Increasing Ages of Inga punctata Tree Soils Facilitate Greater Fungal Community Abundance and Successional Development, and Efficiency of Microbial Organic Carbon Utilization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Inga punctata Reforestation Site Description and Soil Sample Collection
2.2. Soil Respiration, Biomass C, and qCO2
2.3. DNA Extraction, Sequencing, and Bioinformatics
2.4. Differences in Fungal Community Compositions
2.5. Indicators of Fungal Community Successional Development
2.6. Potential Influence of Fungal Taxa on the Carbon Metrics
3. Results
3.1. Differences in C Metrics
3.2. Differences in Fungal Community Compositions
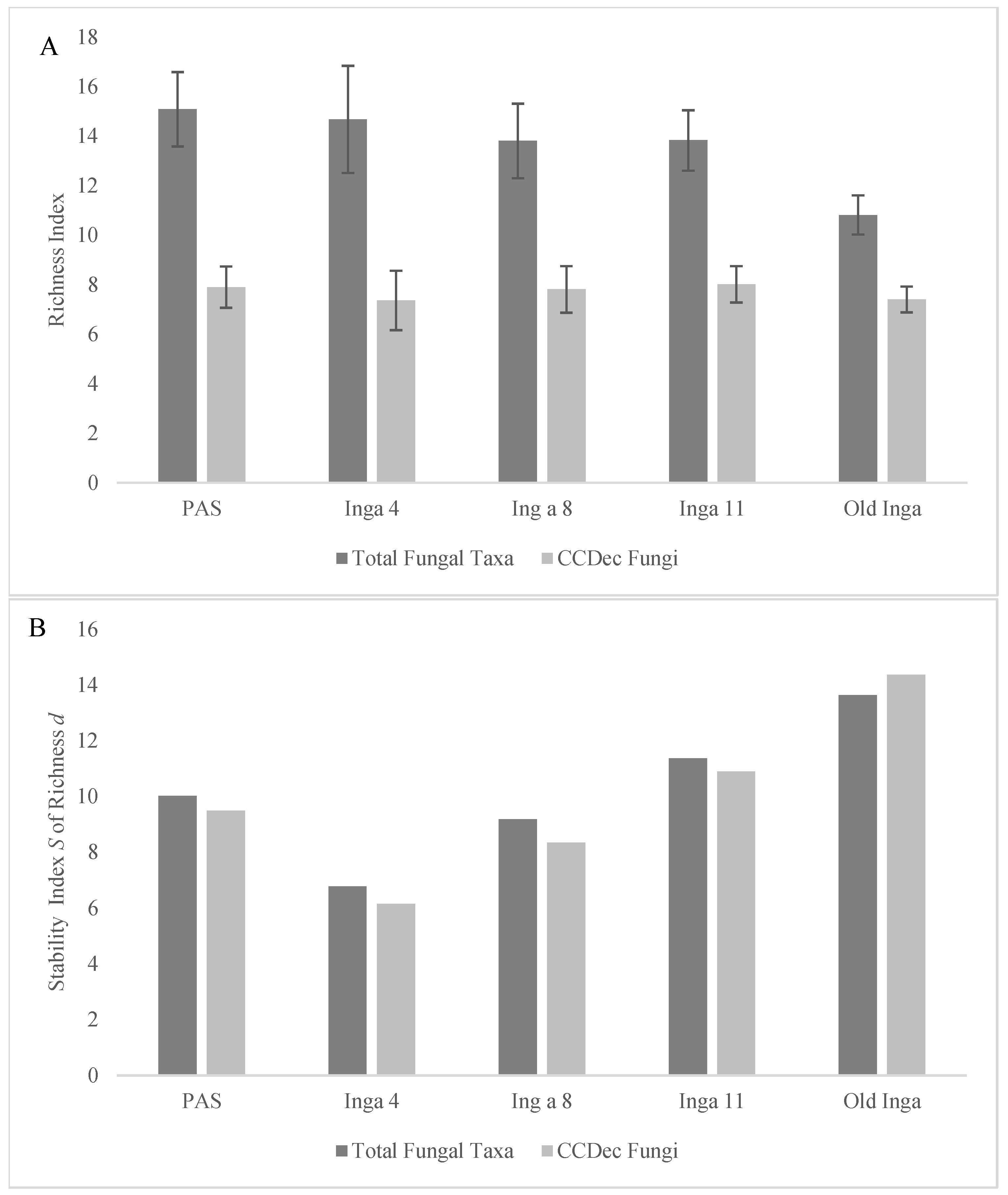
3.3. Indicators of Fungal Community Successional Development
3.4. Potential Influence of Fungal Taxa on the Carbon Metrics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arai, H.; Hadi, A.; Darung, U.; Limin, S.H. Land use change affects microbial biomass and fluxes of carbon dioxide and nitrous oxide in tropical peatlands. Soil Sci. Plant Nutr. 2014, 60, 423–434. [Google Scholar] [CrossRef]
- Lammel, D.R.; Feigl, B.J.; Cerri, C.C.; Nüsslein, K. Specific microbial gene abundances and soil parameters contribute to C, N, and greenhouse gas process rates after land use change in Southern Amazonian Soils. Front. Microbiol. 2015, 6, 1057. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.L.M.; Pellizari, V.H.; Mueller, R.; Baek, K.; Jesus, E.D.C.; Paula, F.S.; Mirza, B.; Hamaoui, G.S.; Tsai, S.M.; Feigl, B.; et al. Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2013, 110, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Eaton, W.D.; McGee, K.M.; Donnelly, R.; Lemenze, A.; Karas, O.; Hajibabaei, M. Differences in the soil microbial community and carbon-use efficiency following development of Vochysia guatemalensis tree plantations in unproductive pastures in Costa Rica. Restor. Ecol. 2019, 27, 1263–1273. [Google Scholar] [CrossRef]
- Eaton, W.D.; McGee, K.M.; Alderfer, K.; Jimenez, A.R.; Hajibabaei, M. Increase in abundance and decrease in richness of soil microbes following Hurricane Otto in three primary forest types in the Northern Zone of Costa Rica. PLoS ONE 2020, 15, e0231187. [Google Scholar] [CrossRef] [PubMed]
- Kardol, P.; Wardle, D.A. How understanding aboveground-belowground linkages can assist restoration ecology. Trends Ecol. Evol. 2010, 25, 670–679. [Google Scholar] [CrossRef]
- Aide, T.M.; Clark, M.L.; Ricardo Grau, H.; Lopez-Carr, D.; Levy, M.A.; Redo, D.; Bonilla-Moheno, M.; Riner, G.; Andrade-Núñez, M.J.; Muñiz, M. Deforestation and reforestation of Latin America and the Caribbean (2001–2010). Biotropica 2013, 45, 262–271. [Google Scholar] [CrossRef]
- Schelhas, J.; Sánchez-Azofeifa, G.A. Post-frontier forest changes adjacent to Braulio Carrillo National Park Costa Rica. Hum. Ecol. 2006, 34, 407–431. [Google Scholar] [CrossRef]
- Chazdon, R.L. Beyond Deforestation: Restoring Forests and Ecosystem Services on Degraded Lands. Science 2008, 320, 1458–1460. [Google Scholar] [CrossRef]
- van der Heijden, M.G.A.; Bardgett, R.D.; van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere: Plant species, soil type and rhizosphere communities. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mendes, M.S.; Latawiec, A.E.; Sansevero, J.B.B.; Crouzeilles, R.; Moraes, L.F.D.; Castro, A.; Alves-Pinto, H.N.; Brancalion, P.H.; Rodrigues, R.R.; Chazdon, R.L.; et al. Look down-there is a gap-the need to include soil data in Atlantic Forest restoration. Restor. Ecol. 2018, 27, 361–370. [Google Scholar] [CrossRef]
- Eaton, W.D.; Hamilton, D.A. Enhanced carbon, nitrogen and associated bacterial community compositional complexity, stability, evenness, and differences within the tree-soils of Inga punctata along an age gradient of planted trees in reforestation plots. Plant Soil 2022, 484, 327–346. [Google Scholar] [CrossRef]
- Wallenstein, M.D.; Burns, R.G. Ecology of extracellular enzyme activities and organic matter degradation in soil: A complex community-driven process. Methods Soil Enzymol. 2011, 9, 35–55. [Google Scholar]
- Xu, C.Y.; Du, C.; Jian, J.S.; Hou, L.; Wang, Z.K.; Wang, Q.; Geng, Z.C. The interplay of labile organic carbon, enzyme activities and microbial communities of two forest soils across seasons. Sci. Rep. 2021, 11, 5002. [Google Scholar] [CrossRef]
- Brown, M.E.; Chang, M.C. Exploring bacterial lignin degradation. Curr. Op. Chem. Biol. 2014, 19, 1–7. [Google Scholar] [CrossRef]
- Datta, R.A.; Kelkar, A.; Baraniya, D.; Molaei, A.; Moulick, A.; Meena, R.S.; Formanek, P.; Baraniya, D.; Molaei, A.; Moulick, A.; et al. Enzymatic Degradation of Lignin in Soil: A Review. Sustainability 2017, 9, 1163. [Google Scholar] [CrossRef]
- Kamimura, N.; Takahashi, K.; Mori, K.; Araki, T.; Fujita, M.; Higuchi, Y.; Masai, E. Bacterial catabolism of lignin-derived aromatics: New findings in a recent decade: Update on bacterial lignin catabolism. Environ. Microb. Rep. 2017, 9, 679–705. [Google Scholar] [CrossRef]
- Silva, J.P.; Ticona, A.R.P.; Hamann, P.R.V.; Quirino, B.F.; Noronha, E.F. Deconstruction of Lignin: From Enzymes to Microorganisms. Molecules 2021, 26, 2299. [Google Scholar] [CrossRef]
- Větrovský, T.; Steffen, K.T.; Baldrian, P. Potential of Cometabolic Transformation of Polysaccharides and Lignin in Lignocellulose by Soil Actinobacteria. PLoS ONE 2014, 9, e89108. [Google Scholar] [CrossRef]
- Wilhelm, R.C.; Singh, R.; Eltis, L.D.; Mohn, W.W. Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J. 2019 13, 413–429. [CrossRef]
- Hatakka, A.; Hammel, K.E. Fungal biodegradation of lignocelluloses. The Mycota: A comprehensive treatise on fungi as experimental systems for basic and applied research. In X, Industrial Applications, 2nd ed.; Esser, K., Hofrichter, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 319–340. [Google Scholar] [CrossRef]
- Rahman, M.M.; Tsukamoto, J.; Tokumoto, Y.; Shuvo, M.A.R. The role of quantitative traits of leaf litter on decomposition and nutrient cycling of the forest ecosystems. J. For. Sci. 2013, 29, 38–48. [Google Scholar] [CrossRef]
- Thevenot, M.; Dignac, M.F.; Rumpel, C. Fate of lignins in soils: A review. Soil Biol. Biochem. 2010, 42, 1200–1211. [Google Scholar] [CrossRef]
- Amazonas, N.T.; Martinelli, L.A.; Piccolo, M.D.C.; Rodrigues, R.R. Nitrogen dynamics during ecosystem development in tropical forest restoration. For. Ecol. Manag. 2011, 262, 1551–1557. [Google Scholar] [CrossRef]
- Batterman, S.A.; Hedin, L.O.; van Breugel, M.; Ransijn, J.; Craven, D.J.; Hall, J.S. Key role of symbiotic dinitrogen fixation in tropical forest secondary succession. Nature 2013, 502, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Davidson, E.A.; de Carvalho, C.J.R.; Figueira, A.M.; Ishida, F.Y.; Ometto, J.P.H.B.; Nardoto, G.B.; Sabá, R.T.; Hayashi, S.N.; Leal, E.C.; Vieira, I.C.G.; et al. Recuperation of nitrogen cycling in Amazonian forests following agricultural abandonment. Nature 2007, 447, 995–998. [Google Scholar] [CrossRef]
- Groppo, J.D.; Lins, S.R.M.; Camargo, P.B.; Assad, E.D.; Pinto, H.S.; Martins, S.C.; Salgado, P.R.; Evangelista, B.; Vasconcellos, E.; Sano, E.E.; et al. Changes in soil carbon, nitrogen, and phosphorus due to land-use changes in Brazil. Biogeosciences 2015, 12, 4765–4780. [Google Scholar] [CrossRef]
- Lojka, B.; Dumas, L.; Preininger, D.; Polesny, Z.; Banout, J. The use and integration of Inga edulis in agroforestry systems in the amazon–review article. Agric. Trop. Subtrop. 2010, 43, 352–359. [Google Scholar]
- Macedo, M.O.; Resende, A.S.; Garcia, P.C.; Boddey, R.M.; Jantalia, C.P.; Urquiaga, S.; Campello, E.F.C.; Franco, A.A. Changes in soil C and N stocks and nutrient dynamics 13 years after recovery of degraded land using leguminous nitrogen-fixer trees. For. Ecol. Manag. 2008, 255, 1516–1524. [Google Scholar] [CrossRef]
- Piotto, D.; Montagnini, F.; Ugalde, L.; Kanninen, M. Performance of forest plantations in small and medium-sized farms in the Atlantic lowlands of Costa Rica. For. Ecol. Manag. 2003, 175, 195–204. [Google Scholar] [CrossRef]
- Leblanc, H.A.; McGraw, R.L.; Nygren, P.; Roux, C.L. Neotropical legume tree Inga edulis forms N2-fixing symbiosis with fast-growing Bradyrhizobium strains. Plant Soil 2005, 275, 123–133. [Google Scholar] [CrossRef]
- Hamilton, D. Offsetting Destruction: The Important Functional Contribution of Carbon Sequestration in the Restoration of a Tropical Forest in Monteverde, Costa Rica; Reference Module in Earth Systems and Environmental Sciences; Elsevier: Amsterdam, The Netherlands, 2022; ISBN 9780124095489. [Google Scholar] [CrossRef]
- Ettema, C. Spatial soil ecology. Trends Ecol. Evol. 2002, 17, 177–183. [Google Scholar] [CrossRef]
- Turbé, A.; De Toni, A.; Benito, P.; Lavelle, P.; Lavelle, P.; Camacho, N.R.; van Der Putten, W.H.; Labouze, E.; Mudgal, S. Soil Biodiversity: Functions, Threats and Tools for Policy Makers. Bio Intelligence Service, IRD, and NIOO, Technical Report 2010-49 for European Commission (DG Environment); Bio Intelligenence Service SAS: Paris, France, 2010. [Google Scholar]
- Bardgett, R.D.; van der Putten, W.H. Belowground biodiversity and ecosystem functioning. Nature 2014, 515, 505–511. [Google Scholar] [CrossRef]
- Addison, S.; Daley, K. Getting to the root of Tree Soil Microbiome Sampling. Phytobiomes 2023, 7, 352–364. [Google Scholar] [CrossRef]
- Lucas, Y.; Santin, R.C.; Da Silva, W.T.L.; Merdy, P.; Melfi, A.J.; Pereira, O.J.R.; Montes, C.R. Soil sample conservation from field to lab for heterotrophic respiration assessment. MethodsX 2020, 7, 101039. [Google Scholar] [CrossRef] [PubMed]
- Alef, K.; Nannipieri, P. Methods in Applied Soil Microbiology and Biochemistry; Elsevier: Amsterdam, The Netherlands; Academic Press: Cambridge, MA, USA, 1995. [Google Scholar]
- Anderson, T.H.; Domsch, K.H. Soil microbial biomass: The eco-physiological approach. Soil Biol. Biochem. 2010, 42, 2039–2043. [Google Scholar]
- He, H.; Liu, Y.; Hu, Y.; Zhang, M.; Wang, G.; Shen, W. Soil microbial community and its interaction with soil carbon dynamics following a wetland drying process in mu us sandy land. Int. J. Environ. Res. Public Health 2020, 17, 4199. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, L.B.; Korthals, G.; Brussaard, L.; Jørgensen, H.B.; De Deyn, G.B. Organic management and cover crop species steer soil microbial community structure and functionality along with soil organic matter properties. Agric. Ecosyst. Environ. 2018, 263, 7–17. [Google Scholar] [CrossRef]
- Six, J.; Frey, S.D.; Thiet, R.K.; Batten, K.M. Bacterial and fungal contributions to carbon sequestration in agroecosystems. Soil Sci. Soc. Am. J. 2006, 70, 555–569. [Google Scholar] [CrossRef]
- Spohn, M. Microbial respiration per unit microbial biomass depends on litter layer carbon-to-nitrogen ratio. Biogeosciences 2015, 12, 817–823. [Google Scholar] [CrossRef]
- Wallenstein, M.D.; Hall, E.K. A trait-based framework for predicting when and where microbial adaptation to climate change will affect ecosystem functioning. Biogeochem. 2011, 109, 35–47. [Google Scholar] [CrossRef]
- McGee, K.M.; Eaton, W.D.; Porter, T.M.; Shokralla, S.; Hajibabaei, M. Soil microbiomes associated with two dominant Costa Rican tree species, and implications for remediation—A case study from a Costa Rican conservation area. Appl. Soil Ecol. 2019, 137, 139–153. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.-L.; Porras-Alfaro, A.; Kuske, C.R.; Eichorst, S.A.; Xie, G. Accurate, rapid taxonomic classification of fungal large-subunit rRNA genes. Appl. Environ. Microbiol. 2012, 78, 1523–1533. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 27. [Google Scholar] [CrossRef]
- Põlme, S.; Abarenkov, K.; Henrik, N.R.; Lindah, B.D.; Clemmensen, K.E.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.T.; Baldrian, P.; et al. FungalTraits: A user-friendly traits database of fungi and funguslike stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]
- Eaton, W.D.; McGee, K.M.; Hoke, E.; Lemenze, A.; Hajibabaei, M. Influence of Two Important Leguminous Trees on Their Soil Microbiomes and Nitrogen Cycle Activities in a Primary and Recovering Secondary Forest in the Northern Zone of Costa Rica. Soil Syst. 2020, 4, 65. [Google Scholar] [CrossRef]
- Eaton, W.D.; McGee, K.M.; Larimer, M.; Hoke, E.; Karas, O.; Hernandez, B.; Wayland, N.A. Changes in soil bacterial communities, and carbon and nitrogen metrics as potential indicators of land use effects in a humid tropical forest. Pedobiol. Soil Ecol. J. 2021, 85–86, 150730. [Google Scholar]
- Eaton, W.D.; McGee, K.M.; Donnelly, R.; Lemenze, A.; Larimer, M.; Hajibabaei, M. The impacts of a logging road on the soil microbial communities, and carbon and nitrogen components in a Northern Zone Costa Rican Forest. Appl. Soil Ecol. 2021, 164, 103937. [Google Scholar] [CrossRef]
- Eaton, W.D.; Hamilton, D.A.; Chen, W.; Lemenze, A.; Soteropoulos, P. Use of high throughput DNA analysis to characterize the nodule-associated bacterial community from four ages of Inga punctata trees in a Costa Rican cloud forest. AIMS Microbiol. 2024, 10, 572–595. [Google Scholar] [CrossRef]
- Anderson, M.J.; Gorley, R.N.; Clarke, K.R. PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods; PRIMER-E Ltd.: Plymouth, UK, 2008. [Google Scholar]
- Clarke, K.R.; Gorley, R.N. PRIMER v6: User Manual; PRIMER-E Ltd.: Plymouth, UK, 2006. [Google Scholar]
- Anderson, M.J.; Willis, T.J. Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology. Ecology 2003, 84, 511–525. [Google Scholar] [CrossRef]
- Allison, S.D.; Martiny, J.B.H. Colloquium paper: Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512–11519. [Google Scholar] [CrossRef]
- Atkinson, J.; Brudvig, L.A.; Mallen-Cooper, M.; Nakagawa, S.; Moles, A.T.; Bonser, S.P. Terrestrial ecosystem restoration increases biodiversity and reduces its variability, but not to reference levels: A global meta-analysis. Ecol. Lett. 2022, 25, 1725–1737. [Google Scholar] [CrossRef]
- Galand, P.E.; Lucas, S.; Fagervold, S.K.; Peru, E.; Pruski, A.M.; Vétion, G.; Dupuy, C.; Guizien, K. Disturbance Increases Microbial Community Diversity and Production in Marine Sediments. Front. Microbiol. 2016, 7, 1950. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Doebeli, M. Transient dynamics of competitive exclusion in microbial communities. Environ. Microbiol. 2016, 18, 1863–1874. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Jacques, S.M.S.; Pires, A.P.F.; Leal, J.S.; Srivastava, D.S.; Parfrey, L.W.; Farjalla, V.F.; Doebeli, M. High taxonomic variability despite stable functional structure across microbial communities. Nat. Ecol. Evol. 2016, 1, 0015. [Google Scholar] [CrossRef]
- Grimm, V.; Wissel, C. Babel, or the ecological stability discussions: An inventory and analysis of terminology and a guide for avoiding confusion. Oecologia 1997, 109, 323–334. [Google Scholar] [CrossRef]
- Tilman, D.; Reich, P.B.; Knops, J.M. Biodiversity and ecosystem stability in a decade-long grassland experiment. Nature 2006, 441, 629–632. [Google Scholar] [CrossRef]
- Ruijven, J.; Berendse, F. Contrasting Effects of Diversity on the Temporal Stability of Plant Populations. Oikos 2007, 116, 1323–1330. [Google Scholar] [CrossRef]
- Isbell, F.I.; Polley, H.W.; Wilsey, B.J. Biodiversity, productivity and the temporal stability of productivity: Patterns and Processes. Ecol. Lett. 2009, 12, 443–451. [Google Scholar] [CrossRef]
- Liu, J.; Jia, X.; Yan, W.; Zhong, Y.; Shangguan, Z. Changes in microbial community structure during secondary succession. Land Degrad. Dev. 2020, 31, 1151–1166. [Google Scholar] [CrossRef]
- Shade, A.; Peter, H.; Allison, S.D.; Baho, D.L.; Berga, M.; Bürgmann, H.; Huber, D.H.; Langenheder, S.; Lennon, J.T.; Martiny, J.B.H.; et al. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 2012, 3, 417. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, B.S.; Philippot, L. Insights into the resistance and resilience of the soil microbial community. FEMS Microbiol. Rev. 2013, 37, 112–129. [Google Scholar] [CrossRef] [PubMed]
- Luis, P.; Walther, G.; Kellner, H.; Martin, F.; Buscot, F. Diversity of laccase genes from basidiomycetes in a forest soil. Soil Biol. Biochem. 2004, 36, 1025–1036. [Google Scholar] [CrossRef]
- Peng, J.; Liu, H.; Hu, Y.; Sun, Y.; Liu, Q.; Li, J.; Dong, Y. Shift in soil bacterial communities from K- to r-strategists facilitates adaptation to grassland degradation. Land Degrad. Develop. 2022, 33, 2076–2091. [Google Scholar] [CrossRef]
- Pinto-Figueroa, E.A.; Seddon, E.; Yashiro, E.; Buri, A.; Niculita-Hirzel, H.; van der Meer, J.R.; Guisan, A. Archaeorhizomycetes spatial distribution in soils along wide elevational and environmental gradients reveal co-abundance patterns with other fungal saprobes and potential weathering capacities. Front. Microbiol. 2019, 10, 656. [Google Scholar] [CrossRef]
- Rosling, A.; Cox, F.; Cruz-Martinez, K.; Ihrmark, K.; Grelet, G.-A.; Lindahl, B.D.; Menkis, A.; James, T.Y. Archaeorhizomycetes: Unearthing an ancient class of ubiquitous soil fungi. Science 2011, 333, 876–879. [Google Scholar] [CrossRef]
- Rosling, A.; Timling, I.; Taylor, D.L. Archaeorhizomycetes: Patterns of distribution and abundance in soil. In Genomics of Soil- and Plant-Associated Fungi; Horwitz, B., Mukherjee, P., Mukherjee, M., Kubicek, C., Eds.; Soil Biology; Springer: Berlin/Heidelberg, Germany, 2013; pp. 333–349. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Puusepp, R.; Nilsson, R.H.; James, T.Y. Novel soil-inhabiting clades fill gaps in the fungal tree of life. Microbiome 2017, 5, 42. [Google Scholar] [CrossRef]
- Turatsinze, A.N.; Kang, B.; Zhu, T.; Hou, F.; Bowatte, S. Soil bacterial and fungal composition and diversity responses to seasonal deer grazing in a subalpine meadow. Diversity 2021, 13, 84. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P.; et al. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef]
- Stürmer, S.L.; Kemmelmeier, K. The Glomeromycota in the neotropics. Front. Microbiol. 2021, 11, 553679. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Polz, M.F.; Mazel, F.; Albright, M.B.N.; Huber, J.A.; O’Connor, M.I.; Ackermann, M.; Hahn, A.S.; Srivastava, D.S.; Crowe, S.A.; et al. Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2018, 2, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Mori, A.S.; Isbell, F.; Fujii, S.; Makoto, K.; Matsuoka, S.; Osono, T. Low multifunctional redundancy of soil fungal diversity at multiple scales. Ecol. Lett. 2015, 19, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Kirkby, C.A.; Schmutter, D.; Bissett, A. Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 2016, 97, 188–198. [Google Scholar] [CrossRef]
- Wagg, C.; Jansa, J.; Schmid, B.; van der Heijden, M.G. Belowground biodiversity effects of plant symbionts support aboveground productivity. Ecol. Lett. 2011, 14, 1001–1009. [Google Scholar] [CrossRef]
- Xu, H.; Detto, M.; Li, Y.; He, F.; Fang, S. Do N-fixing legumes promote neighbor diversity in the tropics? J. Ecol. 2019, 107, 229–239. [Google Scholar] [CrossRef]
- Taylor, B.N.; Chazdon, R.L.; Bachelot, B.; Menge, D.N.L. Nitrogen-fixer trees inhibit growth of regenerating Costa Rican rainforests. Proc. Natl. Acad. Sci. USA 2017, 114, 8817–8822. [Google Scholar] [CrossRef]
- Lai, H.R.; Hall, J.S.; Batterman, S.A.; Turner, B.L.; Breugel, M. Nitrogen fixer abundance has no effect on biomass recovery during tropical secondary forest succession. J. Ecol. 2018, 106, 1415–1427. [Google Scholar] [CrossRef]
- Arroyo-Rodríguez, V.; Melo, F.P.; Martínez-Ramos, M.; Bongers, F.; Chazdon, R.L.; Meave, J.A.; Norden, N.; Santos, B.A.; Leal, I.R.; Tabarelli, M. Multiple successional pathways in human-modified tropical landscapes: New insights from forest succession, forest fragmentation and landscape ecology research. Biol. Rev. Camb Philos Soc. 2017, 92, 326–340. [Google Scholar] [CrossRef]
- Abdel-Azeem, A.M.; Gherbawy, Y.A.; Sabry, A.M. Enzyme profiles and genotyping of Chaetomium globosum isolates from various substrates. Plant Biosyst. 2016, 150, 420–428. [Google Scholar] [CrossRef]
- Abdel-Azeem, A.M.; Abu-Elsaoud, A.M.; Abo Nahas, H.H.; Abdel-Azeem, M.A.; Balbool, B.A.; Mousa, M.K.; Ali, N.H.; Darwish, A.M.G. Biodiversity and industrial applications of genus Chaetomium. In Industrially Important Fungi for Sustainable Development; Biodiversity and Ecological Perspectives; Springer International Publishing: Cham, Switzerland, 2021; Volume 1. [Google Scholar] [CrossRef]
- Alimadadi, N.; Soudi, M.R.; Wang SAn QiWang, M.; Talebpour, Z.; Yan Bai, F. Starmerella orientalis f.a., sp. Nov., an ascomycetous yeast species isolated from flowers. Int. J. Syst. Evol. Microbiol. 2016, 66, 1476–1481. [Google Scholar] [CrossRef] [PubMed]
- Aliyu, H.; Gorte, O.; Neumann, A.; Ochsenreither, K. Global Transcriptome Profile of the Oleaginous Yeast Saitozyma podzolica DSM 27192 Cultivated in Glucose and Xylose. J. Fungi 2021, 7, 758. [Google Scholar] [CrossRef] [PubMed]
- Bilański, P.; Grad, B.; Kowalski, T. Pyrenochaeta fraxinina as colonizer of ash and sycamore petioles, its morphology, ecology, and phylogenetic connections. Mycol. Prog. 2022, 21, 74. [Google Scholar] [CrossRef]
- Čadež, N.; Drumonde-Neves, J.; Sipiczki, M.; Dlauchy, D.; Lima, T.; Pais, C.; Schuller, D.; Franco-Duarte, R.; Lachance, M.A.; Péter, G. Starmerella vitis fa, sp. nov., a yeast species isolated from flowers and grapes. Antonie Leeuwenhoek 2020, 113, 1289–1298. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Jeewon, R.; Hyde, K.D. Phylogenetic investigations of Sordariaceae based on multiple gene sequences and morphology. Mycol. Res. 2006, 110, 137–150. [Google Scholar] [CrossRef]
- Dix, N.J.; Webster, J. Fungal Ecology; Chapman and Hall: London, UK, 1995; 556p. [Google Scholar] [CrossRef]
- Gonçalves, P.; Gonçalves, C.; Brito, P.H.; Sampaio, J.P. The Wickerhamiella/Starmerella clade-A treasure trove for the study of the evolution of yeast metabolism. Yeast 2020, 37, 313–320. [Google Scholar] [CrossRef]
- Gong, Z.; Wang, Q.; Shen, H.; Hu, C.; Jin, G.; Zhao, Z.K. Co-fermentation of cellobiose and xylose by Lipomyces starkeyi for lipid production. Bioresour. Technol. 2012, 117, 20–24. [Google Scholar] [CrossRef]
- Holland, H.L. Investigation of the carbon- and sulfur-oxidizing capabilities of microorganisms by active-site modeling. In Advances in Applied Microbiology; Neidleman, S.L., Laskin, A.I., Eds.; Academic Press: Cambridge, MA, USA, 1997; Volume 44, pp. 125–165. ISSN 0065-2164. ISBN 9780120026449. [Google Scholar] [CrossRef]
- Kedves, O.; Kocsubé, S.; Bata, T.; Andersson, M.A.; Salo, J.M.; Mikkola, R.; Salonen, H.; Szűcs, A.; Kedves, A.; Kónya, Z.; et al. Chaetomium and chaetomium-like species from European indoor environments include Dichotomopilus finlandicus sp. nov. Pathogens 2021, 10, 1133. [Google Scholar] [CrossRef]
- Ladevèze, S.; Haon, M.; Villares ACathala, B.; Grisel, S. The yeast Geotrichum candidum encodes functional lytic polysaccharide monooxygenases. Biotechnol. Biofuels 2017, 10, 215. [Google Scholar] [CrossRef]
- Mandyam, K.; Jumpponen, A. Seeking the elusive function of the root-colonising dark septate endophytic fungi. Stud. Mycol. 2005, 53, 173–189. [Google Scholar] [CrossRef]
- Matos, I.T.S.R.; Cassa-Barbosa, L.A.; Costa-Neto, P.Q.; Astolfi-Filho, S. Cultivation of Trichosporon mycotoxinivorans in sugarcane bagasse hemicellulosic hydrolyzate. Electron. J. Biotechnol. 2012, 15, 12–18. [Google Scholar] [CrossRef]
- Menkis, A.; Urbina, H.; James, T.Y.; Rosling, A. Archaeorhizomyces borealis sp. nov. and a sequence-based classification of related soil fungal species. Fungal Biol. 2014, 118, 943–955. [Google Scholar] [CrossRef] [PubMed]
- Molnar, O.; Schatzmayr, G.; Fuchs, E.; Prillinger, H. Trichosporon mycotoxinivorans sp. nov., a new yeast species useful in biological detoxification of various mycotoxins. Syst. Appl. Microbiol. 2004, 27, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Ozimek, E.; Hanaka, A. Mortierella Species as the PlantGrowth-Promoting Fungi Present in the Agricultural Soils. Agriculture 2021, 11, 7. [Google Scholar] [CrossRef]
- Rodriguez, R.J.; White Jr, J.F.; Arnold, A.E.; Redman, R.S. Fungal endophytes: Diversity and functional roles. New Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef]
- Parveen, S.; Ali, M.I.; Aslam, M.; Ali, I.; Jamal, A.; Al-Ansari, M.M.; Al-Humaid, L.; Urynowicz, M.; Huang, Z. Optimizing biocatalytic potential of Dipodascus australiensis M-2 for degrading lignin under laboratory conditions. Microbiol. Res. 2022, 265, 127179. [Google Scholar] [CrossRef]
- Schlegel, M.; Münsterkötter, M.; Güldener, U.; Bruggmann, R.; Duò, A.; Hainaut, M.; Henrissat, B.; Sieber, C.M.; Hoffmeister, D.; Grünig, C.R. Globally distributed root endophyte Phialocephala subalpina links pathogenic and saprophytic lifestyles. BMC Genom. 2016, 17, 1015. [Google Scholar] [CrossRef]
- Špetík, M.; Berraf-Tebbal, A.; Pokluda, R.; Eichmeier, A. Pyrenochaetopsis kuksensis (Pyrenochaetopsidaceae), a new species associated with an ornamental boxwood in the Czech Republic. Phytotaxa 2021, 498, 177–185. [Google Scholar] [CrossRef]
- Vekiru, E.; Hametner, C.; Mitterbauer, R.; Rechthaler, J.; Adam, G.; Schatzmayr, G.; Krska, R.; Schuhmacher, R. Cleavage of zearalenone by Trichosporon mycotoxinivorans to a novel nonestrogenic metabolite. Appl. Environ. Microbiol. 2010, 76, 2353–2359. [Google Scholar] [CrossRef]
- Wang, H.M.D.; Cheng, Y.S.; Huang, C.H.; Huang, C.-W. Optimization of High Solids Dilute Acid Hydrolysis of Spent Coffee Ground at Mild Temperature for Enzymatic Saccharification and Microbial Oil Fermentation. Appl. Biochem. Biotechnol. 2016, 180, 753–765. [Google Scholar] [CrossRef]
- Weiss, M.; Bauer, R.; Sampaio, J.P.; Oberwinkler, F. Systematics and Evolution; McLaughlin, D.J., Spatafora, J.W., Eds.; The Mycota VII Part A; Springer: Berlin/Heidelberg, Germany, 2014; pp. 331–355. [Google Scholar] [CrossRef]

| (A) | |||
| Habitat | Biomass C (µgCO2/g Soil) | Resp (µgCO2/g Soil) | qCO2 (Resp/Biomass C) |
| PAS | 806.32 ± 64.57 | 261. 04 ± 22.58 | 0.33 ± 0.06 |
| Inga 4 | 547.50 ± 31.05 | 180.67 ± 19.49 | 0.33 ± 0.04 |
| Inga 8 | 540.67 ± 17.13 | 181.33 ± 14.17 | 0.34 ± 0.03 |
| Inga 11 | 725.33 ± 58.29 | 201.65 ± 28.90 | 0.28 ± 0.03 |
| Old Inga | 934.83 ± 70.06 | 209.83 ± 30.47 | 0.22 ± 0.04 |
| (B) | |||
| Comparisons | p Values for Comparisons of Means | ||
| Biomass C | Respiration | qCO2 | |
| PAS to Inga 4 | <0.0001 | <0.0001 | 1 |
| PAS to Inga 8 | <0.0001 | <0.0001 | 1 |
| PAS to Inga 11 | 0.0001 | 0.022 | 0.034 |
| PAS to Old Inga | 0.031 | 0.008 | 0.006 |
| Old Inga to Inga 4 | <0.0001 | 0.054 | 0.0002 |
| Old Inga to Inga 8 | <0.0001 | 0.051 | 0.0005 |
| Old Inga to Inga 11 | 0.0002 | 0.644 | 0.015 |
| Inga 11 to Inga 4 | 0.0001 | 0.145 | 0.006 |
| Inga 11 to Inga 8 | 0.0001 | 0.143 | 0.033 |
| Inga 8 to Inga 4 | 0.6472 | 0.942 | 0.635 |
| Taxa | Function | MPS PAS | MPS Inga 4 | MPS Inga 8 | MPS Inga 11 | MPS Old Inga |
|---|---|---|---|---|---|---|
| Apiotrichum | CCDec | 5.37% | 0.19% | 6.16% | 43.61% | 41.96% |
| Archaeorhizomyces | RADec | 32.11% | 3.74% | 5.31% | 2.02% | 3.53% |
| Chaetomium | CCDec | 0.29% | 1.73% | 0.99% | 0.3% | 0.57% |
| Dipodascus | CCDec | 4.21% | 0% | 0.56% | 2.02% | 0.54% |
| Geotrichum | CCDec | 0.18% | 0% | 0.07% | 1.28% | 0.22% |
| Glomeromycota | ARM | 4.8% | 19.29% | 18.41% | 12.65% | 15.93% |
| Leohumicola | SAPDec | 2.46% | 0.05% | 0.17% | 0% | 0.38% |
| Lipomyces | CCDec | 0% | 0.23% | 1.54% | 1.01% | 0.99% |
| Mortierella | CCDec | 0.37% | 1.15% | 0.38% | 0.02% | 0.45% |
| Phialocephala | CCDec | 1.87% | 3.56% | 2.23% | 1.84% | 0.89% |
| Pleosporales | SAPDec | 1.64% | 0.46% | 0.38% | 0.28% | 0.2% |
| Pyrenochaetopsis | CCDec | 0.79% | 0.23% | 1.94% | 0.77% | 0% |
| Rozella | PAR | 7.97% | 0.76% | 1.92% | 1.61% | 0.84% |
| Saitozyma | CCDec | 0.74% | 29.42% | 28.89% | 7.68% | 8.77% |
| Sordariaceae | CCDec | 1.44% | 7.98% | 11.6% | 0.29% | 1.05% |
| Starmerella | SAPDec | 0% | 2.95% | 2.35% | 8.13% | 11.44% |
| Tremella | CCDec | 0.04% | 0.67% | 1.5% | 0.22% | 8.21% |
| MPS of CCDec Taxa | 15.30% | 47.16% | 55.86% | 59.04% | 63.65% |
| (a) | |||||
| ANOSIM of Total Fungal Taxa | CAP of Total Fungal Taxa | ||||
| Global R = 0.404 | CAP Model p Value = 0.0024 | ||||
| Global p Value 0.0001 | |||||
| Pairwise Groups | R Statistic | p Values | Comparisons | R2 Value | Strength of Diff. |
| PAS and Inga 4 | 0.369 | 0.015 | PAS and Inga 4 | 0.568 | moderate |
| PAS and Inga 8 | 0.439 | 0.002 | PAS and Inga 8 | 0.568 | moderate |
| PAS and Inga 11 | 0.576 | 0.002 | PAS and Inga 11 | 0.791 | strong |
| PAS and Old Inga | 0.564 | 0.009 | PAS and Old Inga | 0.791 | strong |
| Inga 4 and Inga 8 | 0.097 | 0.182 | Inga 4 and Inga 8 | 0.044 | no difference |
| Inga 4 and Inga 11 | 0.669 | 0.002 | Inga 4 and Inga 11 | 0.791 | strong |
| Inga 4 and Old Inga | 0.564 | 0.016 | Inga 4 and Old Inga | 0.791 | strong |
| Inga 8 and Inga 11 | 0.323 | 0.019 | Inga 8 and Inga 11 | 0.449 | weak |
| Inga 8 and Old Inga | 0.503 | 0.053 | Inga 8 and Old Inga | 0.568 | moderate |
| Inga 11 and Old Inga | 0.201 | 0.037 | Inga 11 and Old Inga | 0.044 | no difference |
| (b) | |||||
| ANOSIM of CCDec Fungal Taxa | CAP of CCDec Fungal Taxa | ||||
| Global R = 0.473 | CAP model p Value = 0.0006 | ||||
| Global p Value 0.0001 | |||||
| Pairwise Groups | R Value | p Value | Pairwise Groups | R2 Value | Strength of Diff. |
| PAS and Inga 4 | 0.379 | 0.019 | PAS and Inga 4 | 0.582 | moderate |
| PAS and Inga 8 | 0.533 | 0.004 | PAS and Inga 8 | 0.687 | moderate |
| PAS and Inga 11 | 0.654 | 0.002 | PAS and Inga 11 | 0.745 | strong |
| PAS and Old Inga | 0.616 | 0.006 | PAS and Old Inga | 0.745 | strong |
| Inga 4 and Inga 8 | 0.027 | 0.359 | Inga 4 and Inga 8 | 0.019 | no difference |
| Inga 4 and Inga 11 | 0.853 | 0.002 | Inga 4 and Inga 11 | 0.745 | strong |
| Inga 4 and Old Inga | 0.648 | 0.024 | Inga 4 and Old Inga | 0.745 | strong |
| Inga 8 and Inga 11 | 0.368 | 0.039 | Inga 8 and Inga 11 | 0.233 | weak |
| Inga 8 and Old Inga | 0.607 | 0.002 | Inga 8 and Old Inga | 0.582 | moderate |
| Inga 11 and Old Inga | 0.320 | 0.017 | Inga 11 and Old Inga | 0.019 | no difference |
| % Contribution of Fungal Taxa to the Total Fungal Community Composition in PAS Soils | % Contribution of Fungal Taxa to the Total Fungal Community Composition in Inga 4 Soils | ||||
| Species | % Contribution | Function | Species | % Contribution | Function |
| Archaeorhizomyces | 18.22 | RADec | Glomeromycota | 22.63 | ARM |
| Rozella | 13.69 | PAR | Saitozyma | 16.48 | CCDec |
| Apiotrichum | 9.48 | CCDec | Phialocephala | 9.84 | RACCDec |
| Glomeromycota | 9.45 | ARM | Sordariaceae | 9.39 | CCDec |
| Dipodascus | 7.32 | CCDEC | Archaeorhizomyces | 6.25 | RADec |
| Leohumicola | 3.91 | SAPDec | Starmerella | 5.96 | SAPDec |
| Pleosporales | 3.58 | SAPDec | Chaetomium | 3.69 | CCDec |
| Sordariaceae | 3.48 | CCDec | Mortierella | 3.01 | CCDec |
| Phialocephala | 2.98 | CCDec | Rozella | 2.61 | PAR |
| Pyrenochaetopsis | 1.85 | CCDec | Pleosporales | 1.78 | SAPDec |
| Total % Contr CCDec | 25.11 | Total % Contr CCDec | 42.41 | ||
| % Contribution of Fungal Taxa to the Total Fungal Community Composition In Inga 8 Soils | % Contribution of Fungal Taxa to the Total Fungal Community Composition in Inga 11 Soils | ||||
| Species | % Contribution | Function | Species | % Contribution | Function |
| Glomeromycota | 18.65 | ARM | Apiotrichum | 22.38 | CCDec |
| Saitozyma | 15.35 | CCDec | Glomeromycota | 16.28 | ARM |
| Archaeorhizomyces | 11.12 | RADec | Saitozyma | 10.95 | CCDec |
| Sordariaceae | 9.85 | CCDec | Starmerella | 10.45 | SAPDec |
| Apiotrichum | 6.99 | CCDec | Dipodasus | 8.16 | CCDec |
| Phialocephela | 6.06 | CCDec | Archaeorhizomyces | 6.24 | RADec |
| Starmerella | 5.18 | SAPDec | Phialocephala | 5.16 | CCDec |
| Rozella | 5.11 | PAR | Rozella | 4.82 | PAR |
| Pyrenochaetopsis | 4.63 | CCDec | Geotrichum | 4.35 | CCDec |
| Lipomyces | 4.14 | CCDec | Lipomyces | 4.13 | CCDec |
| Total % Contr CCDec | 47.02 | Total % Contr CCDec | 55.13 | ||
| % Contribution of Fungal Taxa to the Total Fungal Community Composition in Old Inga Soils | |||||
| Species | % Contribution | Function | |||
| Apiotrichum | 25.53 | CCDec | Key To Functions: RADec: root-associated decomposers SAPDec: saprobic simple C decomposers CCDec: complex C decomposers PAR: endoparasites | ||
| Glomeromycota | 17.62 | ARM | |||
| Starmerella | 14.77 | SAPDec | |||
| Saitozyma | 10.24 | CCDec | |||
| Tremella | 10.23 | CCDec | |||
| Archaeorhizomyces | 8.19 | RADec | |||
| Sordariaceae | 5.84 | CCDec | |||
| Rozella | 4.24 | PAR | |||
| Lipomyces | 3.85 | CCDec | |||
| Phialocephala | 3.07 | CCDec | |||
| Total % Contr CCDec | 58.76 | ||||
| Effects on All C metrics | AICc | Pseudo-F | p value | Variation | Cumulative Variation |
| Saitozyma | 24.793 | 11.03 | 0.0001 | 29.79% | 29.79% |
| Apiotrichum | 20.996 | 6.3272 | 0.0093 | 14.18% | 43.97% |
| Effects on Biomass C | AICc | Pseudo-F | p value | Variation | Cumulative Variation |
| Saitozyma | 16.549 | 24.656 | 0.0003 | 48.67% | 48.67% |
| Effects on Respiration | AICc | Pseudo-F | p value | Variation | Cumulative Variation |
| Saitozyma | 16.362 | 17.637 | 0.0003 | 40.42% | 40.42% |
| Apiotrichum | 11.137 | 7.9661 | 0.0096 | 14.40% | 54.82% |
| Effects on qCO2 | AICc | Pseudo-F | p value | Variation | Cumulative Variation |
| Apiotrichum | 7.7003 | 9.1392 | 0.0055 | 26.00% | 26.00% |
| Saitozyma | 3.4048 | 6.8898 | 0.0153 | 15.99% | 41.99% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eaton, W.D.; Hamilton, D.A. Increasing Ages of Inga punctata Tree Soils Facilitate Greater Fungal Community Abundance and Successional Development, and Efficiency of Microbial Organic Carbon Utilization. Microorganisms 2024, 12, 1996. https://doi.org/10.3390/microorganisms12101996
Eaton WD, Hamilton DA. Increasing Ages of Inga punctata Tree Soils Facilitate Greater Fungal Community Abundance and Successional Development, and Efficiency of Microbial Organic Carbon Utilization. Microorganisms. 2024; 12(10):1996. https://doi.org/10.3390/microorganisms12101996
Chicago/Turabian StyleEaton, William D., and Debra A. Hamilton. 2024. "Increasing Ages of Inga punctata Tree Soils Facilitate Greater Fungal Community Abundance and Successional Development, and Efficiency of Microbial Organic Carbon Utilization" Microorganisms 12, no. 10: 1996. https://doi.org/10.3390/microorganisms12101996






