Recent Progress in the Detection of Surra, a Neglected Disease Caused by Trypanosoma evansi with a One Health Impact in Large Parts of the Tropic and Sub-Tropic World
Abstract
1. Introduction
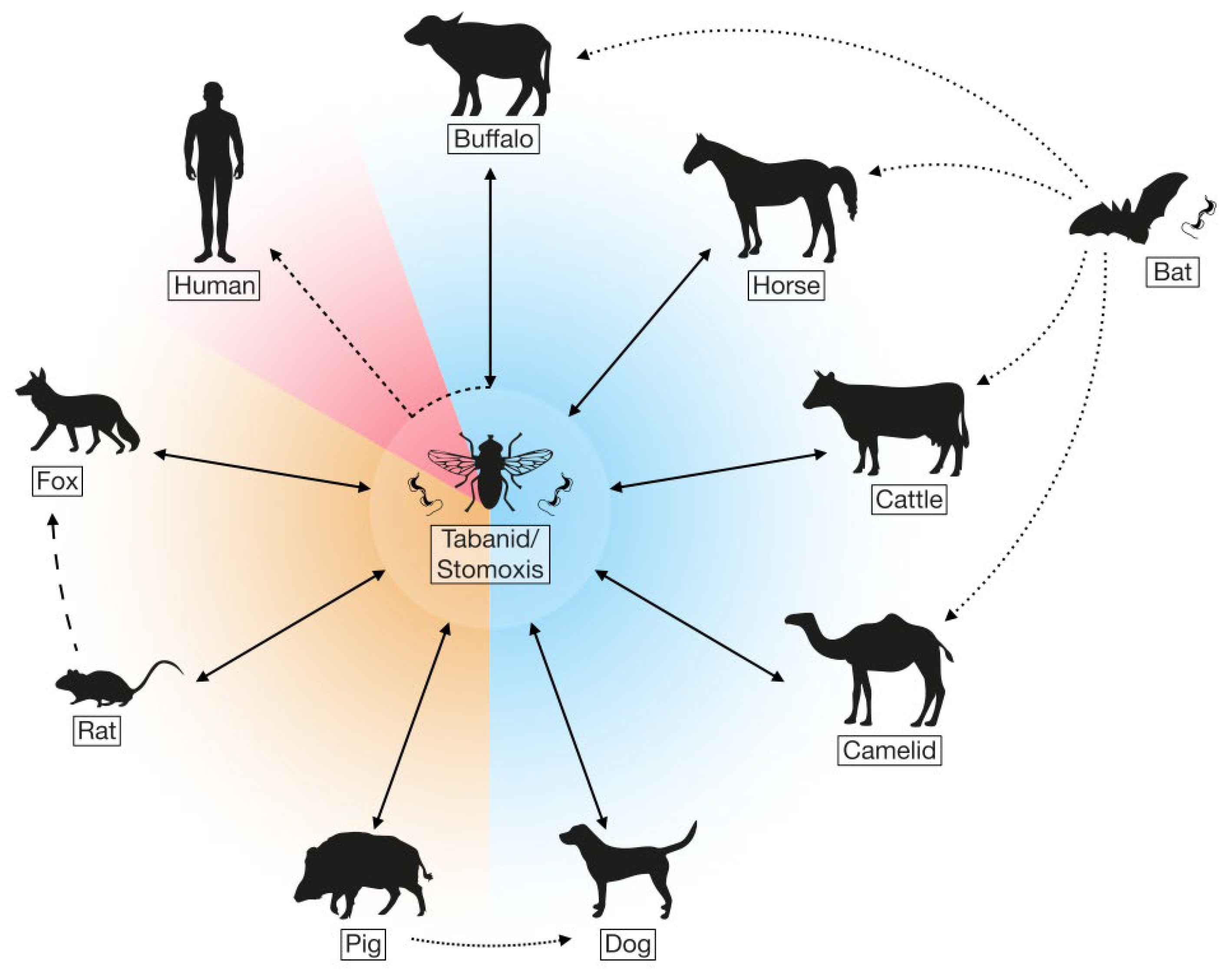
2. Transmission and Pathology
3. Control and Treatment of Surra
4. Parasitology- and Serology-Based Diagnostic Methods
4.1. Microscopy-Based Detection Methods
4.2. Serology-Based Detection Methods
| Species | Name of the Assay | Target | Reference |
|---|---|---|---|
| T. evansi | Double immunodiffusion (DID) test | Whole cell soluble T. evansi antigen | [44] |
| T. evansi Type A | Card agglutination test for trypanosomosis/T. evansi Rotat1.2 (CATT/T. evansi) | VAT T. evansi Rotat1.2 | [44] |
| Trypansomal | IgG-ELISA | Trypanosomal Abs | [45] |
| T. evansi | IgG-ELISA | 2G6 Ag-ELISA (70 kDa antigen) Tr7 Ag-ELISA (15 kDa antigen) | [46] |
| Trypanozoon spp. T. vivax | Ag-ELISA | TeGM6-4r | [47] |
| T. evansi | TeCA-ELISA | T. evansi crude antigens | [47] |
| T. evansi | Suratex | Trypanosome-circulating Ags | [48] |
| T. evansi | IFAT | Anto-T. evansi Abs | [49] |
| T. evansi | LATEX/T. evansi | Rotat 1.2 VSG | [50] |
| T. evansi | ELISA | Rotat 1.2 VSG | [50] |
| T. evansi | ELISA | Excretory-secretory Ags | [51] |
| T. evansi | IgG-ELISA | Anti-T. evansi Abs | [52,53] |
| T. evansi | Ab-ELISA/rISG75 | Anti-T. evansi Abs | [54] |
| T. evansi | Ag-ELISA | TEA 1/23.4.6 | [55] |
| T. evansi | Immune trypanolysis test (TL) | VAT RoTat 1.2 | [56,57] |
| T. evansi type A | Surra Sero K-SeT | VAT RoTat 1.2 | [57] |
| T. evansi | LFA | Anti-T. evansi Abs | [58,59] |
| T. evansi | IgM-ELISA | Anti-T. evansi Abs | [60] |
| T. evansi T. brucei | Ag-ELISA | Circulating Ags | [61] |
5. Molecular Diagnostic Methods
5.1. Polymerase Chain Reaction
5.2. Isothermal Amplification Methods: Loop-Mediated Isothermal Amplification
5.2.1. Loop-Mediated Isothermal Amplification Assay
5.2.2. Recombinase Polymerase Amplification
| Species | Name of the Assay | Primer Name | Oligonucleotide (5′-3′) | Gene Target | Reference |
|---|---|---|---|---|---|
| Trypanosoma spp. | PCR | CF BR | CCGGAAGTTCACCGATATTG TGCTGCGTTCTTCAACGAA | ITS1 | [85] |
| Trypanozoon spp. | PCR | F R | ACATTCCAGCAGGAGTTGGAG CACGTGAATCCTCAATTTTGT | ESAG 6/7 | [86] |
| Trypanozoon spp. | PCR |
21mer 22mer | TGCAGACGACCTGACGCTACT CTCCTAGAAGCTTCGGTGTCCT | Repetitive sequence probe pMUTec 6.258 | [87] |
| Trypanozoon spp. | PCR | TBR1 TBR2 | GAATATTAAACAATGCGCAG CCATTTATTAGCTTTGTTGC | Minisatellite DNA | [88] |
| T. evansi and T. brucei | PCR |
TBS-01 TBS-02 | CGAATGAATAATAAACAATGCGCAGT AGAAGGATTTATTAGCTTTGTTGC | Conserved regions of T. brucei and T. evansi genome | [89] |
| T. evansi | PCR | TR3 TR4 | GCGCGGATTCTTTGCAGACGA TGCAGACACTGGAATGTTACT | Repetitive nucleotide sequences of T. evansi | [90] |
| T. evansi | PCR | TeD-ISGF TeD-ISGR | CAGCCGGTGAGTGAAGAAA CTACGGCCCCTAATAATAAAGAAC | ISG-75 | [91] |
| T. evansi | PCR | NRP1 NRP2 | CGAATGAATATTAAACAATGCGCAGT AGAACCATTTATTAGCTTTGTTGC | Nuclid Repeat | [92] |
| T. evansi | PCR |
MP1 MP2 | CAACGACAAAGAGTCAGT ACGTGTTTTGTGTATGGT | Minicircle DNA | [92] |
| T. evansi | PCR |
EVA1 EVA2 | ACATATCAACAACGACAAAG CCCTAGTATCTCCAATGAAT | Minicircle DNA | [93] |
| T. evansi | PCR |
DITRYF DITRYR | CGACCAGCCAGAACGAGCAGAAT CTTGTCGATCGAGTTGACGGT | VSG | [94] |
| T. evansi (Type A) | PCR |
F R | GCGGGGTGTTTAAAGCAATA ATTAGTGCTGCGTGTGTTCG | Rotat 1.2 VSG | [17] |
| T. evansi (Type B) | PCR |
F R | TTCTACCAACTGACGGAGCG TAGCTCCGGATGCATCGGT | VSG JN 2118Hu | [65] |
| T. evansi (Real-time PCR) | PCR |
TeRoTat920F TeRoTat1070R | CTGAAGAGGTTGGAAATGGAGAAG GTTTCGGTGGTTCTGTTGTTGTTA | RoTat 1.2 VSG | [95] |
| T. evansi (Real-time PCR) | PCR |
TeRTF TeRTR | GGAAGCAAAAGTCGTAACAAGG CCCATGTCAAACGGCATATAG | ITS1 | [2] |
| T. evansi (TaqMan PCR) | PCR |
F R Probe | ATAAATTGCACAGTATGCAACCAAA CATCCCTCATCTCCCATGTCA 6FAM-ACGGCATATAGAAACACA-MGBNFQ | ITS1 internal | [96] |
| T. evansi (Type A) | LAMP |
F R F3 B3 FIP (F1c + F2) BIP (B1c + B2) LF LB | CAAAACTAACAGCCGTTGCAGCG AGTTCCGGTACCTTCTCCATTTC GTAGGAAGCAACACCTGCG TTGATTAGTGCTGCGTGTGT TGCGAGGTGCACCTTGATGTTGAAGCAATAACCGGCAACGAC GAAGGCAAAGTTGACGACCAGCTGTGGTGTGCTTTTCCTTGT GCGATTTTGATCCCGCCG CAGAACGAGCAGAATTTTCCA | Rotat 1.2 VSG | [22] |
| Trypanosoma spp. | LAMP |
F3 B3 FIP (F1c + F2) BIP (B1c + B2) LF LB | CTGTCCGGTGATGTGGAAC CGTGCCTTCGTGAGAGTTTC GGAATACAGCAGATGGGGCGAGGCCAATTGGCATCTTTGGGA AAGGGAGACTCTGCCACAGTCGTCAGCCATCACCGTAGAGC GCCTCCCACCCTGGACTC AGACCGATAGCATCTCAG | RIME | [69] |
| T. evansi (Type A) | LAMP |
F3 B3 FIP (F1c + F2) BIP (B1c + B2) | TCACAACAAGACTCGCACG GGGCTTTGATCTGCTCCTC TCAGAAGCGTCGAGCTGGGATTTTATCGACAATGCCATCGCC CGCAAGTTCCTGTGGCTGCATTTTTTCCCAAGAAGAGCCGTCT | PFR A1 | [67] |
| T. evansi (Type B) | LAMP |
F3 B3 FIP (F1c + F2) BIP (B1c + B2) LF LB | CCAATCAAAGACGAGCGG TGGTTTGTGAGGCCGCAG CGGATGCATCGGTGATGCAATCACTACTGCATCAAGGGAAGC ATCCAGCACCTCGGAACAGCTCTCGGCAACCAGATCGG GTTCACGTGCCTCCGCTTC ACGTAGCGGGAAAATACGC | VSG JN 2118Hu | [80] |
| T. evansi (Type A) | RPA |
TevRPA-Fw TevRPA-Rv | CACCGAAGCAAGCGCAGCAAGAGGGTTAGCA GTAGCTGTCTCCTGGGGCCGAGGTGTCATAG | Rotat 1.2 VSG | [40] |
6. Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Habila, N.; Inuwa, M.H.; Aimola, I.A.; Udeh, M.U.; Haruna, E. Pathogenic Mechanisms of Trypanosoma evansi Infections. Res. Vet. Sci. 2012, 93, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, A.T.; Monje, L.D.; Zurvera, D.A.; Beldomenico, P.M. Detection of Trypanosoma evansi Infection in Wild Capybaras from Argentina Using Smear Microscopy and Real-Time PCR Assays. Vet. Parasitol. 2014, 202, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Desquesnes, M.; Holzmuller, P.; Lai, D.-H.; Dargantes, A.; Lun, Z.-R.; Jittaplapong, S. Trypanosoma evansi and Surra: A Review and Perspectives on Origin, History, Distribution, Taxonomy, Morphology, Hosts, and Pathogenic Effects. BioMed Res. Int. 2013, 2013, 194176. [Google Scholar] [CrossRef] [PubMed]
- Aregawi, W.G.; Agga, G.E.; Abdi, R.D.; Büscher, P. Systematic Review and Meta-Analysis on the Global Distribution, Host Range, and Prevalence of Trypanosoma evansi. Parasit. Vectors 2019, 12, 67. [Google Scholar] [CrossRef]
- Ramírez-Iglesias, J.R.; Eleizalde, M.C.; Gómez-Piñeres, E.; Mendoza, M. Trypanosoma evansi: A Comparative Study of Four Diagnostic Techniques for Trypanosomosis Using Rabbit as an Experimental Model. Exp. Parasitol. 2011, 128, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Luckins, A.G. Trypanosoma evansi in Asia. Parasitol. Today 1988, 4, 137–142. [Google Scholar] [CrossRef]
- Gutierrez, C.; Desquesnes, M.; Touratier, L.; Büscher, P. Trypanosoma evansi: Recent Outbreaks in Europe. Vet. Parasitol. 2010, 174, 26–29. [Google Scholar] [CrossRef]
- Salman, D.; Sivakumar, T.; Otgonsuren, D.; Mahmoud, M.E.; Elmahallawy, E.K.; Khalphallah, A.; Kounour, A.M.E.Y.; Bayomi, S.A.; Igarashi, M.; Yokoyama, N. Molecular Survey of Babesia, Theileria, Trypanosoma, and Anaplasma Infections in Camels (Camelus dromedaries) in Egypt. Parasitol. Int. 2022, 90, 102618. [Google Scholar] [CrossRef]
- Brun, R.; Hecker, H.; Lun, Z.-R. Trypanosoma evansi and T. equiperdum: Distribution, Biology, Treatment and Phylogenetic Relationship (a Review). Vet. Parasitol. 1998, 79, 95–107. [Google Scholar] [CrossRef]
- Inoue, N.; Tafalla Lluz, A.; Mori, T.; Nagasawa, H.; Fujisaki, K.; Mikami, T. Novel Species Specific Antigens of Trypanosoma congolense and Their Different Localization among Life-Cycle Stages. J. Vet. Med. Sci. 2000, 62, 1041–1045. [Google Scholar] [CrossRef][Green Version]
- Stevens, J.R.; Brisse, S. Systematics of Trypanosomes of Medical and Veterinary Importance. In The Trypanosomiases; Maudlin, I., Holmes, P.H., Miles, M.A., Eds.; CABI Publishing: Wallingford, UK, 2004; pp. 1–23. ISBN 978-0-85199-475-8. [Google Scholar]
- Borst, P.; Fase-Fowler, F.; Gibson, W.C. Kinetoplast DNA of Trypanosoma evansi. Mol. Biochem. Parasitol. 1987, 23, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Salim, B.; Hayashida, K.; Mossaad, E.; Nakao, R.; Yamagishi, J.; Sugimoto, C. Development and Validation of Direct Dry Loop Mediated Isothermal Amplification for Diagnosis of Trypanosoma evansi. Vet. Parasitol. 2018, 260, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Lun, Z.-R.; Desser, S.S. Is the Broad Range of Hosts and Geographical Distribution of Trypanosoma evansi Attributable to the Loss of Maxicircle Kinetoplast DNA? Parasitol. Today 1995, 11, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Lai, D.-H.; Hashimi, H.; Lun, Z.-R.; Ayala, F.J.; Lukeš, J. Adaptations of Trypanosoma brucei to Gradual Loss of Kinetoplast DNA: Trypanosoma equiperdum and Trypanosoma evansi Are Petite Mutants of T. brucei. Proc. Natl. Acad. Sci. USA 2008, 105, 1999–2004. [Google Scholar] [CrossRef] [PubMed]
- Njiru, Z.K.; Constantine, C.C.; Ndung’u, J.M.; Robertson, I.; Okaye, S.; Thompson, R.C.A.; Reid, S.A. Detection of Trypanosoma evansi in Camels Using PCR and CATT/T. evansi Tests in Kenya. Vet. Parasitol. 2004, 124, 187–199. [Google Scholar] [CrossRef]
- Claes, F.; Radwanska, M.; Urakawa, T.; Majiwa, P.A.; Goddeeris, B.; Büscher, P. Variable Surface Glycoprotein RoTat 1.2 PCR as a Specific Diagnostic Tool for the Detection of Trypanosoma evansi Infections. Kinetoplastid Biol. Dis. 2004, 3, 3. [Google Scholar] [CrossRef]
- Holland, W.G.; My, L.N.; Dung, T.V.; Thanh, N.G.; Tam, P.T.; Vercruysse, J.; Goddeeris, B.M. The Influence of T. evansi Infection on the Immuno-Responsiveness of Experimentally Infected Water Buffaloes. Vet. Parasitol. 2001, 102, 225–234. [Google Scholar] [CrossRef]
- Powar, R.; Shegokar, V.; Joshi, P.; Dani, V.; Tankhiwale, N.; Truc, P.; Jannin, J.; Bhargava, A. A Rare Case of Human Trypanosomiasis Caused By Trypanosoma evansi. Indian J. Med Microbiol. 2006, 24, 72–74. [Google Scholar] [CrossRef]
- Truc, P.; Salkar, H.R.; Joshi, P.P.; Dani, V.S.; Shegokar, V.R.; Bhargava, A.; Powar, R.M.; Herder, S.; Jannin, J.; Katti, R. Human Trypanosomiasis Caused By Trypanosoma evansi in India: The First Case Report. Am. J. Trop. Med. Hyg. 2005, 73, 491–495. [Google Scholar] [CrossRef]
- Van Vinh Chau, N.; Buu Chau, L.; Desquesnes, M.; Herder, S.; Phu Huong Lan, N.; Campbell, J.I.; Van Cuong, N.; Yimming, B.; Chalermwong, P.; Jittapalapong, S.; et al. A Clinical and Epidemiological Investigation of the First Reported Human Infection With the Zoonotic Parasite Trypanosoma evansi in Southeast Asia. Clin. Infect. Dis. 2016, 62, 1002–1008. [Google Scholar] [CrossRef]
- Tong, Q.; Chen, R.; Kong, Q.; Goossens, J.; Radwanska, M.; Lou, D.; Ding, J.; Zheng, B.; Fu, Y.; Wang, T.; et al. DNA Detection of Trypanosoma evansi: Diagnostic Validity of a New Assay Based on Loop-Mediated Isothermal Amplification (LAMP). Vet. Parasitol. 2018, 250, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Herrera, H.M.; Dávila, A.M.R.; Norek, A.; Abreu, U.G.; Souza, S.S.; D’Andrea, P.S.; Jansen, A.M. Enzootiology of Trypanosoma evansi in Pantanal, Brazil. Vet. Parasitol. 2004, 125, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Faria, J.; Briggs, E.M.; Black, J.A.; McCulloch, R. Emergence and Adaptation of the Cellular Machinery Directing Antigenic Variation in the African Trypanosome. Curr. Opin. Microbiol. 2022, 70, 102209. [Google Scholar] [CrossRef] [PubMed]
- Macleod, O.J.S.; Cook, A.D.; Webb, H.; Crow, M.; Burns, R.; Redpath, M.; Seisenberger, S.; Trevor, C.E.; Peacock, L.; Schwede, A.; et al. Invariant Surface Glycoprotein 65 of Trypanosoma brucei Is a Complement C3 Receptor. Nat. Commun. 2022, 13, 5085. [Google Scholar] [CrossRef] [PubMed]
- Bangs, J.D. Evolution of Antigenic Variation in African Trypanosomes: Variant Surface Glycoprotein Expression, Structure, and Function. BioEssays 2018, 40, 1800181. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.T.; Guevarra, R.B.; Magez, S.; Radwanska, M. Single-Cell Transcriptome Profiling and the Use of AID Deficient Mice Reveal That B Cell Activation Combined with Antibody Class Switch Recombination and Somatic Hypermutation Do Not Benefit the Control of Experimental Trypanosomosis. PLoS Pathog. 2021, 17, e1010026. [Google Scholar] [CrossRef]
- Nguyen, H.T.T.; Radwanska, M.; Magez, S. Tipping the Balance between Erythroid Cell Differentiation and Induction of Anemia in Response to the Inflammatory Pathology Associated with Chronic Trypanosome Infections. Front. Immunol. 2022, 13, 1051647. [Google Scholar] [CrossRef]
- Radwanska, M.; Guirnalda, P.; De Trez, C.; Ryffel, B.; Black, S.; Magez, S. Trypanosomiasis-Induced B Cell Apoptosis Results in Loss of Protective Anti-Parasite Antibody Responses and Abolishment of Vaccine-Induced Memory Responses. PLoS Pathog. 2008, 4, e1000078. [Google Scholar] [CrossRef]
- Frenkel, D.; Zhang, F.; Guirnalda, P.; Haynes, C.; Bockstal, V.; Radwanska, M.; Magez, S.; Black, S.J. Trypanosoma brucei Co-Opts NK Cells to Kill Splenic B2 B Cells. PLoS Pathog. 2016, 12, e1005733. [Google Scholar] [CrossRef]
- Radwanska, M.; Nguyen, H.T.T.; Magez, S. African Trypanosomosis Obliterates DTPa Vaccine-Induced Functional Memory So That Post-Treatment Bordetella Pertussis Challenge Fails to Trigger a Protective Recall Response. Vaccines 2021, 9, 603. [Google Scholar] [CrossRef]
- Moon, S.; Janssens, I.; Kim, K.H.; Stijlemans, B.; Magez, S.; Radwanska, M. Detrimental Effect of Trypanosoma brucei brucei Infection on Memory B Cells and Host Ability to Recall Protective B-Cell Responses. J. Infect. Dis. 2022, 226, 528–540. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.T.; Magez, S.; Radwanska, M. From Helping to Regulating—A Transcriptomic Profile of Ifng+ Il10+ Il21+ Cd4+ Th1 Cells Indicates Their Role in Regulating Inflammation during Experimental Trypanosomosis. Front. Trop. Dis. 2023, 4, 1127022. [Google Scholar] [CrossRef]
- Álvarez-Rodríguez, A.; Jin, B.-K.; Radwanska, M.; Magez, S. Recent Progress in Diagnosis and Treatment of Human African Trypanosomiasis Has Made the Elimination of This Disease a Realistic Target by 2030. Front. Med. 2022, 9, 1037094. [Google Scholar] [CrossRef] [PubMed]
- Fernández, D.; González-Baradat, B.; Eleizalde, M.; González-Marcano, E.; Perrone, T.; Mendoza, M. Trypanosoma evansi: A Comparison of PCR and Parasitological Diagnostic Tests in Experimentally Infected Mice. Exp. Parasitol. 2009, 121, 1–7. [Google Scholar] [CrossRef]
- Birhanu, H.; Gebrehiwot, T.; Goddeeris, B.M.; Büscher, P.; Van Reet, N. New Trypanosoma evansi Type B Isolates from Ethiopian Dromedary Camels. PLoS Negl. Trop. Dis. 2016, 10, e0004556. [Google Scholar] [CrossRef] [PubMed]
- Behour, T.S.; Abd El Fattah, E.M. Genotyping of Trypanosoma brucei evansi in Egyptian Camels: Detection of a Different Non-RoTat 1.2 Trypanosoma brucei evansi in Egyptian Camels. Trop. Anim. Health Prod. 2023, 55, 279. [Google Scholar] [CrossRef]
- Crannell, Z.; Castellanos-Gonzalez, A.; Nair, G.; Mejia, R.; White, A.C.; Richards-Kortum, R. Multiplexed Recombinase Polymerase Amplification Assay To Detect Intestinal Protozoa. Anal. Chem. 2016, 88, 1610–1616. [Google Scholar] [CrossRef]
- Giordani, F.; Morrison, L.J.; Rowan, T.G.; De Koning, H.P.; Barrett, M.P. The Animal Trypanosomiases and Their Chemotherapy: A Review. Parasitology 2016, 143, 1862–1889. [Google Scholar] [CrossRef]
- Li, Z.; Pinto Torres, J.E.; Goossens, J.; Stijlemans, B.; Sterckx, Y.G.-J.; Magez, S. Development of a Recombinase Polymerase Amplification Lateral Flow Assay for the Detection of Active Trypanosoma evansi Infections. PLoS Negl. Trop. Dis. 2020, 14, e0008044. [Google Scholar] [CrossRef]
- Verloo, D.; Magnus, E.; Büscher, P. General Expression of RoTat 1.2 Variable Antigen Type in Trypanosoma evansi Isolates from Different Origin. Vet. Parasitol. 2001, 97, 185–191. [Google Scholar] [CrossRef]
- Ahmed, H.A.; Picozzi, K.; Welburn, S.C.; MacLeod, E.T. A Comparative Evaluation of PCR-Based Methods for Species-Specific Determination of African Animal Trypanosomes in Ugandan Cattle. Parasit. Vectors 2013, 6, 316. [Google Scholar] [CrossRef] [PubMed]
- Fikru, R.; Andualem, Y.; Getachew, T.; Menten, J.; Hasker, E.; Merga, B.; Goddeeris, B.M.; Büscher, P. Trypanosome Infection in Dromedary Camels in Eastern Ethiopia: Prevalence, Relative Performance of Diagnostic Tools and Host Related Risk Factors. Vet. Parasitol. 2015, 211, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Pathak, K.M.L.; Singh, Y.; Meirvenne, N.V.; Kapoor, M. Evaluation of Various Diagnostic Techniques for Trypanosoma evansi Infections in Naturally Infected Camels. Vet. Parasitol. 1997, 69, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Luckins, A.G.; Boid, R.; Rae, P.; Mahmoud, M.M.; El Malik, K.H.; Gray, A.R. Serodiagnosis of Infection with Trypanosoma evansi in Camels in the Sudan. Trop. Anim. Health Prod. 1979, 11, 1–12. [Google Scholar] [CrossRef]
- Reid, S.A.; Copeman, D.B. The Development and Validation of an Antibody-ELISA to Detect Trypanosoma evansi Infection in Cattle in Australia and Papua New Guinea. Prev. Vet. Med. 2003, 61, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Mossaad, E.; Salim, B.; Suganuma, K.; Hassan, M.A.; Davaasuren, B.; Elamin, E.A.; Bakhiet, A.O.; Satti, R.A.; Xuan, X.; Musinguzi, S.P.; et al. Utilization of Crude and Recombinant ELISAs for Serodiagnosis of Camel Trypanosomosis in Sudan. Vet. Parasitol. Reg. Stud. Rep. 2019, 16, 100278. [Google Scholar] [CrossRef] [PubMed]
- Ngaira, J.M.; Bett, B.; Karanja, S.M.; Njagi, E.N.M. Evaluation of Antigen and Antibody Rapid Detection Tests for Trypanosoma evansi Infection in Camels in Kenya. Vet. Parasitol. 2003, 114, 131–141. [Google Scholar] [CrossRef]
- Dia, M.L.; Diop, C.; Aminetou, M.; Jacquiet, P.; Thiam, A. Some Factors Affecting the Prevalence of Trypanosoma evansi in Camels in Mauritania. Vet. Parasitol. 1997, 72, 111–120. [Google Scholar] [CrossRef]
- Lejon, V.; Claes, F.; Verloo, D.; Maina, M.; Urakawa, T.; Majiwa, P.A.O.; Büscher, P. Recombinant RoTat 1.2 Variable Surface Glycoprotein as Antigen for Diagnosis of Trypanosoma evansi in Dromedary Camels. Int. J. Parasitol. 2005, 35, 455–460. [Google Scholar] [CrossRef]
- Espino, A.M.; Millán, J.C.; Finlay, C.M. Detection of Antibodies and Circulating Excretory-Secretory Antigens for Assessing Cure in Patients with Fascioliasis. Trans. R. Soc. Trop. Med. Hyg. 1992, 86, 649. [Google Scholar] [CrossRef] [PubMed]
- Reyna-Bello, A.; García, F.A.; Rivera, M.; Sansó, B.; Aso, P.M. Enzyme-Linked Immunosorbent Assay (ELISA) for Detection of Anti-Trypanosoma evansi Equine Antibodies. Vet. Parasitol. 1998, 80, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Holland, W.G.; Thanh, N.G.; Do, T.T.; Sangmaneedet, S.; Goddeeris, B.; Vercruysse, J. Evaluation of Diagnostic Tests for Trypanosoma evansi in Experimentally Infected Pigs and Subsequent Use in Field Surveys in North Vietnam and Thailand. Trop. Anim. Health Prod. 2005, 37, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.; Claes, F.; Verloo, D.; De Greve, H.; Büscher, P. Towards a New Reference Test for Surra in Camels. Clin. Vaccine Immunol. 2009, 16, 999–1002. [Google Scholar] [CrossRef] [PubMed]
- Olaho-Mukani, W.; Munyua, W.K.; Mutugi, M.W.; Njogu, A.R. Comparison of Antibody- and Antigen-Detection Enzyme Immunoassays for the Diagnosis of Trypanosoma evansi Infections in Camels. Vet. Parasitol. 1993, 45, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Verloo, D.; Holland, W.; My, L.N.; Thanh, N.G.; Tam, P.T.; Goddeeris, B.; Vercruysse, J.; Büscher, P. Comparison of Serological Tests for Trypanosoma evansi Natural Infections in Water Buffaloes from North Vietnam. Vet. Parasitol. 2000, 92, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Birhanu, H.; Rogé, S.; Simon, T.; Baelmans, R.; Gebrehiwot, T.; Goddeeris, B.M.; Büscher, P. Surra Sero K-SeT, a New Immunochromatographic Test for Serodiagnosis of Trypanosoma evansi Infection in Domestic Animals. Vet. Parasitol. 2015, 211, 153–157. [Google Scholar] [CrossRef]
- Peng, Y.; Wu, J.; Wang, J.; Li, W.; Yu, S. Study and Evaluation of Wondfo Rapid Diagnostic Kit Based on Nano-Gold Immunochromatography Assay for Diagnosis of Plasmodium Falciparum. Parasitol. Res. 2012, 110, 1421–1425. [Google Scholar] [CrossRef]
- Preechakasedkit, P.; Pinwattana, K.; Dungchai, W.; Siangproh, W.; Chaicumpa, W.; Tongtawe, P.; Chailapakul, O. Development of a One-Step Immunochromatographic Strip Test Using Gold Nanoparticles for the Rapid Detection of Salmonella Typhi in Human Serum. Biosens. Bioelectron. 2012, 31, 562–566. [Google Scholar] [CrossRef]
- Reid, S.A.; Copeman, D.B. Evaluation of an Antibody-ELISA Using Five Crude Antigen Preparations for the Diagnosis of Trypanosoma evansi Infection in Cattle. Vet. Parasitol. 2002, 104, 79–84. [Google Scholar] [CrossRef]
- Monzon, C.M.; Jara, A.; Nantulya, V.M. Sensitivity of Antigen ELISA Test for Detecting Trypanosoma evansi Antigen in Horses in the Subtropical Area of Argentina. J. Parasitol. 1995, 81, 806. [Google Scholar] [CrossRef]
- Mullis, K.; Faloona, F.; Scharf, S.; Saiki, R.; Horn, G.; Erlich, H. Specific Enzymatic Amplification of DNA In Vitro: The Polymerase Chain Reaction. Cold Spring Harb. Symp. Quant. Biol. 1986, 51, 263–273. [Google Scholar] [CrossRef]
- Baticados, W.N.; Castro, D.L.; Baticados, A.M. Parasitological and PCR Detection of Trypanosoma evansi in Buffaloes from Luzon, Philippines. Ceylon J. Sci. Biol. Sci. 2012, 40, 141–146. [Google Scholar] [CrossRef]
- Pruvot, M.; Kamyingkird, K.; Desquesnes, M.; Sarataphan, N.; Jittapalapong, S. A Comparison of Six Primer Sets for Detection of Trypanosoma evansi by Polymerase Chain Reaction in Rodents and Thai Livestock. Vet. Parasitol. 2010, 171, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Ngaira, J.M.; Olembo, N.K.; Njagi, E.N.M.; Ngeranwa, J.J.N. The Detection of Non-RoTat 1.2 Trypanosoma evansi. Exp. Parasitol. 2005, 110, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Keatley, S.; Botero, A.; Fosu-Nyarko, J.; Pallant, L.; Northover, A.; Thompson, R.C.A. Species-Level Identification of Trypanosomes Infecting Australian Wildlife by High-Resolution Melting—Real Time Quantitative Polymerase Chain Reaction (HRM-qPCR). Int. J. Parasitol. Parasites Wildl. 2020, 13, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Thekisoe, O.M.M.; Inoue, N.; Kuboki, N.; Tuntasuvan, D.; Bunnoy, W.; Borisutsuwan, S.; Igarashi, I.; Sugimoto, C. Evaluation of Loop-Mediated Isothermal Amplification (LAMP), PCR and Parasitological Tests for Detection of Trypanosoma evansi in Experimentally Infected Pigs. Vet. Parasitol. 2005, 130, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Njiru, Z.K.; Mikosza, A.S.J.; Matovu, E.; Enyaru, J.C.K.; Ouma, J.O.; Kibona, S.N.; Thompson, R.C.A.; Ndung’u, J.M. African Trypanosomiasis: Sensitive and Rapid Detection of the Sub-Genus Trypanozoon by Loop-Mediated Isothermal Amplification (LAMP) of Parasite DNA. Int. J. Parasitol. 2008, 38, 589–599. [Google Scholar] [CrossRef]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase Polymerase Amplification: Basics, Applications and Recent Advances. TrAC Trends Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef]
- Kuboki, N.; Inoue, N.; Sakurai, T.; Di Cello, F.; Grab, D.J.; Suzuki, H.; Sugimoto, C.; Igarashi, I. Loop-Mediated Isothermal Amplification for Detection of African Trypanosomes. J. Clin. Microbiol. 2003, 41, 5517–5524. [Google Scholar] [CrossRef]
- Joung, J.; Ladha, A.; Saito, M.; Kim, N.-G.; Woolley, A.E.; Segel, M.; Barretto, R.P.J.; Ranu, A.; Macrae, R.K.; Faure, G.; et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N. Engl. J. Med. 2020, 383, 1492–1494. [Google Scholar] [CrossRef] [PubMed]
- Thekisoe, O.M.M.; Kuboki, N.; Nambota, A.; Fujisaki, K.; Sugimoto, C.; Igarashi, I.; Yasuda, J.; Inoue, N. Species-Specific Loop-Mediated Isothermal Amplification (LAMP) for Diagnosis of Trypanosomosis. Acta Trop. 2007, 102, 182–189. [Google Scholar] [CrossRef]
- Hughes, A.L.; Piontkivska, H. Molecular Phylogenetics of Trypanosomatidae: Contrasting Results from 18S rRNA and Protein Phylogenies. Kinetoplastid Biol. Dis. 2003, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Rivero, R.; Bisio, M.; Velázquez, E.B.; Esteva, M.I.; Scollo, K.; González, N.L.; Altcheh, J.; Ruiz, A.M. Rapid Detection of Trypanosoma cruzi by Colorimetric Loop-Mediated Isothermal Amplification (LAMP): A Potential Novel Tool for the Detection of Congenital Chagas Infection. Diagn. Microbiol. Infect. Dis. 2017, 89, 26–28. [Google Scholar] [CrossRef] [PubMed]
- Grab, D.J.; Nikolskaia, O.V.; Courtioux, B.; Thekisoe, O.M.M.; Magez, S.; Bogorad, M.; Dumler, J.S.; Bisser, S. Using Detergent-Enhanced LAMP for African Trypanosome Detection in Human Cerebrospinal Fluid and Implications for Disease Staging. PLoS Negl. Trop. Dis. 2019, 13, e0007631. [Google Scholar] [CrossRef] [PubMed]
- Grab, D.J.; Nikolskaia, O.V.; Inoue, N.; Thekisoe, O.M.M.; Morrison, L.J.; Gibson, W.; Dumler, J.S. Using Detergent to Enhance Detection Sensitivity of African Trypanosomes in Human CSF and Blood by Loop-Mediated Isothermal Amplification (LAMP). PLoS Negl. Trop. Dis. 2011, 5, e1249. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mori, Y.; Nagamine, K.; Tomita, N.; Notomi, T. Detection of Loop-Mediated Isothermal Amplification Reaction by Turbidity Derived from Magnesium Pyrophosphate Formation. Biochem. Biophys. Res. Commun. 2001, 289, 150–154. [Google Scholar] [CrossRef]
- Kumar, B.; Maharana, B.R.; Brahmbhatt, N.N.; Thakre, B.J.; Parmar, V.L. Development of a Loop-Mediated Isothermal Amplification Assay Based on RoTat1.2 Gene for Detection of Trypanosoma evansi in Domesticated Animals. Parasitol. Res. 2021, 120, 1873–1882. [Google Scholar] [CrossRef]
- Njiru, Z.K.; Ouma, J.O.; Enyaru, J.C.; Dargantes, A.P. Loop-Mediated Isothermal Amplification (LAMP) Test for Detection of Trypanosoma evansi Strain B. Exp. Parasitol. 2010, 125, 196–201. [Google Scholar] [CrossRef]
- Castellanos-Gonzalez, A.; White, A.C.; Melby, P.; Travi, B. Molecular Diagnosis of Protozoan Parasites by Recombinase Polymerase Amplification. Acta Trop. 2018, 182, 4–11. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA Detection Using Recombination Proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Liao, C.; Liang, L.; Yi, X.; Zhou, Z.; Wei, G. Recent Advances in Recombinase Polymerase Amplification: Principle, Advantages, Disadvantages and Applications. Front. Cell. Infect. Microbiol. 2022, 12, 1019071. [Google Scholar] [CrossRef] [PubMed]
- Lv, R.; Lu, N.; Wang, J.; Li, Y.; Qi, Y. Recombinase Polymerase Amplification for Rapid Detection of Zoonotic Pathogens: An Overview. Zoonoses 2022, 2, e990. [Google Scholar] [CrossRef]
- Njiru, Z.K.; Constantine, C.C.; Guya, S.; Crowther, J.; Kiragu, J.M.; Thompson, R.C.A.; Dávila, A.M.R. The Use of ITS1 rDNA PCR in Detecting Pathogenic African Trypanosomes. Parasitol. Res. 2005, 95, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Holland, W.G.; Claes, F.; My, L.N.; Thanh, N.G.; Tam, P.T.; Verloo, D.; Büscher, P.; Goddeeris, B.; Vercruysse, J. A Comparative Evaluation of Parasitological Tests and a PCR for Trypanosoma evansi Diagnosis in Experimentally Infected Water Buffaloes. Vet. Parasitol. 2001, 97, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Aradaib, I.E.; Majid, A.A. A Simple and Rapid Method for Detection of Trypanosoma evansi in the Dromedary Camel Using a Nested Polymerase Chain Reaction. Kinetoplastid Biol. Dis. 2006, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Masiga, D.K.; Smyth, A.J.; Hayes, P.; Bromidge, T.J.; Gibson, W.C. Sensitive Detection of Trypanosomes in Tsetse Flies by DNA Amplification. Int. J. Parasitol. 1992, 22, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Ali, N.O.; Croof, H.I.; Abdalla, H.S. Molecular Diagnosis of Trypanosoma evansi Infection in Camelus Dromedarius from Eastern and Western Regions of the Sudan. Emir. J. Food Agric. 2011, 23, 320–329. [Google Scholar]
- Sharma, A.; Singla, L.D.; Tuli, A.; Kaur, P.; Batth, B.K.; Javed, M.; Juyal, P.D. Molecular Prevalence of Babesia bigemina and Trypanosoma evansi in Dairy Animals from Punjab, India, by Duplex PCR: A Step Forward to the Detection and Management of Concurrent Latent Infections. BioMed Res. Int. 2013, 2013, 893862. [Google Scholar] [CrossRef]
- Rudramurthy, G.R.; Sengupta, P.P.; Balamurugan, V.; Prabhudas, K.; Rahman, H. PCR Based Diagnosis of Trypanosomiasis Exploring Invariant Surface Glycoprotein (ISG) 75 Gene. Vet. Parasitol. 2013, 193, 47–58. [Google Scholar] [CrossRef]
- Artama, W.T.; Agey, M.W.; Donelson, J.E. DNA Comparisons of Trypanosoma evansi (Indonesia) and Trypanosoma brucei Spp. Parasitology 1992, 104, 67–74. [Google Scholar] [CrossRef] [PubMed]
- International Atomic Energy Agency. Developing Methodologies for the Use of Ploymerase Chain Reaction in the Diagnosis and Monitoring of Trypanosomosis; Gardners Books: Eastbourne, UK, 2007; ISBN 978-92-0-105507-1. [Google Scholar]
- Sengupta, P.P.; Balumahendiran, M.; Suryanaryana, V.V.S.; Raghavendra, A.G.; Shome, B.R.; Gajendragad, M.R.; Prabhudas, K. PCR-Based Diagnosis of Surra-Targeting VSG Gene: Experimental Studies in Small Laboratory Rodents and Buffalo. Vet. Parasitol. 2010, 171, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Konnai, S.; Mekata, H.; Mingala, C.N.; Abes, N.S.; Gutierrez, C.A.; Herrera, J.R.V.; Dargantes, A.P.; Witola, W.H.; Cruz, L.C.; Inoue, N. Development and Application of a Quantitative Real-Time PCR for the Diagnosis of Surra in Water Buffaloes. Infect. Genet. Evol. 2009, 9, 449–452. [Google Scholar] [CrossRef] [PubMed]
- Taylor, T.K.; Boyle, D.B.; Bingham, J. Development of a TaqMan PCR Assay for the Detection of Trypanosoma evansi, the Agent of Surra. Vet. Parasitol. 2008, 153, 255–264. [Google Scholar] [CrossRef]
- Austen, J.M.; Barbosa, A.D. Diversity and Epidemiology of Bat Trypanosomes: A One Health Perspective. Pathogens 2021, 10, 1148. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Álvarez-Rodríguez, A.; Li, Z.; Radwanska, M.; Magez, S. Recent Progress in the Detection of Surra, a Neglected Disease Caused by Trypanosoma evansi with a One Health Impact in Large Parts of the Tropic and Sub-Tropic World. Microorganisms 2024, 12, 44. https://doi.org/10.3390/microorganisms12010044
Kim J, Álvarez-Rodríguez A, Li Z, Radwanska M, Magez S. Recent Progress in the Detection of Surra, a Neglected Disease Caused by Trypanosoma evansi with a One Health Impact in Large Parts of the Tropic and Sub-Tropic World. Microorganisms. 2024; 12(1):44. https://doi.org/10.3390/microorganisms12010044
Chicago/Turabian StyleKim, Jeongmin, Andrés Álvarez-Rodríguez, Zeng Li, Magdalena Radwanska, and Stefan Magez. 2024. "Recent Progress in the Detection of Surra, a Neglected Disease Caused by Trypanosoma evansi with a One Health Impact in Large Parts of the Tropic and Sub-Tropic World" Microorganisms 12, no. 1: 44. https://doi.org/10.3390/microorganisms12010044
APA StyleKim, J., Álvarez-Rodríguez, A., Li, Z., Radwanska, M., & Magez, S. (2024). Recent Progress in the Detection of Surra, a Neglected Disease Caused by Trypanosoma evansi with a One Health Impact in Large Parts of the Tropic and Sub-Tropic World. Microorganisms, 12(1), 44. https://doi.org/10.3390/microorganisms12010044







