Phenotypic Characterization and Draft Genome Sequence Analyses of Two Novel Endospore-Forming Sporosarcina spp. Isolated from Canada Goose (Branta canadensis) Feces
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Media Culture of Bacteria
2.2. Growth Inhibition Assays for Antibacterial Analyses of Isolates
2.3. Phenotype Analyses of Isolates
2.4. Bacterial DNA Purification and Draft Whole Genome Sequencing
2.5. Genome Sequence Analyses and Phylogenetic Analyses of Isolates’ Genomes
3. Results and Discussion
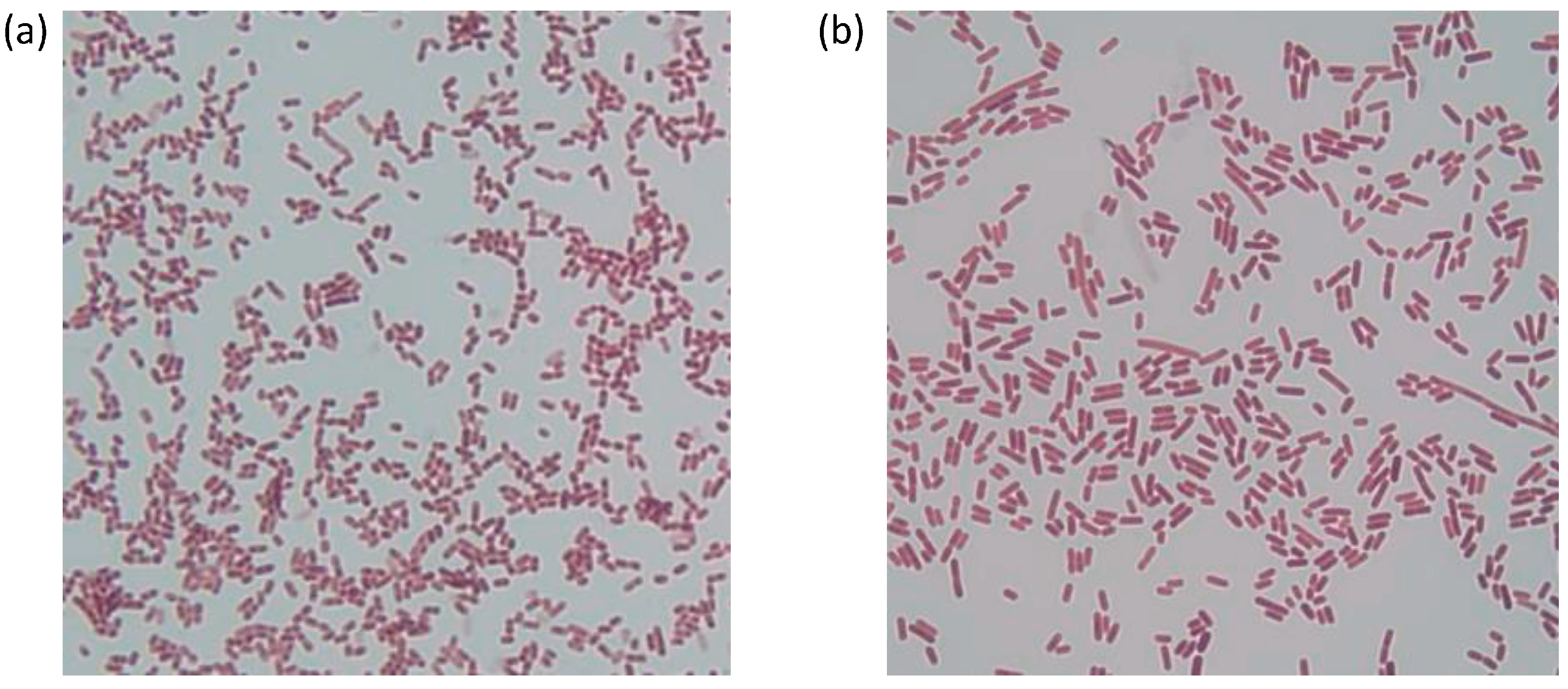
3.1. Phenotypic Characterization of Strains A4 and A15
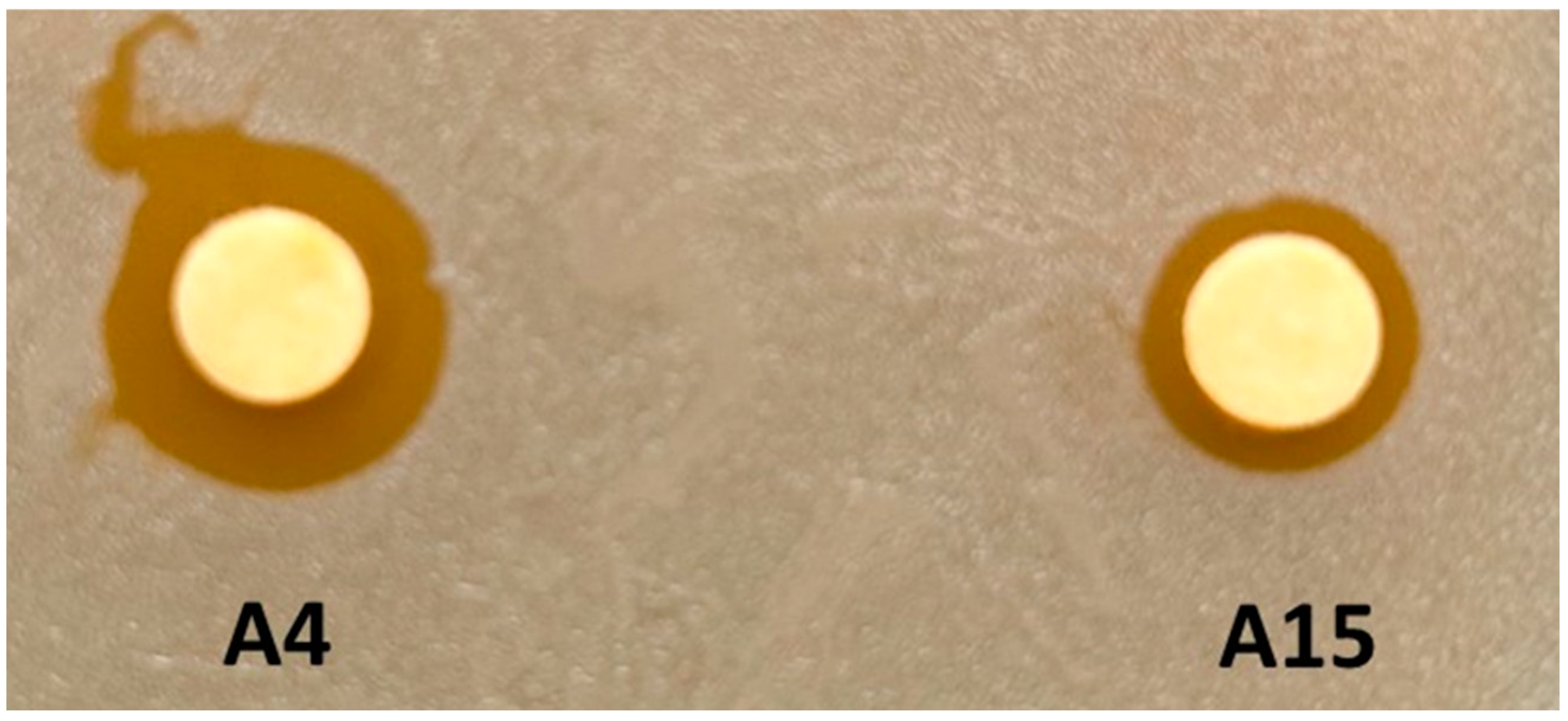
3.2. Growth Inhibition Assays of Target Bacteria for Strains A4 and A15
3.3. Genome Analyses of Strains A4 and A15
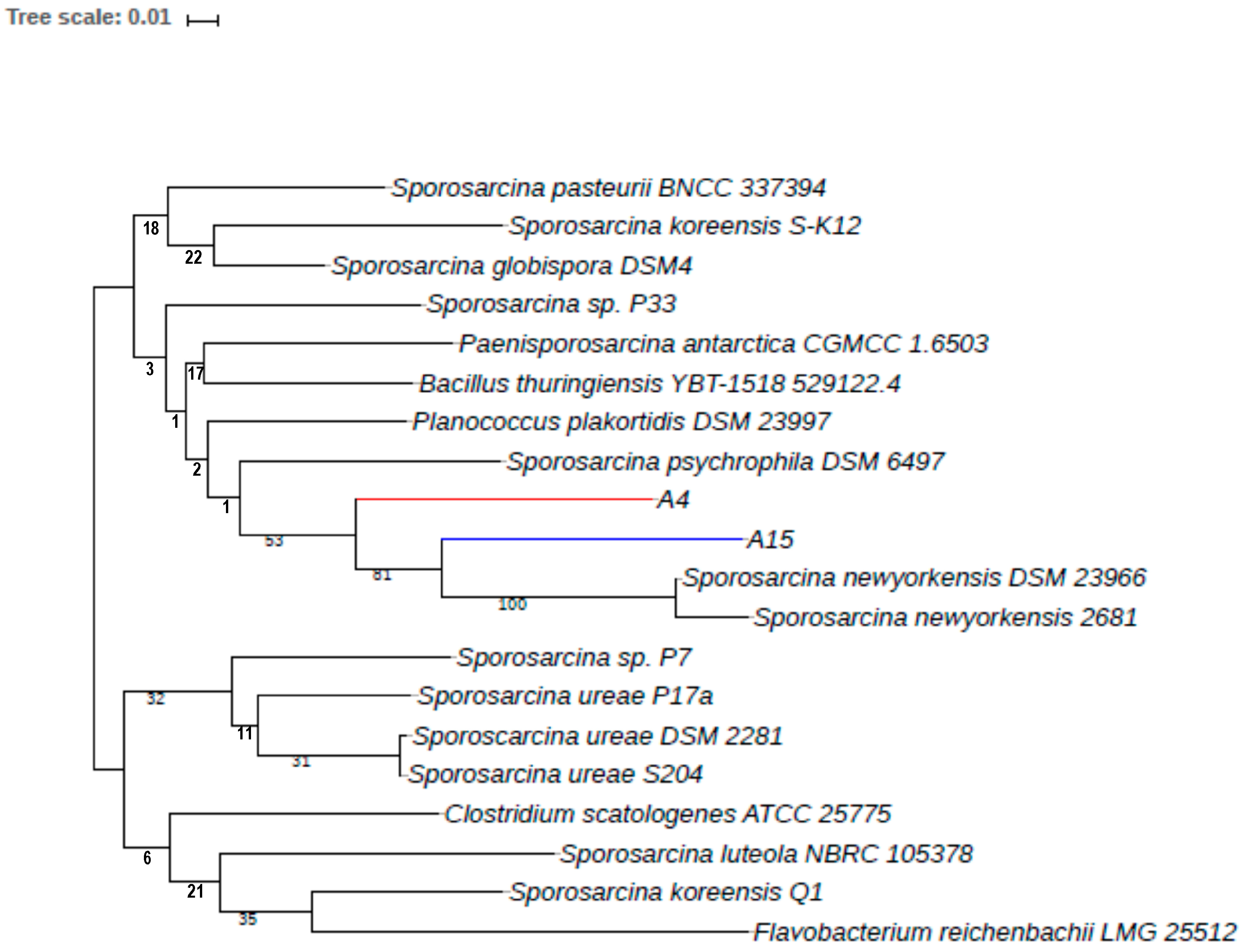
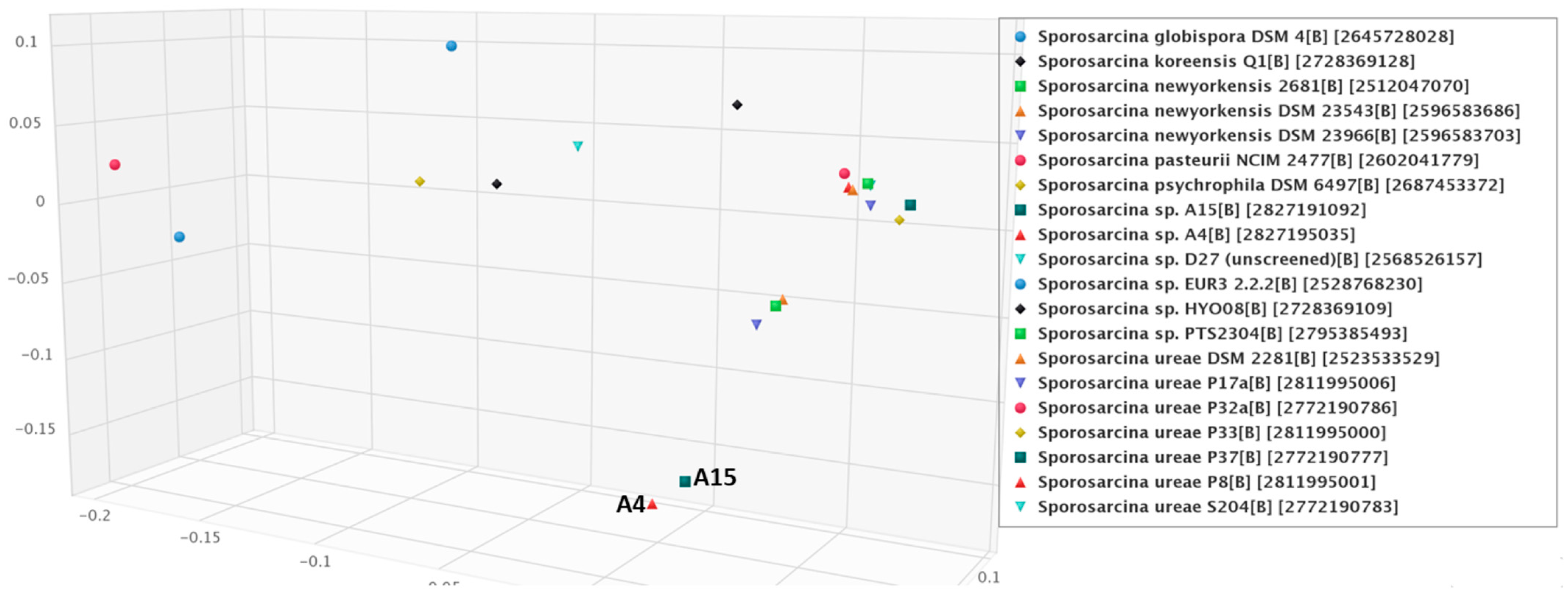
3.4. Phylogenetic Relationships among the Sporosarcina spp. with Isolates A4 and A15
4. Summary and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kluyver, A.J.; van Niel, C.B. Prospects for a natural system of classification of bacteria. Zentralbl. Bakteriol. Parasitenkd. Infektionskr. Hyg. 1936, 94, 369–403. [Google Scholar]
- Kämpfer, P.; Falsen, E.; Lodders, N.; Schumann, P. Sporosarcina contaminans sp. nov. and Sporosarcina thermotolerans sp. nov., two endospore-forming species. Int. J. Syst. Evol. Microbiol. 2010, 60, 1353–1357. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Xin, Y.H.; Liu, H.C.; Chen, B.; Sheng, J.; Chi, Z.M.; Zhou, P.J.; Zhang, D.C. Sporosarcina antarctica sp. nov., a psychrophilic bacterium isolated from the Antarctic. Int. J. Syst. Evol. Microbiol. 2008, 58, 2114–2117. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhao, Q.; Zhi, D.; Wang, Z.; Wang, Y.; Xie, Q.; Wu, Z.; Wang, X.; Li, Y.; Yu, L.; et al. Sporosarcina terrae sp. nov., isolated from orchard soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 2104–2108. [Google Scholar] [CrossRef] [PubMed]
- Claus, D.; Fahmy, F.; Rolf, H.J.; Tosunoglu, N. Sporosarcina halophila sp. nov., an obligate, slightly halophilic bacterium from salt marsh soils. Syst. Appl. Microbiol. 1983, 4, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.W.; Kim, B.Y.; Song, J.; Weon, H.Y.; Schumann, P.; Tindall, B.J.; Stackebrandt, E.; Fritze, D. Sporosarcina koreensis sp. nov. and Sporosarcina soli sp. nov., isolated from soil in Korea. Int. J. Syst. Evol. Microbiol. 2007, 57, 1694–1698. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Ren, H.; Chen, X.; Zhang, Y.; Yang, Y.; Wang, S.; Jiang, Y. Sporosarcina siberiensis sp. nov., isolated from the East Siberian Sea. Antonie Van Leeuwenhoek. 2014, 106, 489–495. [Google Scholar] [CrossRef] [PubMed]
- An, S.Y.; Haga, T.; Kasai, H.; Goto, K.; Yokota, A. Sporosarcina saromensis sp. nov., an aerobic endospore-forming bacterium. Int. J. Syst. Evol. Microbiol. 2007, 57 Pt 8, 1868–1871. [Google Scholar] [CrossRef]
- Yoon, J.H.; Lee, K.C.; Weiss, N.; Kho, Y.H.; Kang, K.H.; Park, Y.H. Sporosarcina aquimarina sp. nov., a bacterium isolated from seawater in Korea, and transfer of Bacillus globisporus (Larkin and Stokes 1967), Bacillus psychrophilus (Nakamura 1984) and Bacillus pasteurii (Chester 1898) to the genus Sporosarcina as Sporosarcina globispora comb. nov., Sporosarcina psychrophila comb. nov. and Sporosarcina pasteurii comb. nov. and emended description of th. Int. J. Syst. Evol. Microbiol. 2001, 51, 1079–1086. [Google Scholar]
- Wolfgang, W.J.; Coorevits, A.; Cole, J.A.; De Vos, P.; Dickinson, M.C.; Hennett, G.E.; Jose, R.; Nazarian, E.J.; Schumann, P.; Van Landschoot, A.; et al. Sporosarcina newyorkensis sp. nov. from clinical specimens and raw cow’s milk. Int. J. Syst. Evol. Microbiol. 2012, 62, 322–329. [Google Scholar] [CrossRef]
- Reddy, G.S.; Matsumoto, G.I.; Shivaji, S. Sporosarcina macmurdoensis sp. nov., from a cyanobacterial mat sample from a pond in the McMurdo Dry Valleys, Antarctica. Int. J. Syst. Evol. Microbiol. 2003, 53, 1363–1367. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, T.; An, S.Y.; Oyaizu, H.; Yokota, A. Sporosarcina luteola sp. nov. isolated from soy sauce production equipment in Japan. J. Gen. Appl. Microbiol. 2009, 55, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Zhang, R.; Li, X.; Gao, C.; Hu, X.; Hussain, S.; Zhang, L.; Wang, M.; Ma, X.; Pan, Q.; et al. Long-term simulated microgravity alters gut microbiota and metabolome in mice. Front. Microbiol. 2023, 14, 1100747. [Google Scholar] [CrossRef]
- Simpson, A.C.; Sengupta, P.; Zhang, F.; Hameed, A.; Parker, C.W.; Singh, N.K.; Miliotis, G.; Rekha, P.D.; Raman, K.; Mason, C.E.; et al. Phylogenomics, phenotypic, and functional traits of five novel (Earth-derived) bacterial species isolated from the International Space Station and their prevalence in metagenomes. Sci. Rep. 2023, 13, 19207. [Google Scholar] [CrossRef] [PubMed]
- Chuo, S.C.; Mohamed, S.F.; Mohd Setapar, S.H.; Ahmad, A.; Jawaid, M.; Wani, W.A.; Yaqoob, A.A.; Mohamad Ibrahim, M.N. Insights into the current trends in the utilization of bacteria for microbially induced calcium carbonate precipitation. Materials 2020, 13, 4993. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, H.; Li, Y. Biomineralization induced by cells of Sporosarcina pasteurii: Mechanisms, applications and challenges. Microorganisms 2021, 9, 2396. [Google Scholar] [CrossRef]
- Saif, A.; Cuccurullo, A.; Gallipoli, D.; Perlot, C.; Bruno, A.W. Advances in enzyme induced carbonate precipitation and application to soil improvement: A review. Materials 2022, 15, 950. [Google Scholar] [CrossRef]
- Ma, L.; Pang, A.P.; Luo, Y.; Lu, X.; Lin, F. Beneficial factors for biomineralization by ureolytic bacterium Sporosarcina pasteurii. Microb. Cell Fact. 2020, 19, 12. [Google Scholar] [CrossRef]
- Garrity, G.M. Bergey’s Manual of Systematic Bacteriology: The Firmicutes. In Bergey’s Manual of Systematic Bacteriology; Vos, P., Garrity, G.M., Jones, D., Krieg, N.R., Ludwig, W., Rainey, F.A., Schleifer, K.-H., Whitman, W.B., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; Volume 3. [Google Scholar]
- Priyodip, P.; Balaji, S. A preliminary study on probiotic characteristics of Sporosarcina spp. for poultry applications. Curr. Microbiol. 2019, 76, 448–461. [Google Scholar] [CrossRef]
- Wise, M.G.; Siragusa, G.R. Quantitative analysis of the intestinal bacterial community in one- to three-week-old commercially reared broiler chickens fed conventional or antibiotic-free vegetable-based diets. J. Appl. Microbiol. 2007, 102, 1138–1149. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Mitsuoka, T. Production of gnotobiotic mice with normal physiological functions. I. Selection of useful bacteria from feces of conventional mice. Z. Versuchstierkd. 1980, 22, 173–178. [Google Scholar] [PubMed]
- Summanen, P.; Baron, E.J.; Citron, D.M.; Strong, C.A.; Wexler, H.M.; Finegold, S.M. Wadsworth Anaerobic Bacteriology Manual, 5th ed.; Star Publishing: Belmont, CA, USA, 1993. [Google Scholar]
- Hayden, R.L. Elective localization in the eye of bacteria from infected teeth. Arch. Int. Med. 1923, 32, 828–849. [Google Scholar] [CrossRef]
- Levine, M. Differentiation of B. coli and B. aerogenes on a simplified eosin-methylene blue agar. J. Infect. Dis. 1918, 23, 43–47. [Google Scholar] [CrossRef]
- Chapman, G.H. The significance of sodium chloride in studies of staphylococci. J. Bacteriol. 1945, 50, 201–203. [Google Scholar] [CrossRef] [PubMed]
- Beveridge, T.J. Mechanism of Gram Variability in Select Bacteria J. Bacteriol. 1990, 172, 1609–1620. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 36, 493–496. [Google Scholar] [CrossRef]
- Star, M.P. Spirit blue agar: A medium for the detection of lipolytic microorganisms. Science 1941, 93, 333–334. [Google Scholar] [CrossRef]
- Flint, S.; Hartley, N. A modified selective medium for the detection of Pseudomonas species that cause spoilage of milk and dairy products. Int. Dairy J. 1996, 2, 223–230. [Google Scholar] [CrossRef]
- Hardy, B.L.; Bansal, G.; Hewlett, K.H.; Arora, A.; Schaffer, S.D.; Kamau, E.; Bennett, J.W.; Merrell, D.S. Antimicrobial activity of clinically isolated bacterial species against Staphylococcus aureus. Front. Microbiol. 2020, 10, 2977. [Google Scholar] [CrossRef]
- Bochner, B.R.; Gadzinski, P.; Panomitros, E. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001, 11, 1246–1255. [Google Scholar] [CrossRef] [PubMed]
- Shea, A.; Wolcott, M.; Daefler, S.; Rozak, D.A. Biolog phenotype microarrays. Methods Mol. Biol. 2012, 881, 331–373. [Google Scholar] [PubMed]
- Mackie, A.M.; Hassan, K.A.; Paulsen, I.T.; Tetu, S.G. Biolog Phenotype Microarrays for phenotypic characterization of microbial cells. Methods Mol. Biol. 2014, 1096, 123–130. [Google Scholar] [PubMed]
- Stackebrandt, E.; Frederiksen, W.; Garrity, G.M.; Grimont, P.A.; Kämpfer, P.; Maiden, M.C.J.; Nesme, X.; Rosselló-Mora, R.; Swings, J.; Trüper, H.G.; et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 2002, 52, 1043–1047. [Google Scholar] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.A.; Chu, K.; Palaniappan, K.; Ratner, A.; Huang, J.; Huntemann, M.; Hajek, P.; Ritter, S.; Varghese, N.; Seshadri, R.; et al. The IMG/M data management and analysis system v.6.0: New tools and advanced capabilities. Nucleic Acids Res. 2021, 47, D666–D677. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Gruber-Vodicka, H.R.; Seah, B.K.B.; Pruesse, E. PhyloFlash: Rapid small-subunit rRNA profiling and targeted assembly from metagenomes. mSystems 2020, 5, e00920-20. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Medlar, A.J.; Törönen, P.; Holm, L. AAI-profiler: Fast proteome-wide exploratory analysis reveals taxonomic identity, misclassification, and contamination. Nucleic Acids Res. 2018, 46, W479–W485. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Janarthine, S.R.S.; Eganathan, P. Plant Growth Promoting of Endophytic Sporosarcina aquimarina SjAM16103 Isolated from the Pneumatophores of Avicennia marina L. Int. J. Microbiol. 2012, 2012, 532060. [Google Scholar] [CrossRef] [PubMed]
- Taylor, W.I.; Achanzar, D. Catalase test as an aid to the identification of Enterobacteriaceae. J. Appl. Microbiol. 1972, 24, 58–61. [Google Scholar] [CrossRef] [PubMed]
- Jurtshuk, P.; McQuitty, D.N. Use of a quantitative oxidase test for characterizing oxidative metabolism in bacteria. Appl. Environ. Microbiol. 1976, 3, 668–679. [Google Scholar] [CrossRef]
- Abdulhamid, M.B.; Costas, L.; Del Valle Loto, F.; Baigorí, M.D.; Pera, L.M. Industrial biotransformations catalyzed by microbial lipases: Screening platform and commercial aspects. Folia Microbiol. 2021, 66, 1009–1022. [Google Scholar] [CrossRef]
- Lapierre, F.M.; Schmid, J.; Ederer, B.; Ihling, N.; Büchs, J.; Huber, R. Revealing nutritional requirements of MICP-relevant Sporosarcina pasteurii DSM33 for growth improvement in chemically defined and complex media. Sci. Rep. 2020, 10, 22448. [Google Scholar] [CrossRef]
- Pei, D.; Liu, Z.; Wu, W.; Hu, B. Transcriptome analyses reveal the utilization of nitrogen sources and related metabolic mechanisms of Sporosarcina pasteurii. PLoS ONE 2021, 16, e0246818. [Google Scholar] [CrossRef] [PubMed]
- Bakermans, C.; Skidmore, M.L. Microbial metabolism in ice and brine at −5 °C. Environ. Microbiol. 2011, 13, 2269–2278. [Google Scholar] [CrossRef] [PubMed]
- Seal, B.S.; Drider, D.; Oakley, B.B.; Brüssow, H.; Bikard, D.; Rich, J.O.; Miller, S.; Devillard, E.; Kwan, J.; Bertin, G.; et al. Microbial-derived products as potential new antimicrobials. Vet. Res. 2018, 49, 66. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Bai, Q.; Li, Y.; Zhan, H.; Liu, Y.; Liu, B.; Wang, J. Sporosarcina aquimarina MS4 Regulates the Digestive Enzyme Activities, Body Wall Nutrients, Gut Microbiota, and Metabolites of Apostichopus japonicus. Fishes 2022, 7, 134. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Yutin, N.; Wolf, Y.I.; Vera Alvarez, R.; Koonin, E.V. Conservation and Evolution of the Sporulation Gene Set in Diverse Members of the Firmicutes. J. Bacteriol. 2022, 204, e0007922. [Google Scholar] [CrossRef]
- Yu, B.; Kanaan, J.; Shames, H.; Wicander, J.; Aryal, M.; Li, Y.; Korza, G.; Brul, S.; Kramer, G.; Li, Y.Q.; et al. Identification and characterization of new proteins crucial for bacterial spore resistance and germination. Front. Microbiol. 2023, 14, 1161604. [Google Scholar] [CrossRef]
- D’Urzo, A.; Santambrogio, C.; Grandori, R.; Ciurli, S.; Zambelli, B. The conformational response to Zn(II) and Ni(II) binding of Sporosarcina pasteurii UreG, an intrinsically disordered GTPase. J. Biol. Inorg. Chem. 2014, 19, 1341–1354. [Google Scholar] [CrossRef]
- Fong, Y.H.; Wong, H.C.; Yuen, M.H.; Lau, P.H.; Chen, Y.W.; Wong, K.B. Structure of UreG/UreF/UreH complex reveals how urease accessory proteins facilitate maturation of Helicobacter pylori urease. PLoS Biol. 2013, 11, e1001678. [Google Scholar] [CrossRef]
- Lehri, B.; Seddon, A.M.; Karlyshev, A.V. Lactobacillus fermentum 3872 genome sequencing reveals plasmid and chromosomal genes potentially involved in a probiotic activity. FEMS Microbiol. Lett. 2015, 362, fnv068. [Google Scholar] [CrossRef]
- Drew, D.; North, R.A.; Nagarathinam, K.; Tanabe, M. Structures and general transport mechanisms by the major facilitator superfamily (MFS). Chem. Rev. 2021, 121, 5289–5335. [Google Scholar] [CrossRef]
- Gupta, R.S.; Patel, S. Robust Demarcation of the Family Caryophanaceae (Planococcaceae) and Its Different Genera Including Three Novel Genera Based on Phylogenomics and Highly Specific Molecular Signatures. Front. Microbiol. 2020, 10, 2821. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef]
- Thompson, R.S.; Leadbetter, E.R. On the isolation of dipicolinic acid from endospores of Sarcina ureae. Arch. Mikrobiol. 1963, 45, 27–32. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Hilpert, M.; Ward, M.J. Detection of a common and persistent tet(L)-carrying plasmid in chicken-waste-impacted farm soil. Appl. Environ. Microbiol. 2012, 78, 3203–3213. [Google Scholar] [CrossRef]
- Wang, J.P.; Liu, B.; Liu, G.H.; Xiao, R.F.; Zheng, X.F.; Shi, H.; Ge, C.B. Draft Genome Sequence of Sporosarcina globispora W 25T (DSM 4), a Psychrophilic Bacterium Isolated from Soil and River Water. Genome Announc. 2015, 3, 5. [Google Scholar] [CrossRef]
- Abdul Majid, S.; Graw, M.F.; Nguyen, H.; Hay, A.G. Draft Whole-Genome Sequence of Urease-Producing Sporosarcina koreensis. Genome Announc. 2016, 4, e00096-16. [Google Scholar] [CrossRef]
- Yan, W.; Xiao, X.; Zhang, Y. Complete genome sequence of the Sporosarcina psychrophila DSM 6497, a psychrophilic Bacillus strain that mediates the calcium carbonate precipitation. J. Biotechnol. 2016, 226, 14–15. [Google Scholar] [CrossRef]
- Oliver, A.; Kay, M.; Cooper, K.K. Comparative genomics of cocci-shaped Sporosarcina strains with diverse spatial isolation. BMC Genom. 2018, 19, 310. [Google Scholar] [CrossRef]
- Priyodip, P.; Balaji, S. An in vitro chicken gut model for the assessment of phytase producing bacteria. 3 Biotech 2019, 9, 294. [Google Scholar] [CrossRef]

| Genome1 | Genome2 | ANI1->2 | ANI2->1 | AF1->2 | AF2->1 |
|---|---|---|---|---|---|
| A4 | Sporosarcina newyorkensis DSM 23543 | 78.71 | 78.71 | 66.96 | 68.03 |
| A4 | Sporosarcina newyorkensis 2681 | 78.67 | 78.67 | 67.05 | 67.49 |
| A4 | Sporosarcina newyorkensis DSM 23966 | 78.65 | 78.65 | 70.14 | 71.90 |
| A4 | Sporosarcina ureae P37 | 77.38 | 77.38 | 58.77 | 65.46 |
| A4 | Sporosarcina ureae P33 | 77.35 | 77.35 | 58.64 | 67.62 |
| A4 | Sporosarcina ureae S204 | 76.52 | 76.52 | 57.93 | 64.57 |
| A4 | Sporosarcina ureae DSM 2281 | 76.49 | 76.49 | 58.85 | 65.85 |
| A4 | Sporosarcina ureae P17a | 76.36 | 76.37 | 59.16 | 64.84 |
| A4 | Sporosarcina ureae P8 | 76.32 | 76.32 | 58.18 | 65.07 |
| A4 | Sporosarcina ureae P32a | 76.23 | 76.23 | 59.58 | 65.83 |
| A4 | Sporosarcina sp. PTS2304 | 76.14 | 76.13 | 57.39 | 62.48 |
| A4 | Sporosarcina pasteurii ATCC 11859 | 72.32 | 72.32 | 0.05 | 73.76 |
| A4 | Sporosarcina koreensis Q1 | 71.27 | 71.26 | 39.81 | 34.46 |
| A4 | Sporosarcina psychrophila DSM 6497 | 70.80 | 70.79 | 40.98 | 34.54 |
| A4 | Sporosarcina sp. HYO08 | 70.79 | 70.77 | 36.45 | 43.88 |
| A4 | Sporosarcina sp. ZBG7A | 70.65 | 70.65 | 36.44 | 38.27 |
| A4 | Sporosarcina koreensis S-K12 | 70.56 | 70.55 | 34.08 | 42.44 |
| A4 | Sporosarcina sp. D27 (unscreened) | 70.53 | 70.53 | 36.94 | 38.24 |
| A4 | Sporosarcina sp. EUR3 2.2.2 | 69.03 | 69.04 | 24.35 | 26.61 |
| A4 | Planococcus plakortidis DSM 23997 | 68.84 | 68.84 | 23.73 | 26.88 |
| A4 | Sporosarcina globispora DSM 4 | 68.20 | 68.20 | 20.48 | 13.96 |
| A4 | Sporosarcina pasteurii NCIM 2477 | 67.62 | 67.62 | 15.48 | 13.72 |
| A4 | Bacillus thuringiensis YBT-1518 | 67.20 | 67.21 | 16.29 | 9.72 |
| A4 | Clostridium scatologenes ATCC 25775 | 65.38 | 65.38 | 4.98 | 2.54 |
| A4 | Flavobacterium reichenbachii LMG 25512 | 63.94 | 63.94 | 0.48 | 0.31 |
| A4 | A15 | 78.08 | 78.08 | 68.39 | 69.72 |
| A15 | Sporosarcina newyorkensis 2681 | 81.08 | 81.08 | 67.89 | 67.04 |
| A15 | Sporosarcina newyorkensis DSM 23966 | 81.06 | 81.06 | 69.65 | 69.99 |
| A15 | Sporosarcina newyorkensis DSM 23543 | 81.06 | 81.06 | 68.18 | 67.92 |
| A15 | Sporosarcina sp. PTS2304 | 77.27 | 77.27 | 60.90 | 64.83 |
| A15 | Sporosarcina ureae S204 | 77.09 | 77.10 | 59.43 | 64.98 |
| A15 | Sporosarcina ureae P8 | 77.08 | 77.08 | 60.34 | 66.20 |
| A15 | Sporosarcina ureae DSM 2281 | 77.07 | 77.07 | 60.28 | 66.16 |
| A15 | Sporosarcina ureae P17a | 76.96 | 76.96 | 59.87 | 64.34 |
| A15 | Sporosarcina ureae P32a | 76.95 | 76.95 | 60.72 | 65.85 |
| A15 | Sporosarcina ureae P33 | 76.89 | 76.91 | 59.64 | 67.56 |
| A15 | Sporosarcina ureae P37 | 76.88 | 76.88 | 60.14 | 65.70 |
| A15 | Sporosarcina pasteurii ATCC 11859 | 72.88 | 72.88 | 0.05 | 74.11 |
| A15 | Sporosarcina sp. HYO08 | 71.23 | 71.23 | 38.19 | 45.08 |
| A15 | Sporosarcina psychrophila DSM 6497 | 71.07 | 71.07 | 42.06 | 34.78 |
| A15 | Sporosarcina koreensis Q1 | 71.06 | 71.05 | 40.48 | 34.37 |
| A15 | Sporosarcina sp. ZBG7A | 70.75 | 70.75 | 37.78 | 38.91 |
| A15 | Sporosarcina sp. D27 (unscreened) | 70.68 | 70.68 | 37.84 | 38.42 |
| A15 | Sporosarcina sp. EUR3 2.2.2 | 69.64 | 69.64 | 26.63 | 28.59 |
| A15 | Sporosarcina koreensis S-K12 | 69.02 | 69.02 | 32.93 | 40.24 |
| A15 | Bacillus thuringiensis YBT-1518 | 67.88 | 67.89 | 17.47 | 10.25 |
| A15 | Planococcus plakortidis DSM 23997 | 67.87 | 67.87 | 23.39 | 25.99 |
| A15 | Sporosarcina globispora DSM 4 | 67.65 | 67.65 | 20.85 | 13.93 |
| A15 | Sporosarcina pasteurii NCIM 2477 | 67.22 | 67.22 | 15.66 | 13.61 |
| A15 | Clostridium scatologenes ATCC 25775 | 65.99 | 65.99 | 5.91 | 2.96 |
| A15 | Flavobacterium reichenbachii LMG 25512 | 64.45 | 64.45 | 0.47 | 0.30 |
| Query Strain | Subject Strain | dDDH (d4, in %) | G+C Content Difference (in %) |
|---|---|---|---|
| A4 | Sporosarcina newyorkensis 2681a | 23.5 | 5.85 |
| A4 | Sporosarcina sp. P33 | 20.5 | 1.64 |
| A4 | Sporosarcina ureae P17a | 20 | 4.67 |
| A4 | Sporosarcina ureae S204 | 19.4 | 4.66 |
| A4 | Sporosarcina ureae DSM 2281a | 19.2 | 5.6 |
| A4 | Sporosarcina sp. P7 | 19.2 | 3.68 |
| A4 | Sporosarcina newyorkensis DSM 23966 | 15 | 3.99 |
| A4 | Sporosarcina luteola NBRC 105378 | 13.5 | 2.77 |
| A4 | Sporosarcina koreensis Q1a | 13.4 | 2.95 |
| A4 | Sporosarcina koreensis S-K12 | 13.4 | 7.81 |
| A4 | Sporosarcina psychrophila DSM 6497 | 13.3 | 5.84 |
| A4 | Sporosarcina pasteurii BNCC 337394 | 13.3 | 7.01 |
| A4 | Bacillus thuringiensis YBT-1518 529122.4 | 13.1 | 10.76 |
| A4 | Planococcus plakortidis DSM 23997 | 13.1 | 3.8 |
| A4 | Paenisporosarcina aarctica CGMCC 1.6503 | 13.1 | 9.18 |
| A4 | Sporosarcina globispora DSM4 | 13.1 | 6.62 |
| A4 | Clostridium scatologenes ATCC 25775 | 13 | 16.57 |
| A4 | Flavobacterium reichenbachii LMG 25512a | 12.9 | 8.8 |
| A4 | A15 | 24.2 | 0.36 |
| A15 | Sporosarcina newyorkensis 2681a | 27.4 | 5.49 |
| A15 | Sporosarcina sp. P33 | 20.7 | 1.28 |
| A15 | Sporosarcina ureae S204 | 20.1 | 4.3 |
| A15 | Sporoscarcina ureae DSM 2281a | 19.9 | 5.25 |
| A15 | Sporosarcina sp. P7 | 19.8 | 3.32 |
| A15 | Sporosarcina ureae P17a | 19.5 | 4.31 |
| A15 | Sporosarcina newyorkensis DSM 23966 | 16.2 | 3.64 |
| A15 | Sporosarcina luteola NBRC 105378 | 13.6 | 2.41 |
| A15 | Sporosarcina psychrophila DSM 6497 | 13.5 | 5.48 |
| A15 | Sporosarcina koreensis Q1a | 13.5 | 3.31 |
| A15 | Sporosarcina pasteurii BNCC 337394 | 13.3 | 6.65 |
| A15 | Sporosarcina koreensis S-K12 | 13.3 | 8.17 |
| A15 | Bacillus thuringiensis YBT-1518 529122.4 | 13.2 | 10.4 |
| A15 | Paenisporosarcina aarctica CGMCC 1.6503 | 13.2 | 8.82 |
| A15 | Planococcus plakortidis DSM 23997 | 13.2 | 4.16 |
| A15 | Sporosarcina globispora DSM4 | 13.1 | 6.26 |
| A15 | Clostridium scatologenes ATCC 25775 | 13 | 16.21 |
| A15 | Flavobacterium reichenbachii LMG 25512a | 12.9 | 8.45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keshri, J.; Smith, K.M.; Svendsen, M.K.; Keillor, H.R.; Moss, M.L.; Jordan, H.J.; Larkin, A.M.; Garrish, J.K.; Line, J.E.; Ball, P.N.; et al. Phenotypic Characterization and Draft Genome Sequence Analyses of Two Novel Endospore-Forming Sporosarcina spp. Isolated from Canada Goose (Branta canadensis) Feces. Microorganisms 2024, 12, 70. https://doi.org/10.3390/microorganisms12010070
Keshri J, Smith KM, Svendsen MK, Keillor HR, Moss ML, Jordan HJ, Larkin AM, Garrish JK, Line JE, Ball PN, et al. Phenotypic Characterization and Draft Genome Sequence Analyses of Two Novel Endospore-Forming Sporosarcina spp. Isolated from Canada Goose (Branta canadensis) Feces. Microorganisms. 2024; 12(1):70. https://doi.org/10.3390/microorganisms12010070
Chicago/Turabian StyleKeshri, Jitendra, Kristina M. Smith, Molly K. Svendsen, Haley R. Keillor, Madeline L. Moss, Haley J. Jordan, Abigail M. Larkin, Johnna K. Garrish, John Eric Line, Patrick N. Ball, and et al. 2024. "Phenotypic Characterization and Draft Genome Sequence Analyses of Two Novel Endospore-Forming Sporosarcina spp. Isolated from Canada Goose (Branta canadensis) Feces" Microorganisms 12, no. 1: 70. https://doi.org/10.3390/microorganisms12010070
APA StyleKeshri, J., Smith, K. M., Svendsen, M. K., Keillor, H. R., Moss, M. L., Jordan, H. J., Larkin, A. M., Garrish, J. K., Line, J. E., Ball, P. N., Oakley, B. B., & Seal, B. S. (2024). Phenotypic Characterization and Draft Genome Sequence Analyses of Two Novel Endospore-Forming Sporosarcina spp. Isolated from Canada Goose (Branta canadensis) Feces. Microorganisms, 12(1), 70. https://doi.org/10.3390/microorganisms12010070







