Identification of Mycoplasma Species in Cattle Associated with Bovine Respiratory Disease Mortality
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Identification of Etiological Agents
2.2.1. Microbiological Investigations
2.2.2. DNA Extraction and PCR Conditions for Mycoplasma spp.
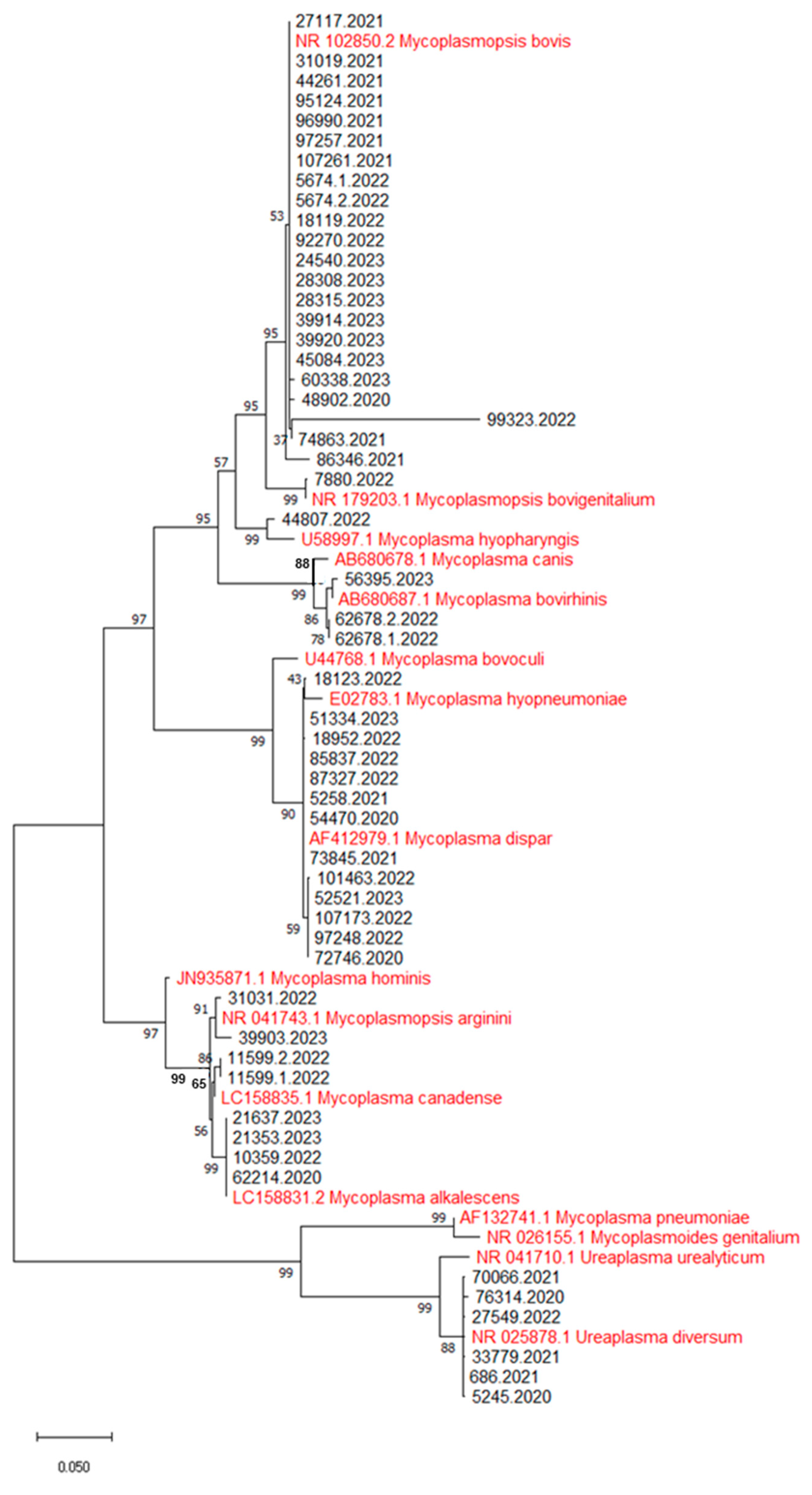
2.2.3. Identification of Mycoplasma Species and Phylogenetic Analysis
2.2.4. DNA Extraction and PCR Conditions for Bovine Herpesvirus 1 (BoHV-1)
2.2.5. RNA Extraction and PCR Conditions for Bovine Parainfluenza Virus 3 (BPIV-3)
2.2.6. RNA Extraction and PCR Conditions for Bovine Viral Diarrhea Virus (BVDV)
2.2.7. RNA Extraction and Real-Time PCR Conditions for Bovine Respiratory Syncytial Virus (BRSV)
2.3. Histopathological Investigations
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Santos Junior, M.N.; Macedo Neres, N.S.; Campos, G.B.; Bastos, B.L.; Timenetsky, J.; Marques, L.M. A Review of Ureaplasma diversum A Representative of the Mollicute Class Associated with Reproductive and Respiratory Disorders in Cattle. Front. Vet. Sci. 2021, 8, 572171. [Google Scholar] [CrossRef] [PubMed]
- Deeney, A.S.; Collins, R.; Ridley, A.M. Identification of Mycoplasma species and related organisms from ruminants in England and Wales during 2005–2019. BMC Vet. Res. 2021, 17, 325. [Google Scholar] [CrossRef] [PubMed]
- Catania, S.; Gastaldelli, M.; Schiavon, E.; Matucci, A.; Tondo, A.; Merenda, M.; Nicholas, R.A.J. Infection Dynamics of Mycoplasma bovis and Other Respiratory Mycoplasmas in Newly Imported Bulls on Italian Fattening Farms. Pathogens 2020, 9, 537. [Google Scholar] [CrossRef] [PubMed]
- Gabinaitiene, A.; Siugzdaite, J.; Zilinskas, H. Laboratory diagnosis of Mycoplasma infection in young cattle. Pol. J. Vet. Sci. 2011, 14, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Burki, S.; Frey, J.; Pilo, P. Virulence, persistence and dissemination of Mycoplasma bovis. Vet. Microbiol. 2015, 179, 15–22. [Google Scholar] [CrossRef]
- Tawab, A.A.A.E.; El-Hofy, F.I.; Hassan, N.I.; Ramadan, M.R. Identification and Genetic Characterization of Mycoplasma Species Affecting Respiratory System in Egyptian Cattle. BVMJ 2021, 40, 21–26. [Google Scholar] [CrossRef]
- Sultana, R.; Cordeiro, R.P.; Timsit, E.; McAllister, T.A.; Alexander, T.W. Prevalence and antimicrobial susceptibility of Mycoplasma bovis from the upper and lower respiratory tracts of healthy feedlot cattle and those diagnosed with bovine respiratory disease. Vet. Microbiol. 2023, 285, 109838. [Google Scholar] [CrossRef]
- Dudek, K.; Nicholas, R.A.J.; Szacawa, E.; Bednarek, D. Mycoplasma bovis Infections—Occurrence, Diagnosis and Control. Pathogens 2020, 9, 640. [Google Scholar] [CrossRef]
- Hewicker-Trautwein, M.; Feldmann, M.; Kehler, W.; Schmidt, R.; Thiede, S.; Seeliger, F.; Wohlsein, P.; Ball, H.J.; Buchenau, I.; Spergser, J.; et al. Outbreak of pneumonia and arthritis in beef calves associated with Mycoplasma bovis and Californicum. Vet. Rec. 2002, 151, 699–703. [Google Scholar] [CrossRef]
- Parker, A.M.; Sheehy, P.A.; Hazelton, M.S.; Bosward, K.L.; House, J.K. A review of mycoplasma diagnostics in cattle. J. Vet. Intern. Med. 2018, 32, 1241–1252. [Google Scholar] [CrossRef]
- Valeris-Chacin, R.; Powledge, S.; McAtee, T.; Morley, P.S.; Richeson, J. Mycoplasma bovis is associated with Mannheimia haemolytica during acute bovine respiratory disease in feedlot cattle. Front. Microbiol. 2022, 13, 946792. [Google Scholar] [CrossRef] [PubMed]
- Mehinagic, K.; Pilo, P.; Vidondo, B.; Stokar-Regenscheit, N. Coinfection of Swiss cattle with bovine parainfluenza virus 3 and Mycoplasma bovis at acute and chronic stages of bovine respiratory disease complex. J. Vet. Diagn. Investig. 2019, 31, 674–680. [Google Scholar] [CrossRef] [PubMed]
- Oucheriah, Y.; Heleili, N.; Colin, A.; Mottet, C.; Tardy, F.; Becker, C.A.M. Prevalence of Mycoplasma bovis in Algeria and Characterisation of the Isolated Clones. Front. Vet. Sci. 2022, 9, 910799. [Google Scholar] [CrossRef] [PubMed]
- Radaelli, E.; Luini, M.; Loria, G.R.; Nicholas, R.A.J.; Scanziani, E. Bacteriological, serological, pathological and immunohistochemical studies of Mycoplasma bovis respiratory infection in veal calves and adult cattle at slaughter. Res. J. Vet. Sci. 2008, 85, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Aebi, M.; van den Borne, B.H.P.; Raemy, A.; Steiner, A.; Pilo, P.; Bodmer, M. Mycoplasma bovis infections in Swiss dairy cattle a clinical investigation. Acta. Vet. Scand. 2015, 57, 10. [Google Scholar] [CrossRef]
- Szacawa, E.; Niemczuk, K.; Dudek, K.; Bednarek, D.; Rosales, R.; Ayling, R. Mycoplasma bovis infections and co-infections with other Mycoplasma spp. with different clinical manifestations in affected cattle herds in eastern region of Poland. Bull. Vet. Inst. Pulawy. 2015, 59, 331–337. [Google Scholar] [CrossRef]
- Gioia, G.; Severgnini, M.; Cremonesi, P.; Castiglioni, B.; Freeman, J.; Sipka, A.; Santisteban, C.; Wieland, M.; Gallardo, V.A.; Scott, J.G.; et al. Genomic Characterization of Mycoplasma arginini Isolated from a Housefly on a Dairy Farm and Comparison with Isolates from Bovine Milk and Lung Tissue. Microbiol. Spectr. 2023, 11, 3. [Google Scholar] [CrossRef]
- Topić Popović, N.; Kazazić, S.P.; Bojanić, K.; Strunjak-Perović, I.; Čož-Rakovac, R. Sample preparation and culture condition effects on MALDI-TOF MS identification of bacteria: A review. Mass. Spec. Rev. 2023, 42, 1589–1603. [Google Scholar] [CrossRef]
- Van Kuppeveld, F.J.M.; Van Der Logt, J.T.M.; Angulo, A.F.; Van Zoest, M.J.; Quint, W.G.V.; Niesters, H.G.M.; Galama, J.M.D.; Melchers, W.J.G. Genus-and species-specific identification of mycoplasmas by 16S rRNA amplification. Appl. Environ. Microbiol. 1992, 58, 2606–2615. [Google Scholar] [CrossRef]
- Yoshida, T.; Maeda, S.I.; Deguchi, T.; Ishiko, H. Phylogeny-based rapid identification of mycoplasmas and ureaplasmas from urethritis patients. J. Clin. Microbiol. 2002, 40, 105–110. [Google Scholar] [CrossRef]
- Abril, C.; Engels, M.; Liman, A.; Hilbe, M.; Albini, S.; Franchini, M.; Suter, M.; Ackermann, M. Both viral and host factors contribute to neurovirulence of bovine herpesvirus1 and 5 in interferon receptor-deficient mice. J. Virol. 2004, 78, 3644–3653. [Google Scholar] [CrossRef] [PubMed]
- Horwood, P.F.; Mahony, T.J. Multiplex real-time RT-PCR detection of three viruses associated with the bovine respiratory disease complex. J. Virol. Methods 2011, 171, 360–363. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, B.; Depner, K.; Schirrmeier, H.; Beer, M. A universal heterologous internal control system for duplex real-time RT PCR assays used in a detection system for pestiviruses. J. Virol. Methods 2006, 136, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Letellier, C.; Kerkhofs, P. Real-time PCR for simultaneous detection and genotyping of bovine viral diarrhea virus. J. Virol. Methods 2003, 7, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Boxus, M.; Letellier, C.; Kerkhofs, P. Real Time RT-PCR for the detection and quantitation of bovine respiratory syncytial virus. J. Virol. Methods 2005, 125, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, A.; Cirilli, M.; Lucente, M.S.; Zarea, A.A.K.; Buonavoglia, D.; Tempesta, M.; Greco, G. Fatal Calf Pneumonia Outbreaks in Italian Dairy Herds Involving Mycoplasma bovis and Other Agents of BRD Complex. Front. Vet. Sci. 2021, 8, 742785. [Google Scholar] [CrossRef]
- Loria, G.R.; Monteverde, V.; La Barbera, E.; Caracappa, S.; Scanziani, E.; Grieco, V.; Ayling, R.; Nicholas, R.A.J. Isolamento di Mycoplasma bovis e patologia respiratoria. Large Anim. Rev. 2004, 5, 17–21. [Google Scholar]
- Manfrin, A.; Friso, S.; Perin, R.; Girelli, L. Isolation of Mycoplasma spp. from cattle in the north-east of Italy. In Mycoplasmas of Ruminants: Pathogenicity, Diagnostics, Epidemiology and Molecular Genetics; Leori, G., Santini, F., Scanziani, E., Frey, J., Eds.; European Commission: Brussels, Belgium, 1998; Volume 2, pp. 84–87. [Google Scholar]
- Barnewall, R.J.; Marsh, I.B.; Cusack, P.M.V.; Galea, F.; Salesc, N.; Quinn, J.C. Detection of Ureaplasma diversum in the upper airways of Australian feedlot cattle. Aust. Vet. J. 2023, 101, 254–257. [Google Scholar] [CrossRef]
- Ayling, R.D.; Bashiruddin, S.E.; Nicholas, R.A.J. Mycoplasma species and related organisms isolated from ruminants in Britain between 1990 and 2000. Vet. Rec. 2004, 155, 413–416. [Google Scholar] [CrossRef]
- Arcangioli, M.A.; Chazel, M.; Sellal, E.; Botrel, M.A.; Bèzille, P.B.; Poumarat, F.; Calavas, D.; Le Grand, D. Prevalence of Mycoplasma bovis udder infection in dairy cattle Preliminary field investigation in southeast France. NZVJ 2011, 59, 75–78. [Google Scholar] [CrossRef]
- Poumarat, F.; Jarrige, N.; Tardy, F. Purpose and overview of results of the Vigimyc Network for the epidemiological surveillance of mycoplasmoses in ruminants in France. Euro. Ref. 2014, 12, 22–27. Available online: https://euroreference.anses.fr/sites/default/files/ER12-RESEAUX-VigimycEN.pdf (accessed on 6 October 2023).
- Gabinaitiene, A.; Siugzdaite, J.; Zilinskas, H.; Siugzda, R.; Petkevicius, S. Mycoplasma bovis and bacterial pathogens in the bovine respiratory tract. Vet. Med. 2011, 56, 28–34. [Google Scholar] [CrossRef]
- Härtel, H.; Nikunen, S.; Neuvonen, E.; Tanskanen, R.; Kivelä, S.L.; Aho, P.; Soveri, T.; Saloniemi, H. Viral and bacterial pathogens in bovine respiratory disease in Finland. Acta Vet. Scand. 2004, 45, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Kokotovic, B.; Friis, N.F.; Ahrens, P. Mycoplasma alkalescens demonstrated in bronchoalveolar lavage of cattle in Denmark. Acta Vet. Scand. 2007, 49, 1–3. [Google Scholar] [CrossRef] [PubMed]
- El-Shabiny, L.M.; Abouel-Makarem, M.M.; Nada, H.S. Mycoplasma isolated from cattle lungs and their pathogenicity study. Egypt. J. Agric. Res. 1999, 77, 421–431. [Google Scholar] [CrossRef]
- Autio, T.; Pohjanvirta, T.; Holopainen, R.; Rikula, U.; Pentikainen, J.; Huovilainen, A.; Rusanen, H.; Soveri, T.; Sihvonen, L.; Pelkonen, S. Etiology of respiratory disease in non-vaccinated, non-medicated calves in rearing herds. Vet. Microbiol. 2007, 119, 256–265. [Google Scholar] [CrossRef]
- Chen, S.; Hao, H.; Zhao, P.; Liu, Y.; Chu, Y. Genome-Wide Analysis of Mycoplasma bovirhinis GS01 Reveals Potential Virulence Factors and Phylogenetic Relationships. Genes Genomes Genet. 2018, 8, 1417–1424. [Google Scholar] [CrossRef]
- Abdelazeem, W.M.; Zolnikov, T.R.; Mohammed, Z.R.; Saad, A.; Osmand, K.M. Virulence, antimicrobial resistance and phylogenetic analysis of zoonotic walking pneumonia Mycoplasma arginini in the one-humped camel (Camelus dromedarius). Acta Trop. 2020, 207, 105500. [Google Scholar] [CrossRef]
- Chazel, M.; Tardy, F.; Le Grand, D.; Calavas, D.; Poumarat, F. Mycoplasmoses of ruminants in France recent data from the national surveillance network. BMC Vet. Res. 2010, 6, 32. [Google Scholar] [CrossRef]
- Lysnyansky, I.; Brenner, J.; Alpert, N.; Benjamin, A.; Bernstein, M.; Elad, D.; Blum, S.; Friedgut, O.; Rotenberg, D. Identification of Mycoplasma bovigenitalium and Mycoplasma canadense from outbreaks of granulopapular vulvovaginitis in dairy cattle in Israel. Vet. Rec. 2009, 165, 319–322. [Google Scholar] [CrossRef]
- Gioia, G.; Addis, M.F.; Santisteban, C.; Gross, B.; Nydam, D.V.; Sipka, A.S.; Virkler, P.D.; Watters, R.D.; Wieland, M.; Zurakowski, M.J.; et al. Mycoplasma species isolated from bovine milk collected from US dairy herds between 2016 and 2019. J. Dairy. Sci. 2020, 104, 4813–4821. [Google Scholar] [CrossRef] [PubMed]
- Blank, W.A.; Erickson, B.Z.; Stemke, G.W. Phylogenetic Relationships of the Porcine Mycoplasmas Mycoplasma hyosynoviae and Mycoplasma hyopharyngis. Int. J. Syst. Bacteriol. 1996, 46, 1181–1182. [Google Scholar] [CrossRef] [PubMed]
- Erickson, B.Z.; Ross, R.F.; Rose, D.L.; Tully, J.G.; Bove, J.M. Mycoplasma hyopharyngis, a New Species from Swine. Int. J. Syst. Bacteriol. 1986, 36, 55–59. [Google Scholar] [CrossRef]
- Mamuad, L.L.; Seo, B.J.; Al Faruk, M.S.; Espiritu, H.M.; Jin, S.J.; Kim, W.I.; Lee, S.S.; Cho, Y.I. Treponema spp., the dominant pathogen in the lesion of bovine digital dermatitis and its characterization in dairy cattle. Vet. Microbiol. 2020, 245, 108696. [Google Scholar] [CrossRef] [PubMed]
- Földi, D.; Nagy, E.Z.; Tóth, G.; Makrai, L.; Gombos, L.; Kreizinger, Z.; Gyuranecz, M. Mycoplasma hyopharyngis isolated from the joint of a weaner: A case report. Acta Vet. Hung. 2024, 72, 115–160. [Google Scholar] [CrossRef]
- Gagea, M.I.; Bateman, K.G.; Shanahan, R.A.; van Dreumel, T.; McEwen, B.J.; Carman, S.; Archambault, M.; Caswell, J.L. Naturally Occurring Mycoplasma Bovis Associated Pneumonia and Polyarthritis in Feedlot Beef Calves. J. Vet. Diagn. Investig. 2006, 18, 29–40. [Google Scholar] [CrossRef]
- Byrne, W.J.; McCormack, R.; Egan, J.; Brice, N.; Ball, H.J.; Markey, B. Isolation of Mycoplasma bovis from bovine clinical samples in the Republic of Ireland. Vet. Rec. 2001, 148, 331–333. [Google Scholar] [CrossRef]
- Oliveira, T.E.S.; Pelaquim, I.F.; Flores, E.F.; Massi, R.P.; Jiménez-Valdiviezo, M.J.; Pretto-Giordano, L.G.; Alfieri, A.A.; Saut, J.P.E.; Headley, S.A. Mycoplasma bovis and viral agents associated with the development of bovine respiratory disease in adult dairy cows. Transbound. Emerg. Dis. 2020, 67, 82–93. [Google Scholar] [CrossRef]

| Animal ID | Biological Matrix | Mycoplasma Species | Other Pathogens | Histopathological Results |
|---|---|---|---|---|
| 48902/20 | Lung | Mycoplasma bovis | Severe and diffuse catarrhal bronchopneumonia | |
| 76314/20 | Lung | Ureaplasma diversum | BPIV-3 | Diffuse catarrhal bronchopneumonia |
| 62214/20 | Lung | Mycoplasma alkalescens | Mannheimia haemolytica | Severe fibrinonecrotizing hemorrhagic pneumonia |
| 50470/20 | Lung | Mycoplasma dispar | Pasteurella multocida | Severe and chronic bronchointerstitial pneumonia |
| 5245/20 | Lung | Ureaplasma diversum | Escherichia coli | Severe fibrinopurulent catarrhal bronchopneumonia and enteritis |
| 72746/20 | Lung | Mycoplasma dispar | Corynebacterium kutscheri | Severe catarrhal bronchopneumonia |
| 44261/21 | Lung | Mycoplasma bovis | Acute and diffuse catarrhal bronchopneumonia and foamy trachea | |
| 686/21 | Lung | Ureaplasma diversum | BoHV-1 | Severe fibrinopurulent catarrhal bronchopneumonia |
| 96990/21 | Lung | Mycoplasma bovis | Trueperella pyogenes | Severe and subchronic fibrinopurulent catarrhal bronchopneumonia |
| 33779/21 | Lung | Ureaplasma diversum | BRSV and Aspergillus fumigatus | Severe fibrinopurulent catarrhal bronchopneumonia and pleurisy |
| 74863/21 | Lung | Mycoplasma bovis | Pasteurella multocida | Severe catarrhal bronchopneumonia |
| 97257/21 | Lung | Mycoplasma bovis | polymicrobism | Severe catarrhal bronchopneumonia |
| 70066/21 | Lung | Ureaplasma diversum | Escherichia. Coli and Acinetobacter schindleri | Severe catarrhal bronchopneumonia |
| 107261/ 21 | Lung | Mycoplasma bovis | Clostridium perfringens | Chronic catarrhal bronchopneumonia, necrotic hemorrhagic enterocolitis, and clostridial enterotoxemia |
| 31019/21 | Lung | Mycoplasma bovis | BRSV and Pasteurella multocida | Severe bilateral purulent bronchopneumonia |
| 95124/21 | Lung | Mycoplasma bovis | Histophilus somni | Severe and diffuse catarrhal bronchopneumonia and hemorrhagic tracheitis |
| 73845/21 | Lung | Mycoplasma dispar | Escherichia coli and Aspergillus fumigatus | Severe and chronic bronchointerstitial pneumonia |
| 5258/21 | Lung | Mycoplasma dispar | Escherichia coli | Severe fibrinonecrotizing hemorrhagic pneumonia and enteritis |
| 27117/21 | Lung | Mycoplasma bovis | Severe catarrhal bronchopneumonia | |
| 86346/21 | Lung | Mycoplasma bovis | Pasteurella multocida, BRSV and BPIV-3 | Diffuse catarrhal bronchopneumonia |
| 11599.1/22 | Lung | Mycoplasma canadense | Trueperella pyogenes and BoHV-1 | Subchronic purulent catarrhal bronchopneumonia |
| 11599.2/22 | Lung | Mycoplasma canadense | Mannheimia haemolytica | Severe and chronic purulent bronchopneumonia |
| 10395/22 | Lung and trachea | Mycoplasma alkalescens | BoHV-1 | Moderate bronchointerstitial pneumonia and a severe and diffuse fibrinopurulent and hemorrhagic tracheitis |
| 18123/22 | Lung | Mycoplasma dispar | Escherichia coli | Chronic fibrinopurulent pleurisy and chronic peritonitis |
| 27549/22 | Lung | Ureaplasma diversum | Pasteurella multocida | Severe catarrhal bronchopneumonia with a subcutaneous emphysema and pleurisy |
| 92270/22 | Lung | Mycoplasma bovis | Severe and diffuse purulent catarrhal bronchopneumonia with a subpleural emphysema and pleurisy | |
| 5674.1/ 22 | Lung | Mycoplasma bovis | BoHV-1 | Purulent catarrhal bronchopneumonia and subchronic pleurisy |
| 5674.2/ 22 | Lung | Mycoplasma bovis | BoHV-1 | Severe purulent catarrhal bronchopneumonia |
| 62678.1/22 | Lung and trachea | Mycoplasma bovirhinis | Severe and diffuse purulent catarrhal bronchopneumonia | |
| 62678.2/22 | Lung and trachea | Mycoplasma bovirhinis | Severe and diffuse purulent catarrhal bronchopneumonia | |
| 18119/22 | Lung | Mycoplasma bovis | Mannheimia haemolytica and Escherichia coli | Severe purulent catarrhal bronchopneumonia and peritonitis |
| 18952/22 | Lung | Mycoplasma dispar | Escherichia coli | Diffuse catarrhal bronchopneumonia and enteritis |
| 85837/22 | Lung | Mycoplasma dispar | Escherichia coli | Acute catarrhal bronchopneumonia and enteritis |
| 44807/22 | Lung | Mycoplasma hyopharyngis | Proteus vulgaris | Severe purulent catarrhal bronchopneumonia |
| 107173/ 22 | Lung | Mycoplasma dispar | Escherichia coli and BRSV | Severe and diffuse purulent catarrhal bronchopneumonia and enteritis |
| 97248/22 | Lung | Mycoplasma dispar | Pasteurella multocida | Severe and subacute catarrhal bronchopneumonia |
| 31031/22 | Lung | Mycoplasma arginini | Pasteurella multocida | Severe subchronic purulent catarrhal bronchopneumonia and pleurisy |
| 87327/22 | Lung | Mycoplasma dispar | Pasteurella multocida and BPIV-3 | Ssevere purulent catarrhal bronchopneumonia and pleurisy |
| 99323/22 | Lung | Mycoplasma bovis | Mannheimia haemolytica | Fibrinonecrotizing hemorrhagic pneumonia |
| 101463/ 22 | Lung | Mycoplasma dispar | Severe catarrhal bronchopneumonia | |
| 7880/22 | Lung | Mycoplasma bovigenitalium | polymicrobism | Fibrinonecrotizing hemorrhagic pneumonia and pleurisy |
| 21353/23 | Lung and trachea | Mycoplasma alkalescens | Pasteurella multocida and BoHV-1 | Purulent bronchopneumonia and congested trachea |
| 24540/23 | Lung | Mycoplasma bovis | Escherichia coli and BRSV | Chronic purulent bronchopneumonia, and emphysema |
| 28308/23 | Lung | Mycoplasma bovis | BRSV | Chronic catarrhal bronchopneumonia |
| 28315/23 | Lung | Mycoplasma bovis | Trueperella pyogenes | Chronic catarrhal bronchopneumonia |
| 56395/23 | Lung and trachea | Mycoplasma bovirhinis | polymicrobism | Congested trachea with foam, emphysema, pleurisy, and catarrhal bronchopneumonia |
| 60338/23 | Lung | Mycoplasma bovis | Escherichia coli | Necrosuppurative bronchopneumonia, fibrinopurulent pleurisy, and enteritis |
| 21637/23 | Lung | Mycoplasma alkalescens | Fibrinopurulent arthrosynovitis | |
| 39903/23 | Lung | Mycoplasma arginini | Pasteurella multocida | Purulent catarrhal bronchopneumonia and pleurisy |
| 39914/23 | Lung | Mycoplasma bovis | Pasteurella multocida | Purulent catarrhal bronchopneumonia and pleurisy |
| 39920/23 | Lung and Pulmonary exudate | Mycoplasma bovis | Catarrhal bronchopneumonia and polyarthritis | |
| 45084/23 | Lung | Mycoplasma bovis | Histophilus somni | Severe and diffuse catarrhal bronchopneumonia and subpleural emphysema |
| 51334/23 | Lung | Mycoplasma dispar | Weissella cibaria | Subchronic catarrhal bronchopneumonia |
| 52521/23 | Lung | Mycoplasma dispar | Histophilus somni | Chronic purulent catarrhal bronchopneumonia and pleurisy |
| Variable | Mycoplasma bovis | Mycoplasma dispar | ||||||
|---|---|---|---|---|---|---|---|---|
| Proportion of Positives to M. bovis | PI | PIR (95% CI) | p-Value | Proportion of Positives to M. dispar | PI | PIR (95% CI) | p-Value | |
| Sampling season | ||||||||
| Cold season | 14/33 | 42.4 | 1.1 (0.6–2.2) | 0.76 | 9/33 | 27.3 | 1.4 (0.5–4.1) | 0.50 |
| Warm season | 8/21 | 38.1 | 4/21 | 19.0 | ||||
| Sex | ||||||||
| Male | 14/30 | 46.7 | 1.4 (0.7–2.8) | 0.34 | 9/30 | 30.0 | 1.8 (0.6–5.2) | 0.28 |
| Female | 8/24 | 33.3 | 4/24 | 16.7 | ||||
| Age | ||||||||
| ≤5 months | 11/28 | 39.3 | 0.9 (0.5–1.8) | 0.82 | 11/28 | 39.3 | 5.1 (1.2–21.2) | 0.02 |
| >5 months | 11/26 | 42.3 | 2/26 | 7.7 | ||||
| Weigh | ||||||||
| ≤175 Kg | 12/27 | 44.4 | 1.2 (0.6–2.3) | 0.58 | 9/27 | 33.3 | 0.4 (0.2–1.3) | 0.13 |
| >175 Kg | 10/27 | 37.0 | 4/27 | 14.8 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carella, E.; Messana, E.; Mugetti, D.; Biasibetti, E.; Pezzolato, M.; Peletto, S.; Begovoeva, M.; Rossi, F. Identification of Mycoplasma Species in Cattle Associated with Bovine Respiratory Disease Mortality. Microorganisms 2024, 12, 2340. https://doi.org/10.3390/microorganisms12112340
Carella E, Messana E, Mugetti D, Biasibetti E, Pezzolato M, Peletto S, Begovoeva M, Rossi F. Identification of Mycoplasma Species in Cattle Associated with Bovine Respiratory Disease Mortality. Microorganisms. 2024; 12(11):2340. https://doi.org/10.3390/microorganisms12112340
Chicago/Turabian StyleCarella, Emanuele, Erika Messana, Davide Mugetti, Elena Biasibetti, Marzia Pezzolato, Simone Peletto, Mattia Begovoeva, and Francesca Rossi. 2024. "Identification of Mycoplasma Species in Cattle Associated with Bovine Respiratory Disease Mortality" Microorganisms 12, no. 11: 2340. https://doi.org/10.3390/microorganisms12112340
APA StyleCarella, E., Messana, E., Mugetti, D., Biasibetti, E., Pezzolato, M., Peletto, S., Begovoeva, M., & Rossi, F. (2024). Identification of Mycoplasma Species in Cattle Associated with Bovine Respiratory Disease Mortality. Microorganisms, 12(11), 2340. https://doi.org/10.3390/microorganisms12112340









