Proteome Profiling of S. cerevisiae Strains Lacking the Ubiquitin-Conjugating Enzymes Ubc4 and Ubc5 During Exponential Growth and After Heat Shock Treatment
Abstract
:1. Introduction
2. Material and Methods
2.1. Materials
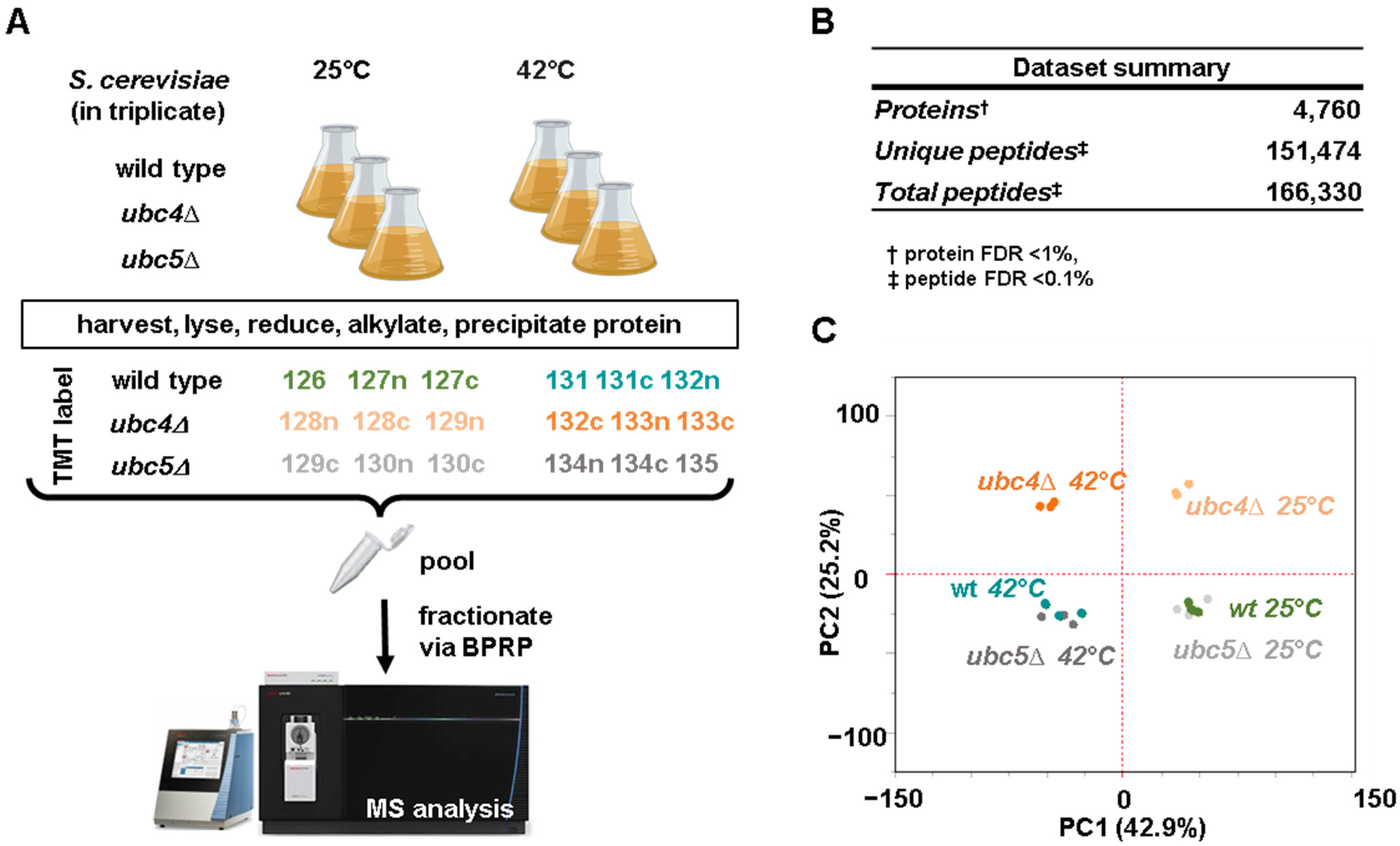
2.2. Yeast Strains, Growth Conditions and Protein Extraction
2.3. Protein Digestion, TMT Labeling, and Sample Processing
2.4. Mass Spectrometry Data Acquisition and Processing
3. Results
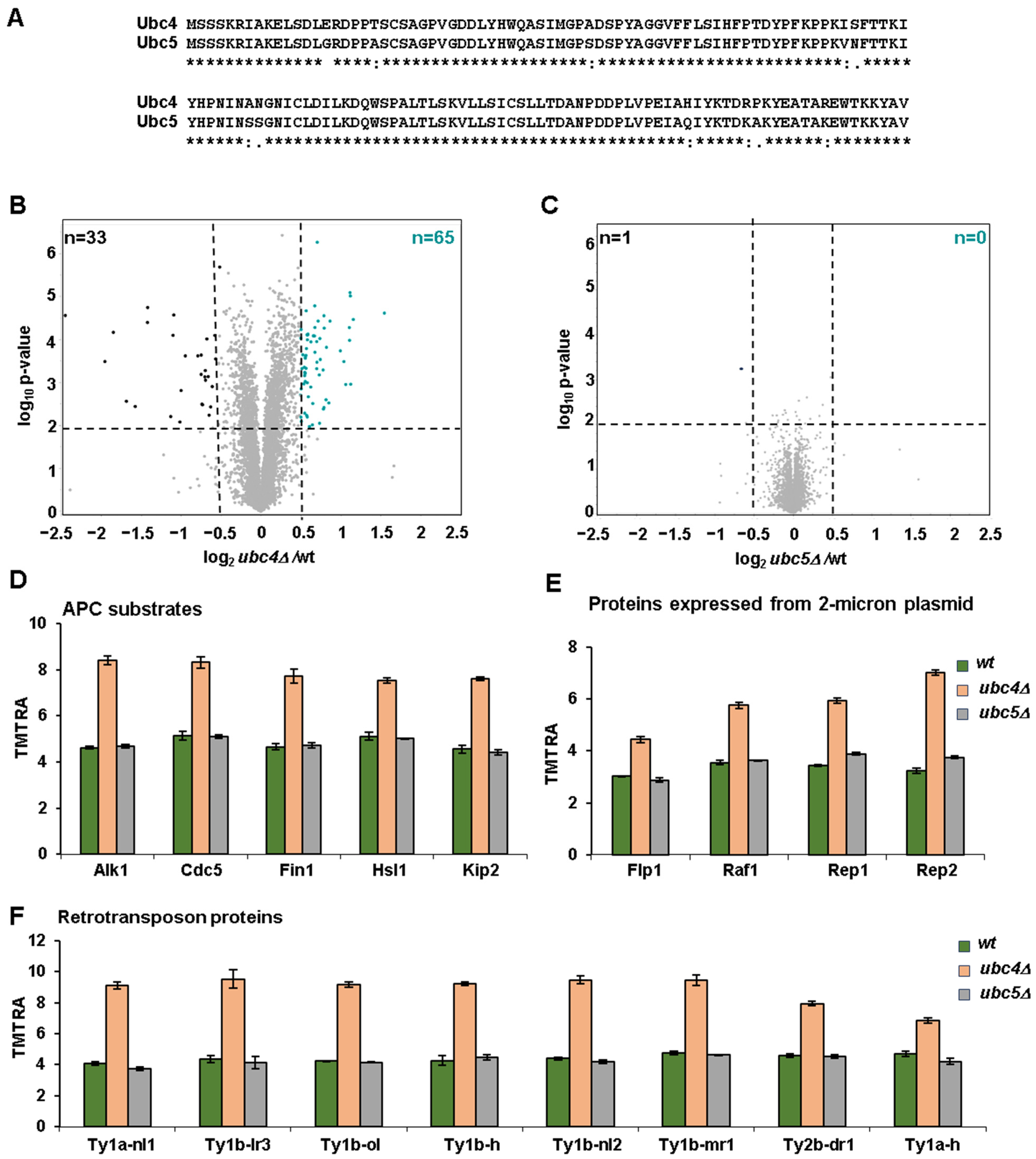
3.1. Comparative Proteome Abundance Profiling of Yeast Deletion Strains of the Homologous Ubiquitin-Conjugating Enzymes UBC4 and UBC5
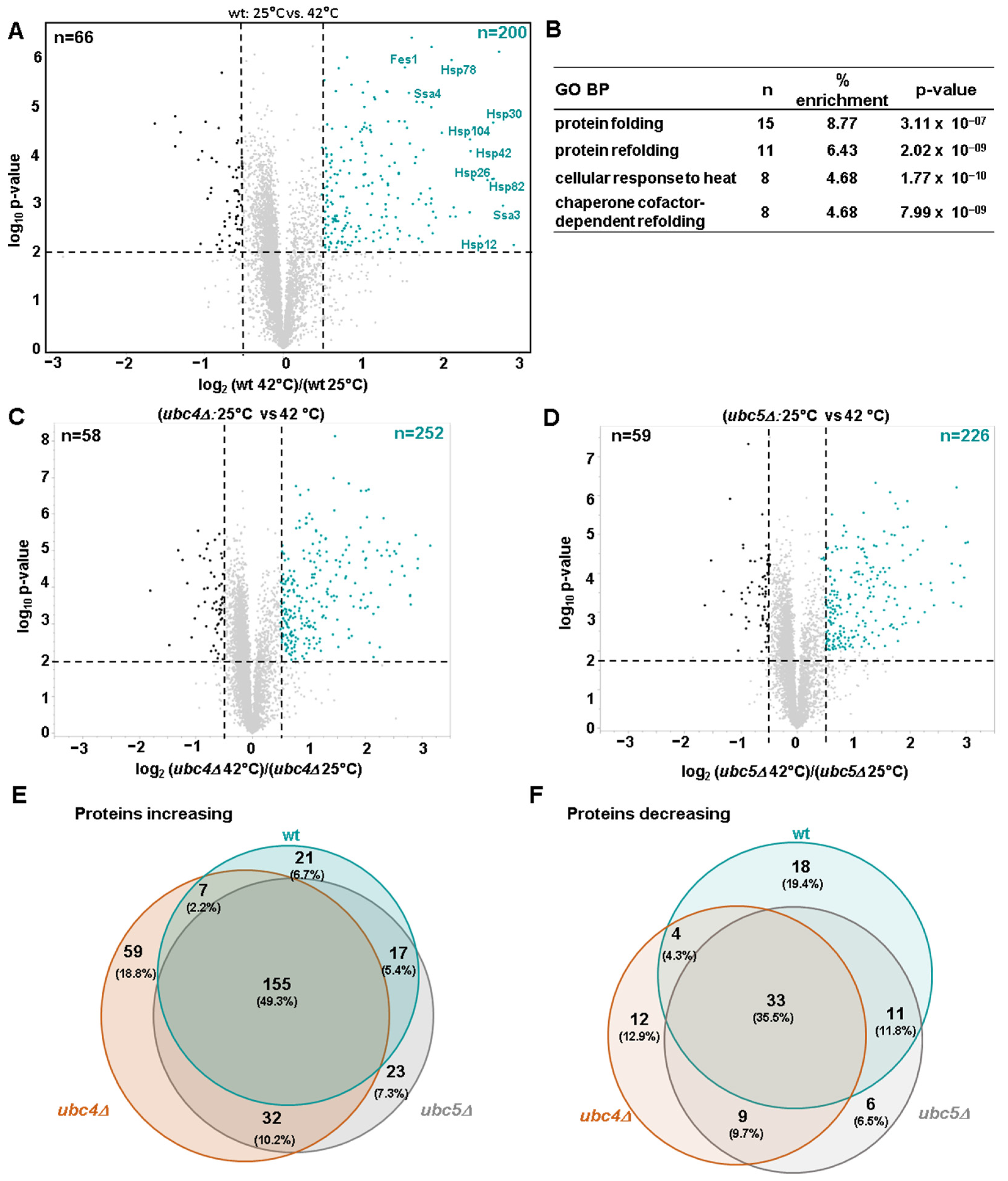
3.2. Proteome-Level Profiling of Heat Shock-Induced Changes in Wild-Type Cells and in Cells Lacking UBC4 or UBC5
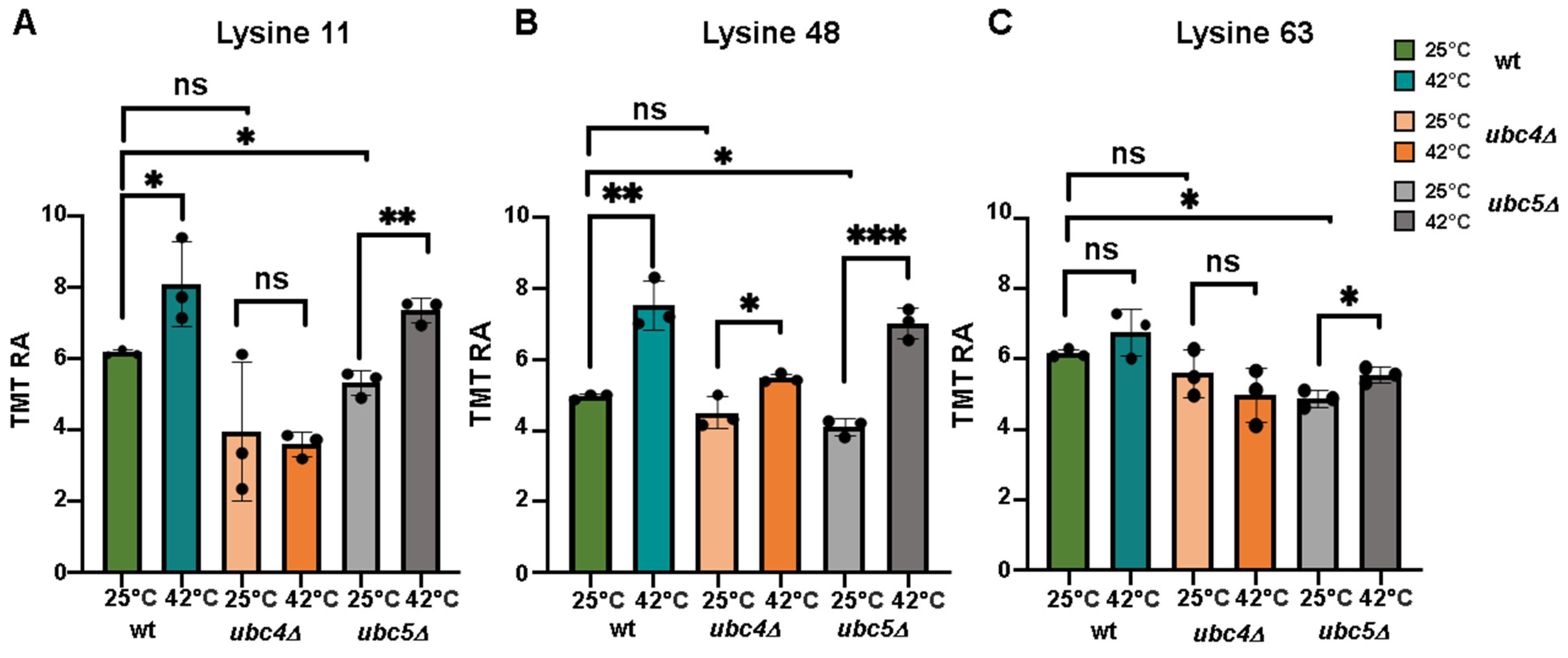
3.3. Profiling of Ubiquitin Linkages Following Heat Shock Treatment in Wild Type, ubc4Δ and ubc5Δ Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hershko, A.; Ciechanover, A. The Ubiquitin System. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Yau, R.; Rape, M. The Increasing Complexity of the Ubiquitin Code. Nat. Cell Biol. 2016, 18, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The Ubiquitin Code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Chau, V.; Tobias, J.W.; Bachmair, A.; Marriott, D.; Ecker, D.J.; Gonda, D.K.; Varshavsky, A. A Multiubiquitin Chain Is Confined to Specific Lysine in a Targeted Short-Lived Protein. Science 1989, 243, 1576–1583. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M. Mechanisms Underlying Ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef]
- Rape, M. Ubiquitylation at the Crossroads of Development and Disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 59–70. [Google Scholar] [CrossRef]
- Grice, G.L.; Nathan, J.A. The Recognition of Ubiquitinated Proteins by the Proteasome. Cell. Mol. Life Sci. 2016, 73, 3497–3506. [Google Scholar] [CrossRef]
- Finley, D.; Sadis, S.; Monia, B.P.; Boucher, P.; Ecker, D.J.; Crooke, S.T.; Chau, V. Inhibition of Proteolysis and Cell Cycle Progression in a Multiubiquitination-Deficient Yeast Mutant. Mol. Cell. Biol. 1994, 14, 5501–5509. [Google Scholar] [CrossRef] [PubMed]
- Erpapazoglou, Z.; Walker, O.; Haguenauer-Tsapis, R. Versatile Roles of K63-Linked Ubiquitin Chains in Trafficking. Cells 2014, 3, 1027–1088. [Google Scholar] [CrossRef]
- Kirkin, V.; McEwan, D.G.; Novak, I.; Dikic, I. A Role for Ubiquitin in Selective Autophagy. Mol. Cell 2009, 34, 259–269. [Google Scholar] [CrossRef]
- Acconcia, F.; Sigismund, S.; Polo, S. Ubiquitin in Trafficking: The Network at Work. Exp. Cell Res. 2009, 315, 1610–1618. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Sumara, I.; Pangou, E. Non-Proteolytic Ubiquitylation in Cellular Signaling and Human Disease. Commun. Biol. 2022, 5, 114. [Google Scholar] [CrossRef] [PubMed]
- Clague, M.J.; Heride, C.; Urbé, S. The Demographics of the Ubiquitin System. Trends Cell Biol. 2015, 25, 417–426. [Google Scholar] [CrossRef]
- Stewart, M.D.; Ritterhoff, T.; Klevit, R.E.; Brzovic, P.S. E2 Enzymes: More than Just Middle Men. Cell Res. 2016, 26, 423–440. [Google Scholar] [CrossRef]
- Roman-Trufero, M.; Dillon, N. The UBE2D Ubiquitin Conjugating Enzymes: Potential Regulatory Hubs in Development, Disease and Evolution. Front. Cell Dev. Biol. 2022, 10, 1058751. [Google Scholar] [CrossRef]
- Michelle, C.; Vourc’h, P.; Mignon, L.; Andres, C.R. What Was the Set of Ubiquitin and Ubiquitin-like Conjugating Enzymes in the Eukaryote Common Ancestor? J. Mol. Evol. 2009, 68, 616–628. [Google Scholar] [CrossRef]
- Seufert, W.; Jentsch, S. Ubiquitin-Conjugating Enzymes UBC4 and UBC5 Mediate Selective Degradation of Short-Lived and Abnormal Proteins. EMBO J. 1990, 9, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Medicherla, B.; Goldberg, A.L. Heat Shock and Oxygen Radicals Stimulate Ubiquitin-Dependent Degradation Mainly of Newly Synthesized Proteins. J. Cell Biol. 2008, 182, 663–673. [Google Scholar] [CrossRef]
- Chuang, S.-M.; Madura, K. Saccharomyces Cerevisiae Ub-Conjugating Enzyme Ubc4 Binds the Proteasome in the Presence of Translationally Damaged Proteins. Genetics 2005, 171, 1477–1484. [Google Scholar] [CrossRef]
- Girard, J.R.; Tenthorey, J.L.; Morgan, D.O. An E2 Accessory Domain Increases Affinity for the Anaphase-Promoting Complex and Ensures E2 Competition. J. Biol. Chem. 2015, 290, 24614–24625. [Google Scholar] [CrossRef]
- Gwizdek, C.; Hobeika, M.; Kus, B.; Ossareh-Nazari, B.; Dargemont, C.; Rodriguez, M.S. The mRNA Nuclear Export Factor Hpr1 Is Regulated by Rsp5-Mediated Ubiquitylation. J. Biol. Chem. 2005, 280, 13401–13405. [Google Scholar] [CrossRef] [PubMed]
- Kus, B.M.; Caldon, C.E.; Andorn-Broza, R.; Edwards, A.M. Functional Interaction of 13 Yeast SCF Complexes with a Set of Yeast E2 Enzymes in Vitro. Proteins 2004, 54, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Mulla, W.A.; Kucharavy, A.; Tsai, H.-J.; Rubinstein, B.; Conkright, J.; McCroskey, S.; Bradford, W.D.; Weems, L.; Haug, J.S.; et al. Targeting the Adaptability of Heterogeneous Aneuploids. Cell 2015, 160, 771–784. [Google Scholar] [CrossRef]
- Stoll, K.E.; Brzovic, P.S.; Davis, T.N.; Klevit, R.E. The Essential Ubc4/Ubc5 Function in Yeast Is HECT E3-Dependent, and RING E3-Dependent Pathways Require Only Monoubiquitin Transfer by Ubc4. J. Biol. Chem. 2011, 286, 15165–15170. [Google Scholar] [CrossRef]
- Janke, C.; Magiera, M.M.; Rathfelder, N.; Taxis, C.; Reber, S.; Maekawa, H.; Moreno-Borchart, A.; Doenges, G.; Schwob, E.; Schiebel, E.; et al. A Versatile Toolbox for PCR-Based Tagging of Yeast Genes: New Fluorescent Proteins, More Markers and Promoter Substitution Cassettes. Yeast 2004, 21, 947–962. [Google Scholar] [CrossRef]
- Mühlhofer, M.; Berchtold, E.; Stratil, C.G.; Csaba, G.; Kunold, E.; Bach, N.C.; Sieber, S.A.; Haslbeck, M.; Zimmer, R.; Buchner, J. The Heat Shock Response in Yeast Maintains Protein Homeostasis by Chaperoning and Replenishing Proteins. Cell Rep. 2019, 29, 4593–4607.e8. [Google Scholar] [CrossRef] [PubMed]
- Rossio, V.; Liu, X.; Paulo, J.A. Comparative Proteomic Analysis of Two Commonly Used Laboratory Yeast Strains: W303 and BY4742. Proteomes 2023, 11, 30. [Google Scholar] [CrossRef]
- Liu, X.; Rossio, V.; Gygi, S.P.; Paulo, J.A. Enriching Cysteine-Containing Peptides Using a Sulfhydryl-Reactive Alkylating Reagent with a Phosphonic Acid Group and Immobilized Metal Affinity Chromatography. J. Proteome Res. 2023, 22, 1270–1279. [Google Scholar] [CrossRef]
- Liu, X.; Rossio, V.; Paulo, J.A. Spin Column-Based Peptide Fractionation Alternatives for Streamlined Tandem Mass Tag (SL-TMT) Sample Processing. J. Proteom. 2023, 276, 104839. [Google Scholar] [CrossRef]
- Rossio, V.; Paulo, J.A. Comparison of the Proteomes and Phosphoproteomes of S. Cerevisiae Cells Harvested with Different Strategies. Proteomes 2023, 11, 28. [Google Scholar] [CrossRef]
- Rossio, V.; Paulo, J.A.; Liu, X.; Gygi, S.P.; King, R.W. Specificity Profiling of Deubiquitylases against Endogenously Generated Ubiquitin-Protein Conjugates. Cell Chem. Biol. 2024, 31, 1349–1362.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liu, X.; Rossio, V.; Dawson, S.L.; Gygi, S.P.; Paulo, J.A. Enhancing Proteome Coverage by Using Strong Anion-Exchange in Tandem with Basic-pH Reversed-Phase Chromatography for Sample Multiplexing-Based Proteomics. J. Proteome Res. 2023, 23, 2870–2881. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Beausoleil, S.A.; Villén, J.; Gerber, S.A.; Rush, J.; Gygi, S.P. A Probability-Based Approach for High-Throughput Protein Phosphorylation Analysis and Site Localization. Nat. Biotechnol. 2006, 24, 1285–1292. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Jedrychowski, M.P.; Elias, J.E.; Goswami, T.; Rad, R.; Beausoleil, S.A.; Villén, J.; Haas, W.; Sowa, M.E.; Gygi, S.P. A Tissue-Specific Atlas of Mouse Protein Phosphorylation and Expression. Cell 2010, 143, 1174–1189. [Google Scholar] [CrossRef] [PubMed]
- Elias, J.E.; Gygi, S.P. Target-Decoy Search Strategy for Increased Confidence in Large-Scale Protein Identifications by Mass Spectrometry. Nat. Methods 2007, 4, 207–214. [Google Scholar] [CrossRef]
- Navarrete-Perea, J.; Yu, Q.; Gygi, S.P.; Paulo, J.A. SL-TMT: A Streamlined Protocol for Quantitative (Phospho)Proteome Profiling Using TMT-SPS-MS3. J. Proteome Res. 2018, 17, 2226–2236. [Google Scholar] [CrossRef]
- Paulo, J.A.; O’Connell, J.D.; Gygi, S.P. A Triple Knockout (TKO) Proteomics Standard for Diagnosing Ion Interference in Isobaric Labeling Experiments. J. Am. Soc. Mass. Spectrom. 2016, 27, 1620–1625. [Google Scholar] [CrossRef]
- Burton, J.L.; Solomon, M.J. Hsl1p, a Swe1p Inhibitor, Is Degraded via the Anaphase-Promoting Complex. Mol. Cell Biol. 2000, 20, 4614–4625. [Google Scholar] [CrossRef]
- Visintin, C.; Tomson, B.N.; Rahal, R.; Paulson, J.; Cohen, M.; Taunton, J.; Amon, A.; Visintin, R. APC/C-Cdh1-Mediated Degradation of the Polo Kinase Cdc5 Promotes the Return of Cdc14 into the Nucleolus. Genes Dev. 2008, 22, 79–90. [Google Scholar] [CrossRef]
- Nespoli, A.; Vercillo, R.; di Nola, L.; Diani, L.; Giannattasio, M.; Plevani, P.; Muzi-Falconi, M. Alk1 and Alk2 Are Two New Cell Cycle-Regulated Haspin-like Proteins in Budding Yeast. Cell Cycle 2006, 5, 1464–1471. [Google Scholar] [CrossRef] [PubMed]
- Zucca, F.; Visintin, C.; Li, J.; Gygi, S.P.; Visintin, R. APC/CCdc20-Mediated Degradation of Clb4 Prompts Astral Microtubule Stabilization at Anaphase Onset. J. Cell Biol. 2022, 222, e202203089. [Google Scholar] [CrossRef] [PubMed]
- Woodbury, E.L.; Morgan, D.O. Cdk and APC Activities Limit the Spindle-Stabilizing Function of Fin1 to Anaphase. Nat. Cell Biol. 2007, 9, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, S.M.A.; Prajapati, H.K.; Ghosh, S.K. The 2 Micron Plasmid: A Selfish Genetic Element with an Optimized Survival Strategy within Saccharomyces Cerevisiae. Curr. Genet. 2018, 64, 25–42. [Google Scholar] [CrossRef]
- Sleep, D.; Finnis, C.; Turner, A.; Evans, L. Yeast 2 Μm Plasmid Copy Number Is Elevated by a Mutation in the Nuclear Gene UBC4. Yeast 2001, 18, 403–421. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Bhat, A.; Ghatage, T.; Bhan, S.; Lahane, G.P.; Dhar, A.; Kumar, R.; Pandita, R.K.; Bhat, K.M.; Ramos, K.S.; Pandita, T.K. Role of Transposable Elements in Genome Stability: Implications for Health and Disease. Int. J. Mol. Sci. 2022, 23, 7802. [Google Scholar] [CrossRef]
- Hulsen, T.; de Vlieg, J.; Alkema, W. BioVenn—A Web Application for the Comparison and Visualization of Biological Lists Using Area-Proportional Venn Diagrams. BMC Genom. 2008, 9, 488. [Google Scholar] [CrossRef]
- Peng, J.; Schwartz, D.; Elias, J.E.; Thoreen, C.C.; Cheng, D.; Marsischky, G.; Roelofs, J.; Finley, D.; Gygi, S.P. A Proteomics Approach to Understanding Protein Ubiquitination. Nat. Biotechnol. 2003, 21, 921–926. [Google Scholar] [CrossRef]
- Xu, P.; Duong, D.M.; Seyfried, N.T.; Cheng, D.; Xie, Y.; Robert, J.; Rush, J.; Hochstrasser, M.; Finley, D.; Peng, J. Quantitative Proteomics Reveals the Function of Unconventional Ubiquitin Chains in Proteasomal Degradation. Cell 2009, 137, 133–145. [Google Scholar] [CrossRef]
- Krastanova, O.; Hadzhitodorov, M.; Pesheva, M. Ty Elements of the Yeast Saccharomyces cerevisiae. Biotechnol. Biotechnol. Equip. 2005, 19 (Suppl. S2), 19–26. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossio, V.; Liu, X.; Paulo, J.A. Proteome Profiling of S. cerevisiae Strains Lacking the Ubiquitin-Conjugating Enzymes Ubc4 and Ubc5 During Exponential Growth and After Heat Shock Treatment. Microorganisms 2024, 12, 2235. https://doi.org/10.3390/microorganisms12112235
Rossio V, Liu X, Paulo JA. Proteome Profiling of S. cerevisiae Strains Lacking the Ubiquitin-Conjugating Enzymes Ubc4 and Ubc5 During Exponential Growth and After Heat Shock Treatment. Microorganisms. 2024; 12(11):2235. https://doi.org/10.3390/microorganisms12112235
Chicago/Turabian StyleRossio, Valentina, Xinyue Liu, and Joao A. Paulo. 2024. "Proteome Profiling of S. cerevisiae Strains Lacking the Ubiquitin-Conjugating Enzymes Ubc4 and Ubc5 During Exponential Growth and After Heat Shock Treatment" Microorganisms 12, no. 11: 2235. https://doi.org/10.3390/microorganisms12112235
APA StyleRossio, V., Liu, X., & Paulo, J. A. (2024). Proteome Profiling of S. cerevisiae Strains Lacking the Ubiquitin-Conjugating Enzymes Ubc4 and Ubc5 During Exponential Growth and After Heat Shock Treatment. Microorganisms, 12(11), 2235. https://doi.org/10.3390/microorganisms12112235







