenile Idiopathic Arthritis in Pediatric Patients. Microorganisms 2020, 8, 1540. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bittinger, K.; Charlson, E.S.; Hoffmann, C.; Lewis, J.; Wu, G.D.; Collman, R.G.; Bushman, F.D.; Li, H. Associating Microbiome Composition with Environmental Covariates Using Generalized UniFrac Distances. Bioinformatics 2012, 28, 2106–2113. [Google Scholar] [CrossRef] [PubMed]
- Welham, Z.; Déjean, S.; Lê Cao, K.-A. Multivariate Analysis with the R Package mixOmics. In Statistical Analysis of Proteomic Data; Burger, T., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2023; Volume 2426, pp. 333–359. ISBN 978-1-07-161966-7. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps. Cran.R-Project. Available online: https://cran.r-project.org/package=pheatmap (accessed on 28 October 2019).
- Wang, J.; Zhang, H.; He, J.; Xiong, X. The Role of the Gut Microbiota in the Development of Ischemic Stroke. Front. Immunol. 2022, 13, 845243. [Google Scholar] [CrossRef]
- Honarpisheh, P.; Bryan, R.M.; McCullough, L.D. Aging Microbiota-Gut-Brain Axis in Stroke Risk and Outcome. Circ. Res. 2022, 130, 1112–1144. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Kong, X.; Wang, H.; Li, Y.; Luo, Y. Ischemic Stroke and Intestinal Flora: An Insight into Brain–Gut Axis. Eur. J. Med. Res. 2022, 27, 73. [Google Scholar] [CrossRef] [PubMed]
- Bonnechère, B.; Amin, N.; Van Duijn, C. What Are the Key Gut Microbiota Involved in Neurological Diseases? A Systematic Review. Int. J. Mol. Sci. 2022, 23, 13665. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.; Moore, R.J.; Wong, C.H.Y. An Insight into Intestinal Mucosal Microbiota Disruption after Stroke. Sci. Rep. 2018, 8, 568. [Google Scholar] [CrossRef] [PubMed]
- Suganya, K.; Koo, B.-S. Gut–Brain Axis: Role of Gut Microbiota on Neurological Disorders and How Probiotics/Prebiotics Beneficially Modulate Microbial and Immune Pathways to Improve Brain Functions. Int. J. Mol. Sci. 2020, 21, 7551. [Google Scholar] [CrossRef] [PubMed]
- Rondanelli, M.; Gasparri, C.; Peroni, G.; Faliva, M.A.; Naso, M.; Perna, S.; Bazire, P.; Sajoux, I.; Maugeri, R.; Rigon, C. The Potential Roles of Very Low Calorie, Very Low Calorie Ketogenic Diets and Very Low Carbohydrate Diets on the Gut Microbiota Composition. Front. Endocrinol. 2021, 12, 662591. [Google Scholar] [CrossRef]
- Kurowska, A.; Ziemichód, W.; Herbet, M.; Piątkowska-Chmiel, I. The Role of Diet as a Modulator of the Inflammatory Process in the Neurological Diseases. Nutrients 2023, 15, 1436. [Google Scholar] [CrossRef]
- Chen, R.; Wu, P.; Cai, Z.; Fang, Y.; Zhou, H.; Lasanajak, Y.; Tang, L.; Ye, L.; Hou, C.; Zhao, J. Puerariae Lobatae Radix with Chuanxiong Rhizoma for Treatment of Cerebral Ischemic Stroke by Remodeling Gut Microbiota to Regulate the Brain–Gut Barriers. J. Nutr. Biochem. 2019, 65, 101–114. [Google Scholar] [CrossRef]
- Liu, C.-Z.; Chen, W.; Wang, M.-X.; Wang, Y.; Chen, L.-Q.; Zhao, F.; Shi, Y.; Liu, H.-J.; Dou, X.-B.; Liu, C.; et al. Dendrobium officinale Kimura et Migo and American Ginseng Mixture: A Chinese Herbal Formulation for Gut Microbiota Modulation. Chin. J. Nat. Med. 2020, 18, 446–459. [Google Scholar] [CrossRef]
- Gerhardt, S.; Mohajeri, M. Changes of Colonic Bacterial Composition in Parkinson’s Disease and Other Neurodegenerative Diseases. Nutrients 2018, 10, 708. [Google Scholar] [CrossRef]
- López-Montoya, P.; Cerqueda-García, D.; Rodríguez-Flores, M.; López-Contreras, B.; Villamil-Ramírez, H.; Morán-Ramos, S.; Molina-Cruz, S.; Rivera-Paredez, B.; Antuna-Puente, B.; Velázquez-Cruz, R.; et al. Association of Gut Microbiota with Atherogenic Dyslipidemia, and Its Impact on Serum Lipid Levels after Bariatric Surgery. Nutrients 2022, 14, 3545. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human Genetics Shape the Gut Microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, X.; Sun, C.; Wu, X.; Lu, M.; Si, Y.; Ye, X.; Wang, T.; Yu, X.; Zhao, X.; et al. Change of Intestinal Microbiota in Cerebral Ischemic Stroke Patients. BMC Microbiol. 2019, 19, 191. [Google Scholar] [CrossRef] [PubMed]
- Alamoudi, M.U.; Hosie, S.; Shindler, A.E.; Wood, J.L.; Franks, A.E.; Hill-Yardin, E.L. Comparing the Gut Microbiome in Autism and Preclinical Models: A Systematic Review. Front. Cell. Infect. Microbiol. 2022, 12, 905841. [Google Scholar] [CrossRef] [PubMed]
- Vernocchi, P.; Ristori, M.V.; Guerrera, S.; Guarrasi, V.; Conte, F.; Russo, A.; Lupi, E.; Albitar-Nehme, S.; Gardini, S.; Paci, P.; et al. Gut Microbiota Ecology and Inferred Functions in Children With ASD Compared to Neurotypical Subjects. Front. Microbiol. 2022, 13, 871086. [Google Scholar] [CrossRef] [PubMed]
- Taniya, M.A.; Chung, H.-J.; Al Mamun, A.; Alam, S.; Aziz, M.A.; Emon, N.U.; Islam, M.M.; Hong, S.-T.S.; Podder, B.R.; Ara Mimi, A.; et al. Role of Gut Microbiome in Autism Spectrum Disorder and Its Therapeutic Regulation. Front. Cell. Infect. Microbiol. 2022, 12, 915701. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Jin, C.; Xie, T.; Cheng, Y.; Li, L.; Wu, N. Alterations in the Fecal Microbiota of Patients with HIV-1 Infection: An Observational Study in A Chinese Population. Sci. Rep. 2016, 6, 30673. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Grandhi, R.; Patterson, T.; Nicholson, S. A Review of Traumatic Brain Injury and the Gut Microbiome: Insights into Novel Mechanisms of Secondary Brain Injury and Promising Targets for Neuroprotection. Brain Sci. 2018, 8, 113. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, H.; Wang, X.; Tian, M.; Tian, Y.; Li, Q.; Liang, C.; Peng, X.; Ding, J.; Wu, X.; et al. Combination Effect between Gut Microbiota and Traditional Potentially Modifiable Risk Factors for First-Ever Ischemic Stroke in Tujia, Miao and Han Populations in China. Front. Mol. Neurosci. 2022, 15, 922399. [Google Scholar] [CrossRef]
- Sun, H.; Gu, M.; Li, Z.; Chen, X.; Zhou, J. Gut Microbiota Dysbiosis in Acute Ischemic Stroke Associated With 3-Month Unfavorable Outcome. Front. Neurol. 2022, 12, 799222. [Google Scholar] [CrossRef]
- Li, A.; Yang, Y.; Qin, S.; Lv, S.; Jin, T.; Li, K.; Han, Z.; Li, Y. Microbiome Analysis Reveals Gut Microbiota Alteration of Early-Weaned Yimeng Black Goats with the Effect of Milk Replacer and Age. Microb. Cell Fact. 2021, 20, 78. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.T.; Mohd Ismail, N.I.; Wei, L.K. Microbiome and Ischemic Stroke: A Systematic Review. PLoS ONE 2021, 16, e0245038. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Zhang, H.; Liang, X.; Dai, Y.; Liu, L.; Tan, K.; Ma, R.; Luo, J.; Ding, Y.; Ke, C. Correction to: Application of Metabolomics to the Discovery of Biomarkers for Ischemic Stroke in the Murine Model: A Comparison with the Clinical Results. Mol. Neurobiol. 2022, 59, 2657. [Google Scholar] [CrossRef] [PubMed]
- Sadler, R.; Cramer, J.V.; Heindl, S.; Kostidis, S.; Betz, D.; Zuurbier, K.R.; Northoff, B.H.; Heijink, M.; Goldberg, M.P.; Plautz, E.J.; et al. Short-Chain Fatty Acids Improve Poststroke Recovery via Immunological Mechanisms. J. Neurosci. 2020, 40, 1162–1173. [Google Scholar] [CrossRef] [PubMed]
- Ke, X.; Xiang, Q.; Jiang, P.; Liu, W.; Yang, M.; Yang, Y.; Shi, D.; Chen, L.; Tao, J. Effect of Electroacupuncture on Short-Chain Fatty Acids in Peripheral Blood after Middle Cerebral Artery Occlusion/Reperfusion in Rats Based on Gas Chromatography–Mass Spectrometry. Mediat. Inflamm. 2022, 2022, 3997947. [Google Scholar] [CrossRef] [PubMed]
- Oleskin, A.V.; Shenderov, B.A. Neuromodulatory Effects and Targets of the SCFAs and Gasotransmitters Produced by the Human Symbiotic Microbiota. Microb. Ecol. Health Dis. 2016, 27, 30971. [Google Scholar] [CrossRef] [PubMed]
- Vinolo, M.A.R.; Rodrigues, H.G.; Nachbar, R.T.; Curi, R. Regulation of Inflammation by Short Chain Fatty Acids. Nutrients 2011, 3, 858–876. [Google Scholar] [CrossRef]
- Singh, V.; Roth, S.; Llovera, G.; Sadler, R.; Garzetti, D.; Stecher, B.; Dichgans, M.; Liesz, A. Microbiota Dysbiosis Controls the Neuroinflammatory Response after Stroke. J. Neurosci. 2016, 36, 7428–7440. [Google Scholar] [CrossRef]
- Xu, K.; Gao, X.; Xia, G.; Chen, M.; Zeng, N.; Wang, S.; You, C.; Tian, X.; Di, H.; Tang, W.; et al. Rapid Gut Dysbiosis Induced by Stroke Exacerbates Brain Infarction in Turn. Gut 2021, 70, 1486–1494. [Google Scholar] [CrossRef]
- Macfarlane, S.; Macfarlane, G.T. Regulation of Short-Chain Fatty Acid Production. Proc. Nutr. Soc. 2003, 62, 67–72. [Google Scholar] [CrossRef]
- Salazar, N.; Arboleya, S.; Fernández-Navarro, T.; De Los Reyes-Gavilán, C.G.; Gonzalez, S.; Gueimonde, M. Age-Associated Changes in Gut Microbiota and Dietary Components Related with the Immune System in Adulthood and Old Age: A Cross-Sectional Study. Nutrients 2019, 11, 1765. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; d’Aigle, J.; Atadja, L.; Quaicoe, V.; Honarpisheh, P.; Ganesh, B.P.; Hassan, A.; Graf, J.; Petrosino, J.; Putluri, N.; et al. Gut Microbiota–Derived Short-Chain Fatty Acids Promote Poststroke Recovery in Aged Mice. Circ. Res. 2020, 127, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Henry, N.; Frank, J.; McLouth, C.; Trout, A.L.; Morris, A.; Chen, J.; Stowe, A.M.; Fraser, J.F.; Pennypacker, K. Short Chain Fatty Acids Taken at Time of Thrombectomy in Acute Ischemic Stroke Patients Are Independent of Stroke Severity But Associated With Inflammatory Markers and Worse Symptoms at Discharge. Front. Immunol. 2022, 12, 797302. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Zhang, X.; Zheng, Y.; Yu, B.; Pan, D.; Jiang, X.; Yan, C.; Yu, Q.; Lu, X. Distinctive Gut Microbiota Alteration Is Associated with Poststroke Functional Recovery: Results from a Prospective Cohort Study. Neural Plast. 2021, 2021, 1469339. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.B.; Hetz, R.A.; Xue, H.; Aroom, K.R.; Bhattarai, D.; Johnson, E.; Bedi, S.; Cox, C.S.; Uray, K. Effects of Traumatic Brain Injury on Intestinal Contractility. Neurogastroenterol. Motil. 2013, 25, 593-e463. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A. All Disease Begins in the (Leaky) Gut: Role of Zonulin-Mediated Gut Permeability in the Pathogenesis of Some Chronic Inflammatory Diseases. F1000Res 2020, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, Y.; Li, H.; Dai, Y.; Chen, D.; Wang, M.; Jiang, X.; Huang, Z.; Yu, H.; Huang, J.; et al. Altered Fecal Microbiota Composition in Older Adults With Frailty. Front. Cell. Infect. Microbiol. 2021, 11, 696186. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Chen, Y.; Zhai, Y.; Meng, L.; Liu, H.; Tian, H.; Feng, R.; Wang, J.; Zhang, R.; Sun, K.; et al. Gut Dysbiosis Is Associated With the Severity of Cryptogenic Stroke and Enhanced Systemic Inflammatory Response. Front. Immunol. 2022, 13, 836820. [Google Scholar] [CrossRef]
- Roth, W.; Zadeh, K.; Vekariya, R.; Ge, Y.; Mohamadzadeh, M. Tryptophan Metabolism and Gut-Brain Homeostasis. Int. J. Mol. Sci. 2021, 22, 2973. [Google Scholar] [CrossRef]
- Xie, Y.; Zou, X.; Han, J.; Zhang, Z.; Feng, Z.; Ouyang, Q.; Hua, S.; Liu, Z.; Li, C.; Cai, Y.; et al. Indole-3-Propionic Acid Alleviates Ischemic Brain Injury in a Mouse Middle Cerebral Artery Occlusion Model. Exp. Neurol. 2022, 353, 114081. [Google Scholar] [CrossRef]
- Ganesh, B.P.; Peesh, P.; Blasco, M.P.; Hamamy, A.E.; Khan, R.; Guzman, G.; Honarpisheh, P.; Mohan, E.C.; Goodman, G.W.; Nguyen, J.N.; et al. Restoring a Balanced Pool of Host-Derived and Microbiota-Derived Ligands of the Aryl Hydrocarbon Receptor Is Beneficial after Stroke. Res. Sq. 2023, in press. [CrossRef]
- Williams, B.B.; Van Benschoten, A.H.; Cimermancic, P.; Donia, M.S.; Zimmermann, M.; Taketani, M.; Ishihara, A.; Kashyap, P.C.; Fraser, J.S.; Fischbach, M.A. Discovery and Characterization of Gut Microbiota Decarboxylases That Can Produce the Neurotransmitter Tryptamine. Cell Host Microbe 2014, 16, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Lee, J. Indole as an Intercellular Signal in Microbial Communities. FEMS Microbiol. Rev. 2010, 34, 426–444. [Google Scholar] [CrossRef]
| Blood Biochemical Parameters (BBP) | Acetic Acid | Propanoic Acid | Butanoic Acid | Total SCFA | Zonulin |
|---|---|---|---|---|---|
| ALT (U/L) | −0.396 | −0.463 | −0.492 | −0.543 | 0.402 |
| AST (mU/ mL) | 0.036 | −0.064 | −0.324 | −0.19 | −0.006 |
| GGT (U/L) | −0.257 | 0.337 | −0.238 | −0.204 | −0.198 |
| Tryglicerides (mg/dL) | 0.028 | 0.387 | 0.018 | 0.083 | 0.738 * |
| Cholesterol (mg/dL) | 0.186 | 0.614 | 0.608 | 0.545 | 0.359 |
| Glucose (mg/dL) | −0.152 | 0.556 | 0.033 | 0.04 | 0.568 |
| Albumin (g/mL) | 0.251 | 0.412 | 0.442 | 0.441 | 0.73 * |
| Bilirubin (mg/dL) | −0.516 | −0.168 | −0.255 | −0.406 | 0.125 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marsiglia, R.; Marangelo, C.; Vernocchi, P.; Scanu, M.; Pane, S.; Russo, A.; Guanziroli, E.; Del Chierico, F.; Valeriani, M.; Molteni, F.; et al. Gut Microbiota Ecological and Functional Modulation in Post-Stroke Recovery Patients: An Italian Study. Microorganisms 2024, 12, 37. https://doi.org/10.3390/microorganisms12010037
Marsiglia R, Marangelo C, Vernocchi P, Scanu M, Pane S, Russo A, Guanziroli E, Del Chierico F, Valeriani M, Molteni F, et al. Gut Microbiota Ecological and Functional Modulation in Post-Stroke Recovery Patients: An Italian Study. Microorganisms. 2024; 12(1):37. https://doi.org/10.3390/microorganisms12010037
Chicago/Turabian StyleMarsiglia, Riccardo, Chiara Marangelo, Pamela Vernocchi, Matteo Scanu, Stefania Pane, Alessandra Russo, Eleonora Guanziroli, Federica Del Chierico, Massimiliano Valeriani, Franco Molteni, and et al. 2024. "Gut Microbiota Ecological and Functional Modulation in Post-Stroke Recovery Patients: An Italian Study" Microorganisms 12, no. 1: 37. https://doi.org/10.3390/microorganisms12010037
APA StyleMarsiglia, R., Marangelo, C., Vernocchi, P., Scanu, M., Pane, S., Russo, A., Guanziroli, E., Del Chierico, F., Valeriani, M., Molteni, F., & Putignani, L. (2024). Gut Microbiota Ecological and Functional Modulation in Post-Stroke Recovery Patients: An Italian Study. Microorganisms, 12(1), 37. https://doi.org/10.3390/microorganisms12010037









