Unique Features of Extremely Halophilic Microbiota Inhabiting Solar Saltworks Fields of Vietnam
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Geochemical Analyses and Salinity-Related Measurements
2.2.1. Anion Analysis
2.2.2. Cation Analysis
2.3. DNA Extraction and Purification
2.4. Next-Generation Sequencing
2.5. NGS Data Analysis
2.6. Statistical Analyses
2.7. Establishment of Polysaccharide-Degrading Enrichments and Isolation of an Axenic Xylanolytic Culture
3. Results and Discussion
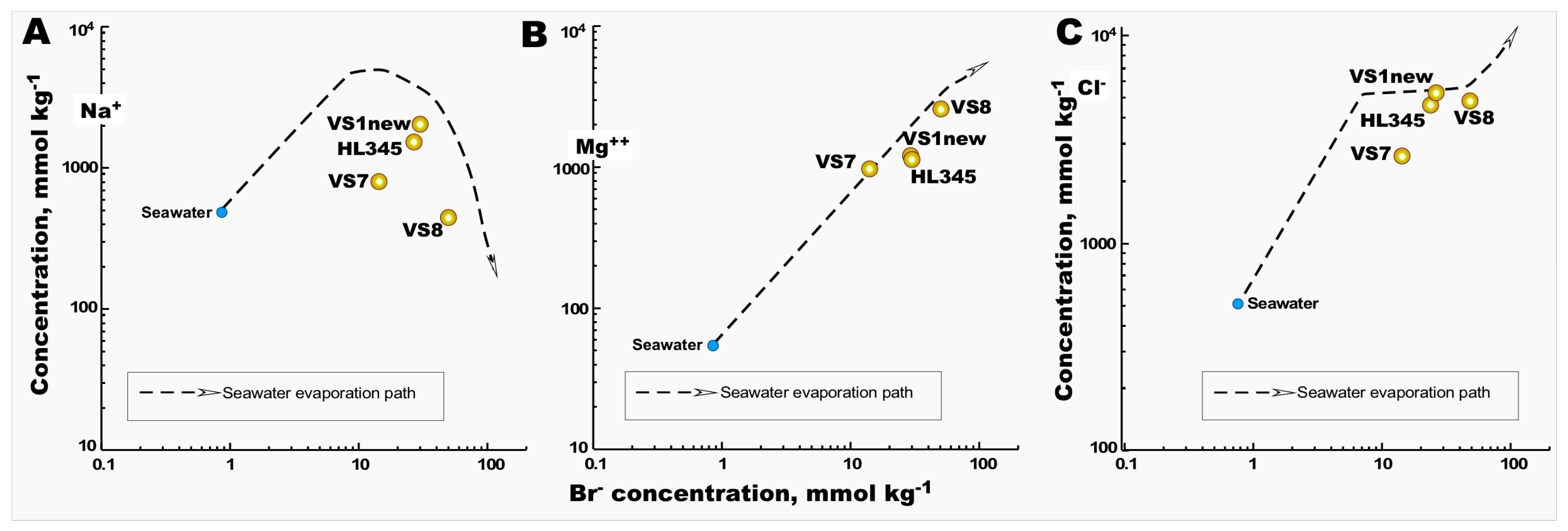
3.1. Chemical Composition of Saltworks Fields
3.2. Alpha and Beta Diversity
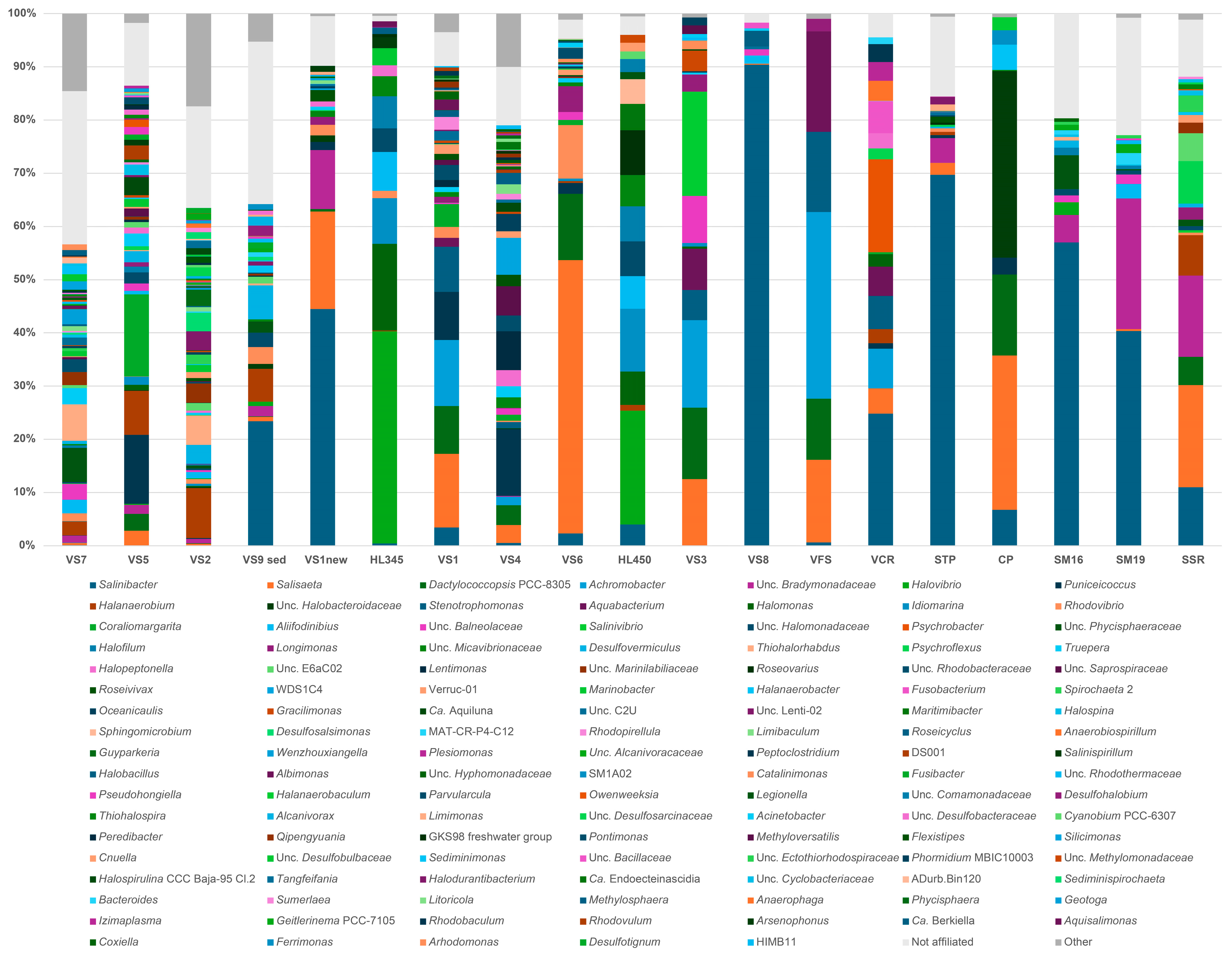
3.3. General Microbiome Profiling
3.4. Bacterial Diversity of Brine, Sediment and Salt (Halite) Samples
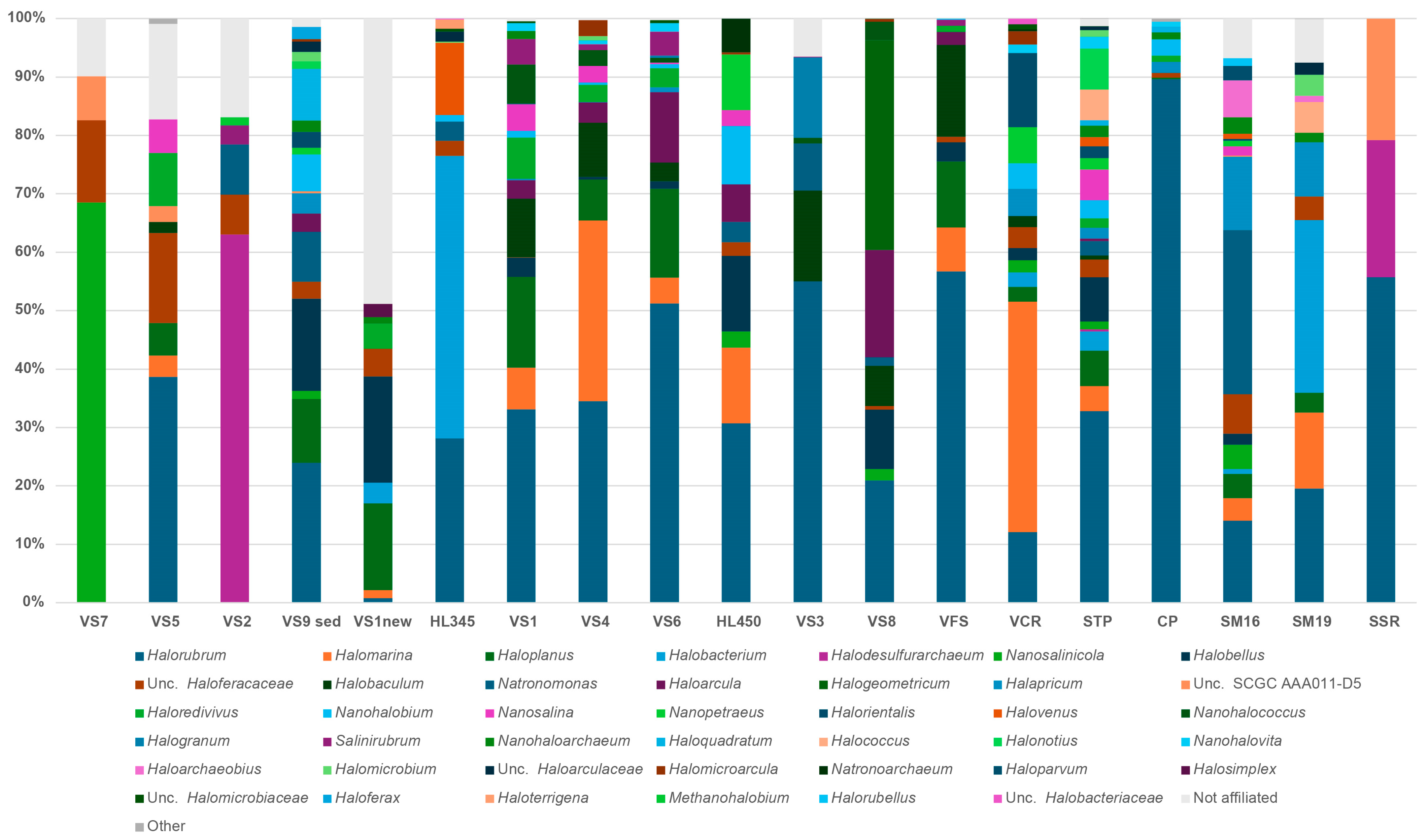
3.5. Archaeal Diversity of Brine, Sediment and Salt (Halite) Samples
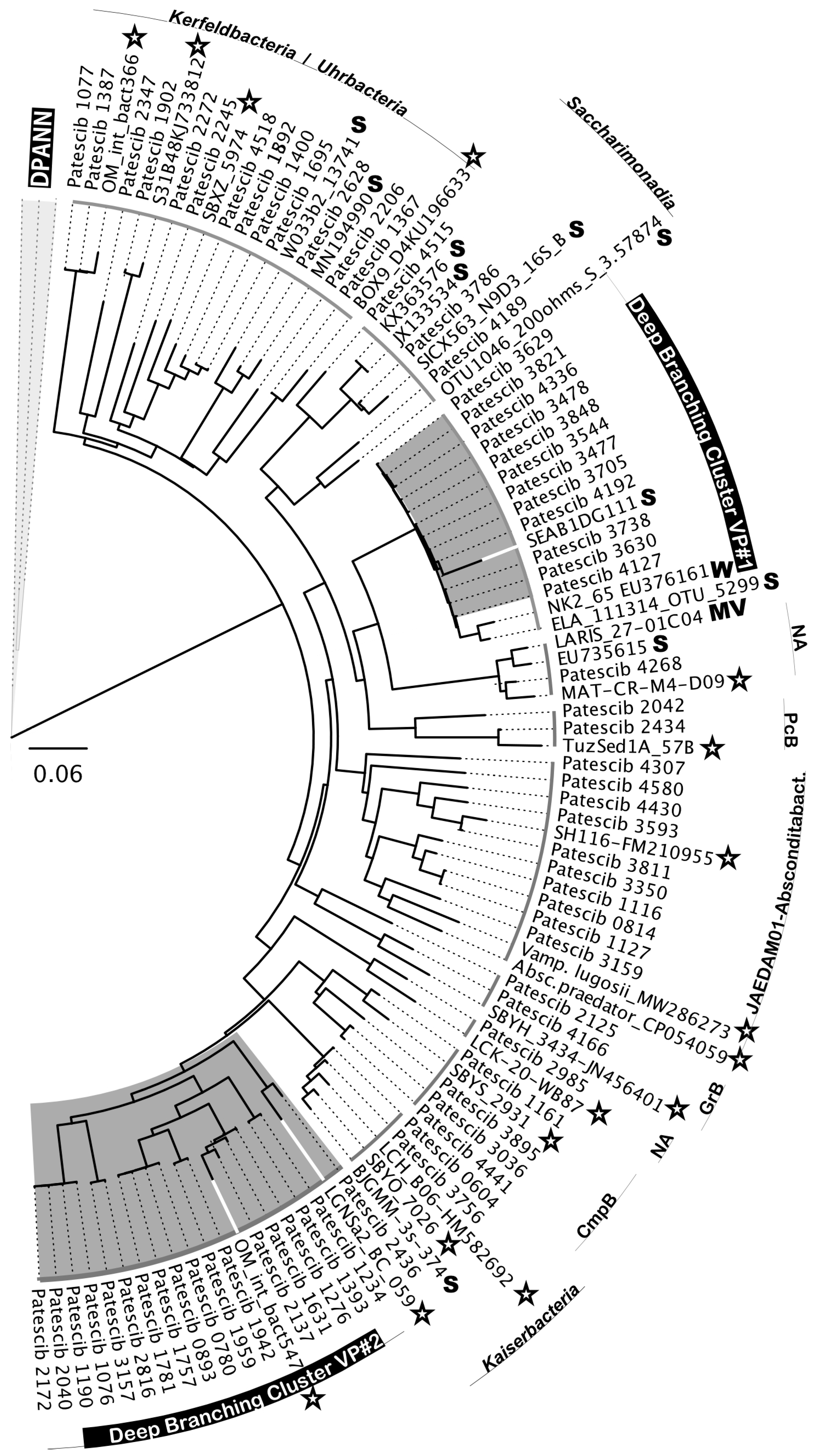
3.6. Diversity of Nano-Sized Microbiota
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wei, Y.-L.; Long, Z.-J.; Ren, M.-X. Microbial community and functional prediction during the processing of salt production in a 1000-year-old marine solar saltern of South China. Sci. Total Environ. 2022, 819, 152014. [Google Scholar] [CrossRef] [PubMed]
- Javor, B.J. Industrial microbiology of solar salt production. J. Ind. Microbiol. Biotechnol. 2002, 28, 42–47. [Google Scholar] [CrossRef] [PubMed]
- De Medeiros Rocha, R.; Costa, D.F.; Lucena-Filho, M.A.; Bezerra, R.M.; Medeiros, D.H.; Azevedo-Silva, A.M.; Araujo, C.N.; Xavier-Filho, L. Brazilian solar saltworks-ancient uses and future possibilities. Aquat. Biosyst. 2012, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Mani, K.; Salgaonkar, B.B.; Braganca, J.M. Culturable halophilic archaea at the initial and crystallization stages of salt production in a natural solar saltern of Goa, India. Aquat. Biosyst. 2012, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Conde-Martinez, N.; Acosta-Gonzalez, A.; Diaz, L.E.; Tello, E. Use of a mixed culture strategy to isolate halophilic bacteria with antibacterial and cytotoxic activity from the Manaure solar saltern in Colombia. BMC Microbiol. 2017, 17, 230. [Google Scholar] [CrossRef]
- Leoni, C.; Volpicella, M.; Fosso, B.; Manzari, C.; Piancone, E.; Dileo, M.C.G.; Arcadi, E.; Yakimov, M.; Pesole, G.; Ceci, L.R. A differential metabarcoding approach to describe taxonomy profiles of bacteria and archaea in the saltern of Margherita di Savoia (Italy). Microorganisms 2020, 8, 936. [Google Scholar] [CrossRef]
- Margesin, R.; Zacke, G.; Schinner, F. Characterization of heterotrophic microorganisms in alpine glacier cryoconite. Arct. Antarct. Alp. Res. 2002, 34, 88–93. [Google Scholar] [CrossRef]
- Kimbrel, J.A.; Ballor, N.; Wu, Y.-W.; David, M.M.; Hazen, T.C.; Simmons, B.A.; Singer, S.W.; Jansson, J.K. Microbial Community Structure and Functional Potential Along a Hypersaline Gradient. Front. Microbiol. 2018, 9, 1492. [Google Scholar] [CrossRef]
- Oren, A. Diversity of halophilic microorganisms: Environments, phylogeny, physiology, and applications. J. Ind. Microbiol. Biotechnol. 2002, 28, 56–63. [Google Scholar] [CrossRef]
- Baati, H.; Guermazi, S.; Amdouni, R.; Gharsallah, N.; Sghir, A.; Ammar, E. Prokaryotic diversity of a Tunisian multipond solar saltern. Extremophiles 2008, 12, 505–518. [Google Scholar] [CrossRef]
- Trigui, H.; Masmoudi, S.; Brochier-Armanet, C.; Barani, A.; Grégori, G.; Denis, M.; Dukan, S.; Maalej, S. Characterization of heterotrophic prokaryote subgroups in the Sfax coastal solar salterns by combining flow cytometry cell sorting and phylogenetic analysis. Extremophiles 2011, 15, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, K.B.; Canfield, D.E.; Teske, A.P.; Oren, A. Community composition of a hypersaline endoevaporitic microbial mat. Appl. Environ. Microbiol. 2005, 11, 7352–7365. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, M.; Kannan, V.; Pašic’, L. Diversity of microorganisms in solar salterns of Tamil Nadu, India. World J. Microbiol. Biotechnol. 2009, 25, 1007–1017. [Google Scholar] [CrossRef]
- Selivanova, E.A.; Poshvina, D.V.; Khlopko, Y.A.; Gogoleva, N.E.; Plotnikov, A.O. Diversity of prokaryotes in planktonic communities of saline Sol-Iletsk lakes (Orenburg Oblast, Russia). Microbiology 2018, 87, 569–582. [Google Scholar] [CrossRef]
- Akpolat, C.; Fernández, A.B.; Caglayan, P.; Calli, B.; Birbir, M.; Ventosa, A. Prokaryotic communities in the Thalassohaline Tuz Lake, deep zone, and Kayacik, Kaldirim and Yavsan Salterns (Turkey) assessed by 16S rRNA amplicon sequencing. Microorganisms 2021, 9, 1525. [Google Scholar] [CrossRef]
- Park, S.J.; Kang, C.H.; Rhee, S.K. Characterization of the microbial diversity in a Korean solar saltern by 16S rRNA gene analysis. J. Microbiol. Biotechnol. 2006, 16, 1640–1645. [Google Scholar]
- Burns, D.G.; Camakaris, H.M.; Janssen, P.H.; Dyall-Smith, M.L. Combined use of cultivation-dependent and cultivation-independent methods indicates that members of most haloarchaeal groups in an Australian crystallizer pond are cultivable. Appl. Environ. Microbiol. 2004, 70, 5258–5265. [Google Scholar] [CrossRef]
- Dillon, J.G.; Carlin, M.; Gutierrez, A.; Nguyen, V.; McLain, N. Patterns of microbial diversity along a salinity gradient in the Guerrero Negro solar saltern, Baja CA Sur, Mexico. Front. Microbiol. 2013, 4, 399. [Google Scholar] [CrossRef]
- Anton, J.; Rossello-Mora, R.; Rodriguez-Valera, F.; Amann, R. Extremely Halophilic Bacteria in Crystallizer Ponds from Solar Salterns. Appl. Environ. Microbiol. 2000, 66, 3052–3057. [Google Scholar] [CrossRef]
- Benlloch, S.; López-López, A.; Casamayor, E.O.; Øvreås, L.; Goddard, V.; Daae, F.L.; Smerdon, G.; Massana, R.; Joint, I.; Thingstad, F.; et al. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ. Microbiol. 2002, 4, 349–360. [Google Scholar] [CrossRef]
- Pašić, L.; Ulrih, N.P.; Crnigoj, M.; Grabnar, M.; Velikonja, B.H. Haloarchaeal communities in the crystallizers of two Adriatic solar salterns. Can. J. Microbiol. 2007, 53, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Ventosa, A.; Fernández, A.B.; León, M.J.; Sánchez-Porro, C.; Rodriguez-Valera, F.J.E. The Santa Pola saltern as a model for studying the microbiota of hypersaline environments. Extremophiles 2014, 18, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Kambourova, M.; Tomova, I.; Boyadzhieva, I.; Radchenkova, N.; Vasileva-Tonkova, E. Unusually high archaeal diversity in a crystallizer pond, Pomorie salterns, Bulgaria, revealed by phylogenetic analysis. Archaea 2016, 16, 7459679. [Google Scholar] [CrossRef] [PubMed]
- Gasol, J.M.; Casamayor, E.O.; Joint, I.; Garde, K.; Gustavson, K.; Benlloch, S.; Díez, B.; Schauer, M.; Massana, R.; Pedrós-Alió, C. Control of heterotrophic prokaryotic abundance and growth rate in hypersaline planktonic environments. Aquat. Microb. Ecol. 2004, 34, 193–206. [Google Scholar] [CrossRef]
- Ghai, R.; Pasic, L.; Fernnandez, A.B.; Martin-Cuadrado, A.B.; Mizuno, C.M.; McMahon, K.D.; Papke, R.T.; Stepanauskas, R.; Rodriguez-Brito, B.; Rohwer, F.; et al. New abundant microbial groups in aquatic hypersaline environments. Sci. Rep. 2011, 1, 135. [Google Scholar] [CrossRef]
- Mani, K.; Taib, N.; Hugoni, M.; Bronner, G.; Bragança, J.M.; Debroas, D. Transient dynamics of archaea and bacteria in sediments and brine across a salinity gradient in a solar saltern of Goa, India. Front. Microbiol. 2020, 11, 1891. [Google Scholar] [CrossRef]
- Casamayor, E.O.; Massana, R.; Benlloch, S.; Øvreås, L.; Díez, B.; Goddard, V.J.; Gasol, J.M.; Joint, I.; Rodriguez-Valera, F.; Pedrós-Alió, C. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ. Microbiol. 2002, 4, 338–348. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Viver, T.; Conrad, R.E.; Venter, S.N.; Rossello-Mora, R. Solar salterns as model systems to study the units of bacterial diversity that matter for ecosystem functioning. Curr. Opin. Biotechnol. 2022, 73, 151–157. [Google Scholar] [CrossRef]
- Norton, C.F.; Grant, W.D. Survival of halobacteria within fluid inclusions in salt crystals. Microbiology 1988, 134, 1365–1373. [Google Scholar] [CrossRef]
- Huby, T.J.C.; Clark, D.R.; McKew, B.A.; McGenity, T.J. Extremely halophilic archaeal communities are resilient to short-term entombment in halite. Environ. Microbiol. 2021, 23, 3370–3383. [Google Scholar] [CrossRef]
- Brown, C.T.; Hug, L.A.; Thomas, B.C.; Sharon, I.; Castelle, C.J.; Singh, A.; Wilkins, M.J.; Wrighton, K.C.; Williams, K.H.; Banfield, J.F. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 2015, 523, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Castelle, C.J.; Banfield, J.F. Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell 2018, 172, 1181–1197. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Ning, D.; He, Z.; Zhang, P.; Spencer, S.J.; Gao, S.; Shi, W.; Wu, L.; Zhang, Y.; Yang, Y.; et al. Small and mighty: Adaptation of superphylum Patescibacteria to groundwater environment drives their genome simplicity. Microbiome 2020, 8, 51. [Google Scholar] [CrossRef] [PubMed]
- Hug, L.A.; Baker, B.J.; Anantharaman, K.; Brown, C.T.; Probst, A.J.; Castelle, C.J.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A new view of the tree of life. Nat. Microbiol. 2016, 1, 1–6. [Google Scholar] [CrossRef]
- La Cono, V.; Messina, E.; Rohde, M.; Arcadi, E.; Ciordia, S.; Crisafi, F.; Denaro, R.; Ferrer, M.; Giuliano, L.; Golyshin, P.N.; et al. Symbiosis between nanohaloarchaeon and haloarchaeon is based on utilization of different polysaccharides. Proc. Natl. Acad. Sci. USA 2020, 117, 20223–20234. [Google Scholar] [CrossRef]
- La Cono, V.; Messina, E.; Reva, O.; Smedile, F.; La Spada, G.; Crisafi, F.; Marturano, L.; Miguez, N.; Ferrer, M.; Selivanova, E.A.; et al. Nanohaloarchaea as beneficiaries of xylan degradation by haloarchaea. Microb. Biotech. 2023, 16, 1803–1822. [Google Scholar] [CrossRef]
- Yakimov, M.M.; Merkel, A.Y.; Gaisin, V.A.; Pilhofer, M.; Messina, E.; Hallsworth, J.E.; Klyukina, A.A.; Tikhonova, E.N.; Gorlenko, V.M. Cultivation of a vampire: ‘Candidatus Absconditicoccus praedator’. Environ. Microbiol. 2022, 24, 30–49. [Google Scholar] [CrossRef]
- Chen, X.; Molenda, O.; Brown, C.T.; Toth, C.R.; Guo, S.; Luo, F.; Howe, J.; Nesbo, C.L.; He, C.; Montabana, E.A.; et al. “Candidatus Nealsonbacteria” are likely biomass recycling ectosymbionts of methanogenic archaea in a stable benzene-degrading enrichment culture. Appl. Environ. Microbiol. 2023, 89, e00025-23. [Google Scholar] [CrossRef]
- Narasingarao, P.; Podell, S.; Ugalde, J.A.; Brochier-Armanet, C.; Emerson, J.B.; Brocks, J.J.; Heidelberg, K.B.; Banfield, J.F.; Allen, E.E. De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J. 2012, 6, 81–93. [Google Scholar] [CrossRef]
- Podell, S.; Emerson, J.B.; Jones, C.M.; Ugalde, J.A.; Welch, S.; Heidelberg, K.B.; Banfield, J.F.; Allen, E.E. Seasonal fluctuations in ionic concentrations drive microbial succession in a hypersaline lake community. ISME J. 2014, 8, 979–990. [Google Scholar] [CrossRef]
- Gornariz, M.; Martínez-García, M.; Santos, F.; Rodriguez, F.; Capella-Gutiérrez, S.; Gabaldón, T.; Rosselló-Móra, R.; Meseguer, I.; Antón, J. From community approaches to single-cell genomics: The discovery of ubiquitous hyperhalophilic Bacteroidetes generalists. ISME J. 2015, 9, 16–31. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Gelsinger, D.R.; Ma, B.; Wierzchos, J.; Ravel, J.; Davila, A.; Casero, M.C.; DiRuggiero, J. Functional interactions of archaea, bacteria and viruses in a hypersaline endolithic community. Environ. Microbiol. 2016, 18, 2064–2077. [Google Scholar] [CrossRef] [PubMed]
- Di Meglio, L.; Santos, F.; Gomariz, M.; Almansa, C.; López, C.; Antón, J.; Nercessian, D. Seasonal dynamics of extremely halophilic microbial communities in three Argentinian salterns. FEMS Microbiol. Ecol. 2016, 92, fiw184. [Google Scholar] [CrossRef] [PubMed]
- Mora-Ruiz, M.D.R.; Cifuentes, A.; Font-Verdera, F.; Pérez-Fernández, C.; Farias, M.E.; González, B.; Orfila, A.; Rosselló-Móra, R. Biogeographical patterns of bacterial and archaeal communities from distant hypersaline environments. Syst. Appl. Microbiol. 2018, 41, 139–150. [Google Scholar] [CrossRef]
- Vavourakis, C.D.; Andrei, A.-S.; Mehrshad, M.; Ghai, R.; Sorokin, D.Y.; Muyzer, G. A metagenomics roadmap to the uncultured genome diversity in hypersaline soda lake sediments. Microbiome 2018, 6, 1–18. [Google Scholar] [CrossRef]
- La Cono, V.; Bortoluzzi, G.; Messina, E.; La Spada, G.; Smedile, F.; Giuliano, L.; Borghini, M.; Stumpp, C.; Schmitt-Kopplin, P.; Harir, M.; et al. The discovery of Lake Hephaestus, the youngest athalassohaline deep-sea formation on Earth. Sci. Rep. 2019, 9, 1679. [Google Scholar] [CrossRef]
- Feng, Y.; Neri, U.; Gosselin, S.; Louyakis, A.S.; Papke, R.T.; Gophna, U.; Gogarten, J.P. The evolutionary origins of extreme halophilic archaeal lineages. Genome Biol. Evol. 2021, 13, evab166. [Google Scholar] [CrossRef]
- Chernyh, N.A.; Merkel, A.Y.; Kondrasheva, K.V.; Alimov, J.E.; Klyukina, A.A.; Bonch-Osmolovskaya, E.A.; Slobodkin, A.I.; Davranov, K.D. At the shores of a vanishing sea: Microbial communities of Aral and southern Aral Sea region. Microbiology 2024, 93, 1–13. [Google Scholar] [CrossRef]
- Gutiérrez-Preciado, A.; Dede, B.; Baker, B.A.; Eme, L.; Moreira, D.; López-García, P. Extremely acidic proteomes and metabolic flexibility in bacteria and highly diversified archaea thriving in geothermal chaotropic brines. Nat. Ecol. Evol. 2024, 1–14. [Google Scholar] [CrossRef]
- Cuadros-Rodrıguez, L.; Bagur-González, M.G.; Sanchez-Viñas, M.; Gonzalez-Casado, A.; Gomez-Saez, A.M. Principles of analytical calibration/quantification for the separation sciences. J. Chromatogr. A 2007, 1158, 33–46. [Google Scholar] [CrossRef]
- Rienitz, O.; Rohker, K.; Schiel, D.; Han, J.; Oeter, D. New equation for the evaluation of standard addition experiments applied to ion chromatography. Microchim. Acta 2006, 154, 21–25. [Google Scholar] [CrossRef]
- U.S. EPA Method 300.1; Determination of Inorganic Anions in Drinking Water by Ion Chromatography. Revision 1.0; U.S. EPA: Cincinnati, OH, USA, 1997.
- U.S. EPA Method 3005A; Acid Digestion of Waters for Total Recoverable or Dissolved Metals for Analyses by Flame Atomic Absorption Spectroscopy. Revision 1.0; U.S. EPA: Cincinnati, OH, USA, 1992.
- U.S. EPA Method 6010D (SW-846); Inductively Coupled Plasma-Atomic Emission Spectrometry. Revision 4; U.S. EPA: Washington, DC, USA, 2014.
- Hurt, R.A.; Qiu, X.; Wu, L.; Roh, Y.; Palumbo, A.V.; Tiedje, J.M.; Zhou, J. Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 2001, 67, 4495–4503. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.; Lauber, C.; Walters, W.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Volume 13. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Crisafi, F.; Smedile, F.; Yakimov, M.M.; Aulenta, F.; Fazi, S.; La Cono, V.; Martinelli, A.; Di Lisio, V.; Denaro, R. Bacterial biofilms on medical masks disposed in the marine environment: A hotspot of biological and functional diversity. Sci. Total Environ. 2022, 837, 155731. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Shetty, S.A.; Lahti, L. Microbiome data science. J. Biosci. 2019, 44, 115. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R. R Package Version 2.3-2. Vegan: Community Ecology Package. Available online: https://cran.r-project.org/package=vegan (accessed on 19 June 2020).
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef]
- Carpenter, A.B. Origin and chemical evolution of brines in sedimentary basins. In Proceedings of the SPE Annual Technical Conference and Exhibition, Dallas, TX, USA, 27–30 September 1978; p. SPE-7504. [Google Scholar]
- De Lange, G.; Middelburg, J.; van der Weijden, C.; Catalano, G.; Luther, G.; Hydes, D.; Woittiez, J.R.W.; Klinkhammer, G.P. Composition of anoxic hypersaline brines in the Tyro and Bannock basins, eastern Mediterranean. Mar. Chem. 1990, 31, 63–88. [Google Scholar] [CrossRef]
- Wallmann, K.; Suess, E.; Westbrook, G.H.; Winckler, G.; Cita, M.B. Salty brines on the Mediterranean Sea floor. Nature 1997, 387, 31–32. [Google Scholar] [CrossRef]
- Wallmann, K.; Aghib, F.S.; Castradori, D.; Cita, M.B.; Suess, E.; Greinert, J.; Rickert, D. Sedimentation and formation of secondary minerals in the hypersaline Discovery Basin, eastern Mediterranean. Mar. Geol. 2002, 186, 9–28. [Google Scholar] [CrossRef]
- Fernandez, A.B.; Ghai, R.; Martin-Cuadrado, A.B.; Sanchez-Porro, C.; Rodriguez-Valera, F.; Ventosa, A. Prokaryotic taxonomic and metabolic diversity of an intermediate salinity hypersaline habitat assessed by metagenomics. FEMS Microbiol. Ecol. 2014, 88, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, A.B.; Vera-Gargallo, B.; Sánchez-Porro, C.; Ghai, R.; Papke, R.T.; Rodriguez-Valera, F.; Ventosa, A. Comparison of prokaryotic community structure from Mediterranean and Atlantic saltern concentrator ponds by a metagenomic approach. Front. Microbiol. 2014, 5, 196. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; D’Agostino McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Schliep, K. phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef]
- Galili, T. dendextend: An R package for visualizing, adjusting, and comparing trees of hierarchical clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef]
- de Vries, A.; Ripley, B.D. ggdendro: Create Dendrograms and Tree Diagrams Using ‘ggplot2’. R Package Version 0.2.0. 2024. Available online: https://andrie.github.io/ggdendro/ (accessed on 14 July 2024).
- Xiao, N. ggsci: Scientific Journal and Sci-Fi Themed Color Palettes for ‘ggplot2’. R Package Version 3.2.0. 2024. Available online: https://CRAN.R-project.org/package=ggsci/ (accessed on 14 July 2024).
- Wilke, C. cowplot: Streamlined Plot Theme and Plot Annotations for ‘ggplot2’. R Package Version 1.1.3. 2024. Available online: https://CRAN.R-project.org/package=cowplot (accessed on 14 July 2024).
- Oren, A. Novel insights into the diversity of halophilic microorganisms and their functioning in hypersaline ecosystems. Npj Biodivers. 2024, 3, 18. [Google Scholar] [CrossRef]
- Takai, K.; Moser, D.P.; DeFlaun, M.; Onstott, T.C.; Fredrickson, J.K. Archaeal diversity in waters from deep South African gold mines. Appl. Environ. Microbiol. 2001, 67, 5750–5760. [Google Scholar] [CrossRef]
- Hollister, E.; Engledow, A.; Hammett, A.; Provin, T.L.; Wilkinson, H.H.; Gentry, T.J. Shifts in microbial community structure along an ecological gradient of hypersaline soils and sediments. ISME J. 2010, 4, 829–838. [Google Scholar] [CrossRef]
- Baricz, A.; Chiriac, C.M.; Andrei, A.-Ș.; Bulzu, P.-A.; Levei, E.A.; Cadar, O.; Battes, K.P.; Cîmpean, M.; Șenilă, M.; Cristea, A. Spatio-temporal insights into microbiology of the freshwater-to-hypersaline, oxic-hypoxic-euxinic waters of Ursu Lake. Environ. Microbiol. 2021, 23, 3523–3540. [Google Scholar] [CrossRef] [PubMed]
- Henriet, O.; Fourmentin, J.; Delincé, B.; Mahillon, J. Exploring the diversity of extremely halophilic archaea in food-grade salts. Int. J. Food Microbiol. 2014, 191, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Koh, H.-W.; Song, H.S.; Song, U.; Yim, K.J.; Roh, S.W.; Park, S.-J. Halolamina sediminis sp. nov., an extremely halophilic archaeon isolated from solar salt. Int. J. Syst. Evol. Microbiol. 2015, 65, 2479–2484. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.R.; Mathieu, M.; Mourot, L.; Dufossé, L.; Underwood, G.J.C.; Dumbrell, A.J.; McGenity, T.J. Biogeography at the limits of life: Do extremophilic microbial communities show biogeographical regionalization? Global Ecol. Biogeogr. 2017, 26, 1435–1446. [Google Scholar] [CrossRef]
- Gibtan, A.; Park, K.; Woo, M.; Shin, J.-K.; Lee, D.-W.; Sohn, J.H.; Song, M.; Roh, S.W.; Lee, S.-J.; Lee, H.-S. Diversity of extremely halophilic archaeal and bacterial communities from commercial salts. Front. Microbiol. 2017, 8, 799. [Google Scholar] [CrossRef]
- McGenity, T.J.; Gemmell, R.T.; Grant, W.D.; Stan-Lotter, H. Origins of halophilic microorganisms in ancient salt deposits. Environ. Microbiol. 2000, 2, 243–250. [Google Scholar] [CrossRef]
- Mormile, M.R.; Biesen, M.A.; Gutierrez, M.C.; Ventosa, A.; Pavlovich, J.B.; Onstott, T.C.; Fredrickson, J.K. Isolation of Halobacterium salinarum retrieved directly from halite brine inclusions. Environ. Microbiol. 2003, 5, 1094–1102. [Google Scholar] [CrossRef]
- Gramain, A.; Díaz, G.C.; Demergasso, C.; Lowenstein, T.K.; McGenity, T.J. Archaeal diversity along a sub- terranean salt core from the Salar Grande (Chile). Environ. Microbiol. 2011, 13, 2105–2121. [Google Scholar] [CrossRef]
- Jaakkola, S.T.; Ravantti, J.J.; Oksanen, H.M.; Bamford, D.H. Buried alive: Microbes from ancient halite. Trends Microbiol. 2016, 24, 148–160. [Google Scholar] [CrossRef]
- Sahl, J.W.; Pace, N.R.; Spear, J.R. Comparative molecular analysis of endoevaporitic microbial communities. Appl. Environ. Microbiol. 2008, 74, 6444–6446. [Google Scholar] [CrossRef]
- Reva, O.N.; La Cono, V.; Crisafi, F.; Smedile, F.; Mudaliyar, M.; Ghosal, D.; Giuliano, L.; Krupovic, M.; Yakimov, M.M. Interplay of intracellular and trans-cellular DNA methylation in natural archaeal consortia. Environ. Microbiol. Rep. 2024, 16, e13258. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Garcia, M.; Santos, F.; Moreno-Paz, M.; Parro, V.; Anton, J. Unveiling viral-host interactions within the ‘microbial dark matter’. Nat. Commun. 2014, 5, 4542. [Google Scholar] [CrossRef] [PubMed]
- Turk, K.; Green, S.J.; Jahnke, L.; Kubo, M.; Parenteau, M.N.; Vogel, M.; Des Marais, D. Phylogenetic analysis of microbial communities from gypsum precipitating environments. Astrobiology 2008, 8, 383. [Google Scholar]
- Vogt, J.C.; Abed, R.M.; Albach, D.C.; Palinska, K.A. Bacterial and archaeal diversity in hypersaline cyanobacterial mats along a transect in the intertidal flats of the Sultanate of Oman. Microb. Ecol. 2018, 75, 331–347. [Google Scholar] [CrossRef]



| Site Name | Label | Latitude | Longitude | Salinity (g/L) * | pH | mV | T (°C) | Sample Type | Date | |
|---|---|---|---|---|---|---|---|---|---|---|
| VIETNAM | HKsf | VS1 | 12°32′7.732′′ N | 109°12′49.244′′ E | 375 | 7.07 | 21 | 40.8 | Salt and brine | 6 June 2022 |
| HKsf | VS2 | 12°32′6.194′′ N | 109°12′49.752′′ E | 220 | 7.75 | −62 | 40.8 | Microbial mat and brine | 6 June 2022 | |
| HKsf | VS3 | 12°32′6.194′′ N | 109°12′49.752′′ E | 1000 | NA ** | NA | NA | Salt | 6 June 2022 | |
| HKsf | VS4 | 12°31′42.2′′ N | 109°13′23.167′′ E | 425 | 6.86 | 2.4 | 44.9 | Brine and salt | 6 June 2022 | |
| HKsf | VS5 | 12°32′5.464′′ N | 109°12′51.347′′ E | 204 | 7.75 | −53 | 38 | Microbial mat and brine | 6 June 2022 | |
| HKsf | VS6 | 12°32′5.464′′ N | 109°12′51.347′′ E | 450 | 6.86 | 29 | 37.8 | Microbial mat and brine | 6 June 2022 | |
| HKsf | VS7 | 12°32′16.735′′ N | 109°12′26.827′′ E | 165 | 7.6 | −50 | 36.5 | Brine and sediment | 15 June 2023 | |
| HKsf | VS1new | 12°32′16.85′′ N | 109°12′26.553′′ E | 340 | 6.9 | 12 | 44 | Brine | 15 June 2023 | |
| HKsf | VS8 | 12°32′20.868′′ N | 109°12′31.428′′ E | 320 | 6.9 | 32 | 42.8 | Brine | 15 June 2023 | |
| HKsf | VS9sed | 12°32′23.496′′ N | 109°12′31.464′′ E | 340 | 6.8 | −243 | 60.2 | Brine, microbial mat and sediment | 15 June 2023 | |
| HKsf | VFS | 12°32′0.164′′ N | 109°12′49.742′′ E | 1000 | NA | NA | NA | Salt | 6 June 2022 | |
| Cam Ranh | VCR | 11°55′17.18′′ N | 109°09′32.870′′ E | 1000 | NA | NA | NA | Salt | 7 June 2022 | |
| Hai Lý | HL450 | 20°07′20.28′′ N | 106°17′45.962′′ E | 450 | 8.5 | 3.9 | 42 | Salt and brine | 19 June 2023 | |
| Hai Lý | HL345 | 20°07′20.64′′ N | 106°17′45.961′′ E | 345 | 8.0 | 8.7 | 40.8 | Salt and brine | 19 June 2023 | |
| ITALY | Trapani | STP | 37°58′49.90′′ N | 12°29′42.000′′ E | 1000 | NA | NA | NA | Salt | 10 September 2022 |
| Motya | CP | 37°51′47.02′′ N | 12°29′4.781′′ E | 1000 | NA | NA | NA | Salt | 23 September 2022 | |
| Motya | SSR | 37°51′46.70′′ N | 12°29′1.263′′ E | 1000 | NA | NA | NA | Salt | 23 September 2022 | |
| Motya | SM16 | 37°51′46.90′′ N | 12°28′59.985′′ E | 1000 | NA | NA | NA | Salt | 23 September 2022 | |
| Motya | SM19 | 37°51′46.90′′ N | 12°28′59.98′′ E | 1000 | NA | NA | NA | Salt | 23 September 2022 |
| Sampling Site Properties | Seawater | VS1new | VS7 | VS8 | HL345 |
|---|---|---|---|---|---|
| Density, kg/m3 | 1.027 | 1.209 | 1.094 | 1.191 | 1.197 |
| Total salinity, g/L | 34.5 | 313.7 | 155.7 | 297.8 | 304.1 |
| Ions, mmol/L | |||||
| Cl− | 563 | 5140 | 2557 | 5160 | 4826 |
| SO42− | 35 | 369 | 191 | 388 | 454 |
| Br− | 1 | 29 | 14 | 52 | 28 |
| Na+ | 478 | 1912 | 796.9 | 433.9 | 1667 |
| K+ | 11 | 48.9 | 20.5 | 11.3 | 44 |
| Mg2+ | 61 | 1979 | 1073 | 2751 | 2031 |
| Ca2+ | 11 | 1.8 | 4.0 | 2.2 | 2.1 |
| Na+/Cl− Ratio | 0.857 | 0.372 | 0.312 | 0.084 | 0.345 |
| Sample ID | Number of Sequences | Number of ASVs 1 | Coverage | Simpson Diversity Index | Shannon Diversity Index |
|---|---|---|---|---|---|
| VS8 | 17,865 | 180 | 1 | 65.82 | 4.54 |
| VS1new | 27,622 | 243 | 0.99 | 132.22 | 5.13 |
| VS3 | 41,356 | 198 | 0.99 | 103.19 | 4.93 |
| VCR | 56,284 | 228 | 1 | 86.72 | 4.77 |
| VFS | 50,992 | 144 | 1 | 63.37 | 4.47 |
| VS9 sed | 56,620 | 542 | 0.99 | 320.05 | 6.00 |
| VS2 | 48,344 | 463 | 0.99 | 292.39 | 5.89 |
| VS5 | 46,810 | 206 | 0.99 | 93.40 | 4.92 |
| VS4 | 56,518 | 434 | 0.99 | 148.66 | 5.41 |
| VS6 | 51,319 | 380 | 0.99 | 179.35 | 5.46 |
| Hai Lý 345 | 10,870 | 128 | 0.98 | 68.79 | 4.43 |
| VS7 | 31,136 | 308 | 1 | 228.86 | 5.56 |
| Hai Lý 450 | 4028 | 82 | 0.99 | 26.59 | 3.60 |
| VS1 | 31,334 | 415 | 0.99 | 268.90 | 5.78 |
| CP | 61,118 | 59 | 1 | 12.12 | 2.99 |
| SM16 | 41,430 | 199 | 0.99 | 133.43 | 5.07 |
| SM19 | 50,621 | 200 | 1 | 73.96 | 4.89 |
| SSR | 26,570 | 133 | 1 | 76.98 | 4.60 |
| STP | 37,020 | 358 | 1 | 180.16 | 5.42 |
| Total | 747,857 | 4900 |
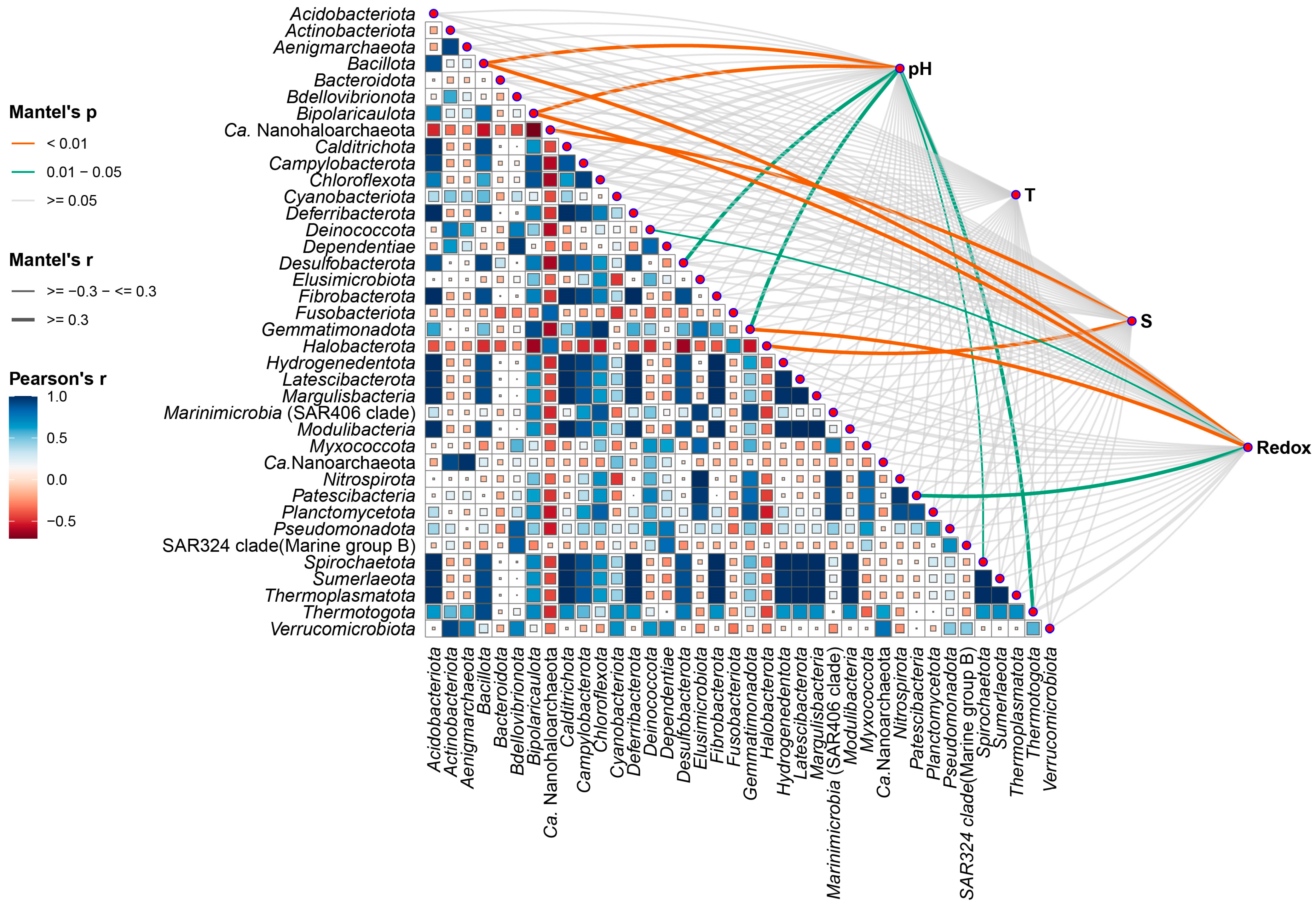
| Phylum | Environmental Factor | Mantel’s r | Mantel’s p | FDR |
|---|---|---|---|---|
| Nanohaloarchaeota | S | 0.899 | 0.001 | 0.013 |
| Halobacterota | S | 0.663 | 0.002 | 0.013 |
| Bacillota | pH | 0.503 | 0.006 | 0.020 |
| Bipolaricaulota | pH | 0.852 | 0.009 | 0.020 |
| Bacillota | Redox | 0.446 | 0.011 | 0.020 |
| Bipolaricaulota | Redox | 0.913 | 0.011 | 0.020 |
| Gemmatimonadota | Redox | 0.615 | 0.011 | 0.020 |
| Patescibacteria | Redox | 0.364 | 0.024 | 0.039 |
| Thermotogota | pH | 0.610 | 0.036 | 0.047 |
| Gemmatimonadota | pH | 0.506 | 0.038 | 0.047 |
| Desulfobacterota | pH | 0.335 | 0.046 | 0.048 |
| Deinococcota | Redox | 0.275 | 0.048 | 0.048 |
| Spirochaetota | pH | 0.253 | 0.049 | 0.047 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
La Cono, V.; La Spada, G.; Smedile, F.; Crisafi, F.; Marturano, L.; Modica, A.; Nhu Khanh, H.H.; Thinh, P.D.; Thuy Hang, C.T.; Selivanova, E.A.; et al. Unique Features of Extremely Halophilic Microbiota Inhabiting Solar Saltworks Fields of Vietnam. Microorganisms 2024, 12, 1975. https://doi.org/10.3390/microorganisms12101975
La Cono V, La Spada G, Smedile F, Crisafi F, Marturano L, Modica A, Nhu Khanh HH, Thinh PD, Thuy Hang CT, Selivanova EA, et al. Unique Features of Extremely Halophilic Microbiota Inhabiting Solar Saltworks Fields of Vietnam. Microorganisms. 2024; 12(10):1975. https://doi.org/10.3390/microorganisms12101975
Chicago/Turabian StyleLa Cono, Violetta, Gina La Spada, Francesco Smedile, Francesca Crisafi, Laura Marturano, Alfonso Modica, Huynh Hoang Nhu Khanh, Pham Duc Thinh, Cao Thi Thuy Hang, Elena A. Selivanova, and et al. 2024. "Unique Features of Extremely Halophilic Microbiota Inhabiting Solar Saltworks Fields of Vietnam" Microorganisms 12, no. 10: 1975. https://doi.org/10.3390/microorganisms12101975
APA StyleLa Cono, V., La Spada, G., Smedile, F., Crisafi, F., Marturano, L., Modica, A., Nhu Khanh, H. H., Thinh, P. D., Thuy Hang, C. T., Selivanova, E. A., Bản, N. K., & Yakimov, M. M. (2024). Unique Features of Extremely Halophilic Microbiota Inhabiting Solar Saltworks Fields of Vietnam. Microorganisms, 12(10), 1975. https://doi.org/10.3390/microorganisms12101975







