Interkingdom Communication via Extracellular Vesicles: Unraveling Plant and Pathogen Interactions and Its Potential for Next-Generation Crop Protection
Abstract
:1. Introduction
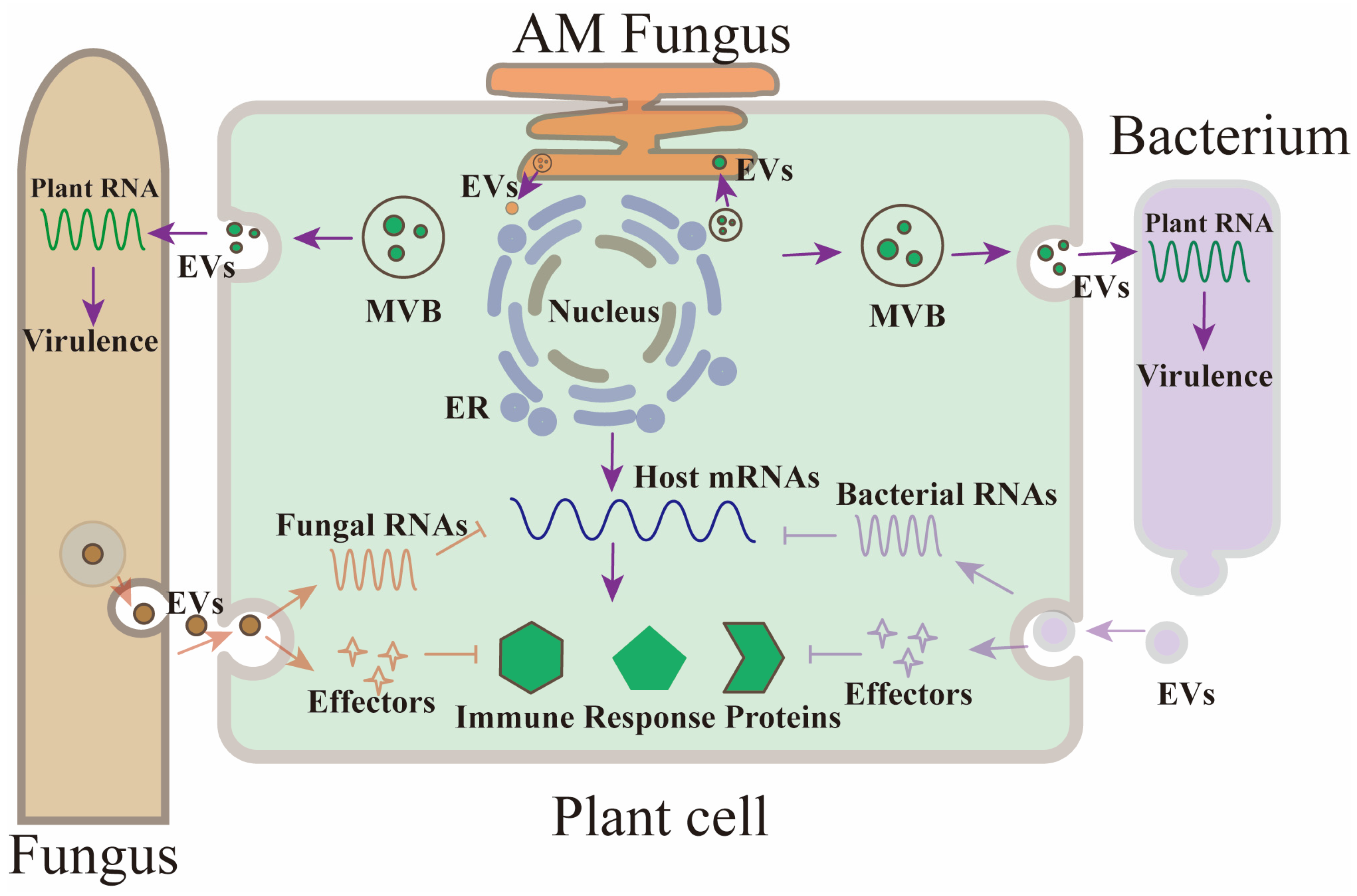
2. Plant Extracellular Vesicles
3. Extracellular Vesicles in Mediating Small RNA Exchange and Plant Defense Mechanisms
3.1. Cross-Kingdom sRNA Trafficking via Plant Extracellular Vesicles
3.2. Plant RNA-Binding Proteins and Their Role in sRNA Loading into Extracellular Vesicles
3.3. Pathogen-to-Host sRNA Trafficking and Its Role in Suppressing Plant Immunity
4. Role of Extracellular Vesicles in Plant Defense Mechanisms
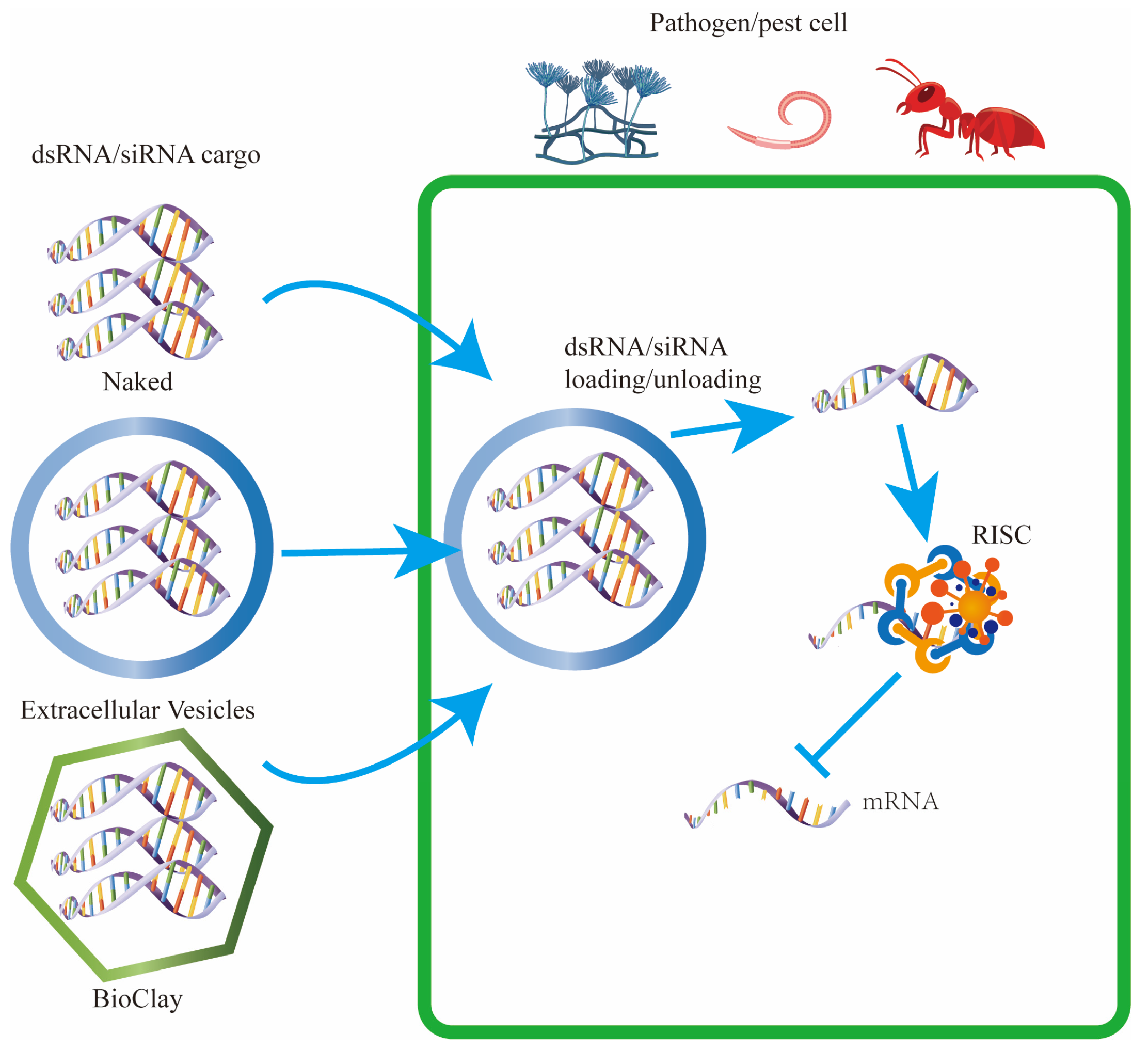
5. Utilizing Extracellular Vesicles for RNA Interference in Crop Protection
5.1. Mechanisms of Environmental RNA Uptake Across Species
5.2. Utilizing Extracellular Vesicles and Nanoparticles for RNA Interference in Agricultural Applications
5.3. Enhancing RNAi Delivery in Plant Protection Using Extracellular Vesicles
5.4. Extracellular Vesicles as Versatile Carriers for Diverse Beneficial Cargos
6. Conclusions: The Dynamic Role of Extracellular Vesicles in Plant–Pathogen Interactions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tabassum, N.; Blilou, I. Cell-to-Cell Communication During Plant-Pathogen Interaction. Mol. Plant-Microbe Interact. 2022, 35, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Halilovic, L.; Shi, T.; Chen, A.; He, B.; Wu, H.; Jin, H. Extracellular Vesicles: Cross-Organismal RNA Trafficking in Plants, Microbes, and Mammalian Cells. Extracell. Vesicles Circ. Nucleic Acids 2023, 4, 262–282. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Xue, Y.; Duan, Y.; Mao, C.; Wan, M. Extracellular Vesicles and Their Engineering Strategies, Delivery Systems, and Biomedical Applications. J. Control. Release 2024, 365, 1089–1123. [Google Scholar] [CrossRef]
- Cai, Q.; He, B.; Weiberg, A.; Buck, A.H.; Jin, H. Small RNAs and Extracellular Vesicles: New Mechanisms of Cross-Species Communication and Innovative Tools for Disease Control. PLoS Pathog. 2019, 15, e1008090. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-Y.; Wang, H.; Hu, P.; Hamby, R.; Jin, H. Small RNAs—Big Players in Plant-Microbe Interactions. Cell Host Microbe 2019, 26, 173–182. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Wang, H.; Liu, G.; Chen, A.; Calvo, A.; Cai, Q.; Jin, H. Fungal Small RNAs Ride in Extracellular Vesicles to Enter Plant Cells through Clathrin-Mediated Endocytosis. Nat. Commun. 2023, 14, 4383. [Google Scholar] [CrossRef]
- Chen, A.; Halilovic, L.; Shay, J.-H.; Koch, A.; Mitter, N.; Jin, H. Improving RNA-Based Crop Protection through Nanotechnology and Insights from Cross-Kingdom RNA Trafficking. Curr. Opin. Plant Biol. 2023, 76, 102441. [Google Scholar] [CrossRef]
- Wang, Z.; Zeng, J.; Deng, J.; Hou, X.; Zhang, J.; Yan, W.; Cai, Q. Pathogen-Derived Extracellular Vesicles: Emerging Mediators of Plant-Microbe Interactions. Mol. Plant-Microbe Interact. 2023, 36, 218–227. [Google Scholar] [CrossRef]
- Kusch, S.; Singh, M.; Thieron, H.; Spanu, P.D.; Panstruga, R. Site-specific Analysis Reveals Candidate Cross-kingdom Small RNAs, tRNA and rRNA Fragments, and Signs of Fungal RNA Phasing in the Barley–Powdery Mildew Interaction. Mol. Plant Pathol. 2023, 24, 570–587. [Google Scholar] [CrossRef]
- Liu, G.; Kang, G.; Wang, S.; Huang, Y.; Cai, Q. Extracellular Vesicles: Emerging Players in Plant Defense Against Pathogens. Front. Plant Sci. 2021, 12, 757925. [Google Scholar] [CrossRef]
- Van Niel, G.; Carter, D.R.F.; Clayton, A.; Lambert, D.W.; Raposo, G.; Vader, P. Challenges and Directions in Studying Cell–Cell Communication by Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2022, 23, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef] [PubMed]
- Crescitelli, R.; Lässer, C.; Szabó, T.G.; Kittel, A.; Eldh, M.; Dianzani, I.; Buzás, E.I.; Lötvall, J. Distinct RNA Profiles in Subpopulations of Extracellular Vesicles: Apoptotic Bodies, Microvesicles and Exosomes. J. Extracell. Vesicles 2013, 2, 20677. [Google Scholar] [CrossRef] [PubMed]
- Bielska, E.; May, R.C. Extracellular Vesicles of Human Pathogenic Fungi. Curr. Opin. Microbiol. 2019, 52, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Bajan, S.; Hutvagner, G. RNA-Based Therapeutics: From Antisense Oligonucleotides to miRNAs. Cells 2020, 9, 137. [Google Scholar] [CrossRef]
- Williams, S.; Fernandez-Rhodes, M.; Law, A.; Peacock, B.; Lewis, M.P.; Davies, O.G. Comparison of Extracellular Vesicle Isolation Processes for Therapeutic Applications. J. Tissue Eng. 2023, 14, 204173142311746. [Google Scholar] [CrossRef]
- Zhang, Y.; Dou, Y.; Liu, Y.; Di, M.; Bian, H.; Sun, X.; Yang, Q. Advances in Therapeutic Applications of Extracellular Vesicles. Int. J. Nanomed. 2023, 18, 3285–3307. [Google Scholar] [CrossRef]
- Alzahrani, F.A.; Khan, M.I.; Kameli, N.; Alsahafi, E.; Riza, Y.M. Plant-Derived Extracellular Vesicles and Their Exciting Potential as the Future of Next-Generation Drug Delivery. Biomolecules 2023, 13, 839. [Google Scholar] [CrossRef]
- Freitas, M.S.; Bonato, V.L.D.; Pessoni, A.M.; Rodrigues, M.L.; Casadevall, A.; Almeida, F. Fungal Extracellular Vesicles as Potential Targets for Immune Interventions. mSphere 2019, 4, e00747-19. [Google Scholar] [CrossRef]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.-M.; Palmquist, J.; Huang, S.-D.; Jin, H. Plants Send Small RNAs in Extracellular Vesicles to Fungal Pathogen to Silence Virulence Genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef]
- Jiang, G.; Zhang, Y.; Chen, M.; Ramoneda, J.; Han, L.; Shi, Y.; Peyraud, R.; Wang, Y.; Shi, X.; Chen, X.; et al. Effects of Plant Tissue Permeability on Invasion and Population Bottlenecks of a Phytopathogen. Nat. Commun. 2024, 15, 62. [Google Scholar] [CrossRef] [PubMed]
- Olive, A.J.; Sassetti, C.M. Metabolic Crosstalk between Host and Pathogen: Sensing, Adapting and Competing. Nat. Rev. Microbiol. 2016, 14, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; He, B.; Wu, H.; Cai, Q.; Ramírez-Sánchez, O.; Abreu-Goodger, C.; Birch, P.R.J.; Jin, H. Plant mRNAs Move into a Fungal Pathogen via Extracellular Vesicles to Reduce Infection. Cell Host Microbe 2024, 32, 93–105.e6. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Ma, K.; Hu, H.; Xing, X.; Huang, X.; Gao, H. Extracellular Vesicles: Their Functions in Plant–Pathogen Interactions. Mol. Plant Pathol. 2022, 23, 760–771. [Google Scholar] [CrossRef] [PubMed]
- Ambrosone, A.; Barbulova, A.; Cappetta, E.; Cillo, F.; De Palma, M.; Ruocco, M.; Pocsfalvi, G. Plant Extracellular Vesicles: Current Landscape and Future Directions. Plants 2023, 12, 4141. [Google Scholar] [CrossRef] [PubMed]
- De Palma, M.; Ambrosone, A.; Leone, A.; Del Gaudio, P.; Ruocco, M.; Turiák, L.; Bokka, R.; Fiume, I.; Tucci, M.; Pocsfalvi, G. Plant Roots Release Small Extracellular Vesicles with Antifungal Activity. Plants 2020, 9, 1777. [Google Scholar] [CrossRef]
- Lian, M.Q.; Chng, W.H.; Liang, J.; Yeo, H.Q.; Lee, C.K.; Belaid, M.; Tollemeto, M.; Wacker, M.G.; Czarny, B.; Pastorin, G. Plant-derived Extracellular Vesicles: Recent Advancements and Current Challenges on Their Use for Biomedical Applications. J. Extracell. Vesicles 2022, 11, 12283. [Google Scholar] [CrossRef]
- Rome, S. Biological Properties of Plant-Derived Extracellular Vesicles. Food Funct. 2019, 10, 529–538. [Google Scholar] [CrossRef]
- Liu, N.-J.; Wang, N.; Bao, J.-J.; Zhu, H.-X.; Wang, L.-J.; Chen, X.-Y. Lipidomic Analysis Reveals the Importance of GIPCs in Arabidopsis Leaf Extracellular Vesicles. Mol. Plant 2020, 13, 1523–1532. [Google Scholar] [CrossRef]
- Holland, S.; Roth, R. Extracellular Vesicles in the Arbuscular Mycorrhizal Symbiosis: Current Understanding and Future Perspectives. Mol. Plant-Microbe Interact. 2023, 36, 235–244. [Google Scholar] [CrossRef]
- Roth, R.; Hillmer, S.; Funaya, C.; Chiapello, M.; Schumacher, K.; Lo Presti, L.; Kahmann, R.; Paszkowski, U. Arbuscular Cell Invasion Coincides with Extracellular Vesicles and Membrane Tubules. Nat. Plants 2019, 5, 204–211. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Cai, Q.; Qiao, L.; Huang, C.-Y.; Wang, S.; Miao, W.; Ha, T.; Wang, Y.; Jin, H. RNA-Binding Proteins Contribute to Small RNA Loading in Plant Extracellular Vesicles. Nat. Plants 2021, 7, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Ruf, A.; Oberkofler, L.; Robatzek, S.; Weiberg, A. Spotlight on Plant RNA-Containing Extracellular Vesicles. Curr. Opin. Plant Biol. 2022, 69, 102272. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Jimenez, S.; Hashimoto, K.; Santana, O.; Aguirre, J.; Kuchitsu, K.; Cárdenas, L. Emerging Roles of Tetraspanins in Plant Inter-Cellular and Inter-Kingdom Communication. Plant Signal. Behav. 2019, 14, e1581559. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Qiao, Q.; Sun, Y.; Xu, Y.; Shu, H.; Zhang, Z.; Liu, F.; Wang, H.; Ye, W.; Dong, S.; et al. Divergent Sequences of Tetraspanins Enable Plants to Specifically Recognize Microbe-Derived Extracellular Vesicles. Nat. Commun. 2023, 14, 4877. [Google Scholar] [CrossRef]
- Huang, Y. Effective Methods for Isolation and Purification of Extracellular Vesicles from Plants. J. Integr. Plant Biol. 2021, 63, 2020–2030. [Google Scholar] [CrossRef]
- Koch, B.L.; Rutter, B.D.; Innes, R.W. Arabidopsis Produces Distinct Subpopulations of Extracellular Vesicles That Respond Differentially to Biotic Stress. bioRxiv 2024. [Google Scholar] [CrossRef]
- Mo, Z.; Cheong, J.Y.A.; Xiang, L.; Le, M.T.N.; Grimson, A.; Zhang, D.X. Extracellular Vesicle-associated Organotropic Metastasis. Cell Prolif. 2021, 54, e12948. [Google Scholar] [CrossRef]
- Nielsen, M.E.; Feechan, A.; Böhlenius, H.; Ueda, T.; Thordal-Christensen, H. Arabidopsis ARF-GTP Exchange Factor, GNOM, Mediates Transport Required for Innate Immunity and Focal Accumulation of Syntaxin PEN1. Proc. Natl. Acad. Sci. USA 2012, 109, 11443–11448. [Google Scholar] [CrossRef]
- Wang, J.; Ding, Y.; Wang, J.; Hillmer, S.; Miao, Y.; Lo, S.W.; Wang, X.; Robinson, D.G.; Jiang, L. EXPO, an Exocyst-Positive Organelle Distinct from Multivesicular Endosomes and Autophagosomes, Mediates Cytosol to Cell Wall Exocytosis in Arabidopsis and Tobacco Cells C W. Plant Cell 2010, 22, 4009–4030. [Google Scholar] [CrossRef]
- Poulsen, C.P.; Dilokpimol, A.; Mouille, G.; Burow, M.; Geshi, N. Arabinogalactan Glycosyltransferases Target to a Unique Subcellular Compartment That May Function in Unconventional Secretion in Plants. Traffic 2014, 15, 1219–1234. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Li, D.; Gu, Y.; Liu, R.; Tang, X.; Zhao, Y.; Qi, F.; Wei, J.; Liu, J. Plant-Derived Nanovesicles: Further Exploration of Biomedical Function and Application Potential. Acta Pharm. Sin. B 2023, 13, 3300–3320. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Wang, Y.; Liu, J.; Zhao, J.; Sun, P.; Wang, S. A Fungal Pathogen Deploys a Small Silencing RNA That Attenuates Mosquito Immunity and Facilitates Infection. Nat. Commun. 2019, 10, 4298. [Google Scholar] [CrossRef] [PubMed]
- Dunker, F.; Trutzenberg, A.; Rothenpieler, J.S.; Kuhn, S.; Pröls, R.; Schreiber, T.; Tissier, A.; Kemen, A.; Kemen, E.; Hückelhoven, R.; et al. Oomycete Small RNAs Bind to the Plant RNA-Induced Silencing Complex for Virulence. Elife 2020, 9, e56096. [Google Scholar] [CrossRef] [PubMed]
- Parperides, E.; El Mounadi, K.; Garcia-Ruiz, H. Induction and Suppression of Gene Silencing in Plants by Nonviral Microbes. Mol. Plant Pathol. 2023, 24, 1347–1356. [Google Scholar] [CrossRef]
- Kong, X.; Yang, M.; Le, B.H.; He, W.; Hou, Y. The Master Role of siRNAs in Plant Immunity. Mol. Plant Pathol. 2022, 23, 1565–1574. [Google Scholar] [CrossRef]
- Bilir, Ö.; Göl, D.; Hong, Y.; McDowell, J.M.; Tör, M. Small RNA-Based Plant Protection against Diseases. Front. Plant Sci. 2022, 13, 951097. [Google Scholar] [CrossRef]
- Weiberg, A.; Wang, M.; Lin, F.-M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.-D.; Jin, H. Fungal Small RNAs Suppress Plant Immunity by Hijacking Host RNA Interference Pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef]
- Wang, M.; Weiberg, A.; Dellota, E.; Yamane, D.; Jin, H. Botrytis Small RNA Bc-siR37 Suppresses Plant Defense Genes by Cross-Kingdom RNAi. RNA Biol. 2017, 14, 421–428. [Google Scholar] [CrossRef]
- Wang, B.; Sun, Y.; Song, N.; Zhao, M.; Liu, R.; Feng, H.; Wang, X.; Kang, Z. Puccinia Striiformis f. Sp. Tritici Mi croRNA -like RNA 1 (Pst -milR1), an Important Pathogenicity Factor of Pst, Impairs Wheat Resistance to Pst by Suppressing the Wheat Pathogenesis-related 2 Gene. New Phytol. 2017, 215, 338–350. [Google Scholar] [CrossRef]
- Shahid, S.; Kim, G.; Johnson, N.R.; Wafula, E.; Wang, F.; Coruh, C.; Bernal-Galeano, V.; Phifer, T.; dePamphilis, C.W.; Westwood, J.H.; et al. MicroRNAs from the Parasitic Plant Cuscuta Campestris Target Host Messenger RNAs. Nature 2018, 553, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Mao, H.; Li, S.; Feng, T.; Zhang, Z.; Cheng, L.; Luo, S.; Borkovich, K.A.; Ouyang, S. Fol-milR1, a Pathogenicity Factor of Fusarium Oxysporum, Confers Tomato Wilt Disease Resistance by Impairing Host Immune Responses. New Phytol. 2021, 232, 705–718. [Google Scholar] [CrossRef] [PubMed]
- Mueth, N.A.; Hulbert, S.H. Small RNAs Target Native and Cross-Kingdom Transcripts on Both Sides of the Wheat Stripe Rust Interaction. Genomics 2022, 114, 110526. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Li, G.; Guo, Y.; Gao, Y.; Zhu, L.; Liu, Z.; Tian, R.; Gao, C.; Han, P.; Wang, N.; et al. A Fungal microRNA-like RNA Subverts Host Immunity and Facilitates Pathogen Infection by Silencing Two Host Receptor-like Kinase Genes. New Phytol. 2022, 233, 2503–2519. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, S.; Wang, P.; Nie, W.; Ahmad, I.; Chen, G.; Zhu, B. Potentiation of Host Defense through sRNA Packaged in OMVs of Xanthomonas oryzae pv. oryzicola. bioRxiv 2023. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.-L.; Zhao, J.-H.; Wang, S.; Jin, Y.; Chen, Z.-Q.; Fang, Y.-Y.; Hua, C.-L.; Ding, S.-W.; Guo, H.-S. Cotton Plants Export microRNAs to Inhibit Virulence Gene Expression in a Fungal Pathogen. Nat. Plants 2016, 2, 16153. [Google Scholar] [CrossRef]
- Zhu, C.; Liu, J.-H.; Zhao, J.-H.; Liu, T.; Chen, Y.-Y.; Wang, C.-H.; Zhang, Z.-H.; Guo, H.-S.; Duan, C.-G. A Fungal Effector Suppresses the Nuclear Export of AGO1–miRNA Complex to Promote Infection in Plants. Proc. Natl. Acad. Sci. USA 2022, 119, e2114583119. [Google Scholar] [CrossRef]
- Cheng, W.; Lin, M.; Chu, M.; Xiang, G.; Guo, J.; Jiang, Y.; Guan, D.; He, S. RNAi-Based Gene Silencing of RXLR Effectors Protects Plants Against the Oomycete Pathogen. Mol. Plant-Microbe Interact. 2022, 35, 440–449. [Google Scholar] [CrossRef]
- Baldrich, P.; Rutter, B.D.; Karimi, H.Z.; Podicheti, R.; Meyers, B.C.; Innes, R.W. Plant Extracellular Vesicles Contain Diverse Small RNA Species and Are Enriched in 10- to 17-Nucleotide “Tiny” RNAs. Plant Cell 2019, 31, 315–324. [Google Scholar] [CrossRef]
- Bleackley, M.R.; Samuel, M.; Garcia-Ceron, D.; McKenna, J.A.; Lowe, R.G.T.; Pathan, M.; Zhao, K.; Ang, C.-S.; Mathivanan, S.; Anderson, M.A. Extracellular Vesicles from the Cotton Pathogen Fusarium Oxysporum f. Sp. Vasinfectum Induce a Phytotoxic Response in Plants. Front. Plant Sci. 2020, 10, 1610. [Google Scholar] [CrossRef]
- Jiao, J. Wheat microRNA1023 Suppresses Invasion of Fusarium Graminearum via Targeting and Silencing FGSG_03101. J. Plant Interact. 2018, 13, 514–521. [Google Scholar] [CrossRef]
- Wang, M.; Weiberg, A.; Lin, F.-M.; Thomma, B.P.H.J.; Huang, H.-D.; Jin, H. Bidirectional Cross-Kingdom RNAi and Fungal Uptake of External RNAs Confer Plant Protection. Nat. Plants 2016, 2, 16151. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Jha, S.K.; Prabhu, K.V.; Kumar, M.; Mukhopadhyay, K. Leaf Rust (Puccinia Triticina) Mediated RNAi in Wheat (Triticum Aestivum L.) Prompting Host Susceptibility. Funct. Integr. Genom. 2019, 19, 437–452. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Wang, X.; Duan, J.; Ma, J. Rhizobial tRNA-Derived Small RNAs Are Signal Molecules Regulating Plant Nodulation. Science 2019, 365, 919–922. [Google Scholar] [CrossRef] [PubMed]
- Buck, A.H.; Coakley, G.; Simbari, F.; McSorley, H.J.; Quintana, J.F.; Le Bihan, T.; Kumar, S.; Abreu-Goodger, C.; Lear, M.; Harcus, Y.; et al. Exosomes Secreted by Nematode Parasites Transfer Small RNAs to Mammalian Cells and Modulate Innate Immunity. Nat. Commun. 2014, 5, 5488. [Google Scholar] [CrossRef]
- Qiao, S.A.; Gao, Z.; Roth, R. A Perspective on Cross-kingdom RNA Interference in Mutualistic Symbioses. New Phytol. 2023, 240, 68–79. [Google Scholar] [CrossRef]
- Jokhio, S.; Peng, I.; Peng, C.-A. Extracellular Vesicles Isolated from Arabidopsis Thaliana Leaves Reveal Characteristics of Mammalian Exosomes. Protoplasma 2024, 261, 1025–1033. [Google Scholar] [CrossRef]
- Rutter, B.D.; Innes, R.W. Extracellular Vesicles Isolated from the Leaf Apoplast Carry Stress-Response Proteins1. Plant Physiol. 2017, 173, 728–741. [Google Scholar] [CrossRef]
- Movahed, N.; Cabanillas, D.G.; Wan, J.; Vali, H.; Laliberté, J.-F.; Zheng, H. Turnip Mosaic Virus Components Are Released into the Extracellular Space by Vesicles in Infected Leaves. Plant Physiol. 2019, 180, 1375–1388. [Google Scholar] [CrossRef]
- Wang, M.; Dean, R.A. Movement of Small RNAs in and between Plants and Fungi. Mol. Plant Pathol. 2020, 21, 589–601. [Google Scholar] [CrossRef]
- Koch, A.; Wassenegger, M. Host-induced Gene Silencing—Mechanisms and Applications. New Phytol. 2021, 231, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Bachman, P.M.; Huizinga, K.M.; Jensen, P.D.; Mueller, G.; Tan, J.; Uffman, J.P.; Levine, S.L. Ecological Risk Assessment for DvSnf7 RNA: A Plant-Incorporated Protectant with Targeted Activity against Western Corn Rootworm. Regul. Toxicol. Pharmacol. 2016, 81, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Ricci, A.; Sabbadini, S.; Miozzi, L.; Mezzetti, B.; Noris, E. Host-Induced Gene Silencing and Spray-Induced Gene Silencing for Crop Protection against Viruses. In RNAi for Plant Improvement and Protection; Mezzetti, B., Sweet, J., Burgos, L., Eds.; CABI: Wallingford, UK, 2021; pp. 72–85. ISBN 978-1-78924-889-0. [Google Scholar]
- Dalakouras, A.; Wassenegger, M.; Dadami, E.; Ganopoulos, I.; Pappas, M.L.; Papadopoulou, K. Genetically Modified Organism-Free RNA Interference: Exogenous Application of RNA Molecules in Plants. Plant Physiol. 2020, 182, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Tatineni, S.; Hein, G.L. Plant Viruses of Agricultural Importance: Current and Future Perspectives of Virus Disease Management Strategies. Phytopathol. 2023, 113, 117–141. [Google Scholar] [CrossRef]
- Lim, F.-H. The Future Is Now: Revolution of RNA-Mediated Gene Silencing in Plant Protection against Insect Pests and Diseases. Plant Biotechnol. Rep. 2020, 14, 643–662. [Google Scholar] [CrossRef]
- Chen, X.; Koo, J.; Gurusamy, D.; Mogilicherla, K.; Reddy Palli, S. Caenorhabditis Elegans Systemic RNA Interference Defective Protein 1 Enhances RNAi Efficiency in a Lepidopteran Insect, the Fall Armyworm, in a Tissue-Specific Manner. RNA Biol. 2021, 18, 1291–1299. [Google Scholar] [CrossRef]
- Shen, X.; Peng, Y.; Song, H.; Wang, J.; Zhao, J.; Tang, P.; Han, Z.; Wang, K. Key Factors Determining Competitions between Double-Stranded RNAs in Tribolium Castaneum. Pestic. Biochem. Physiol. 2022, 181, 105009. [Google Scholar] [CrossRef]
- Marzano, S.-Y.L.; Beligala, G.; Mukherjee, S.; Feng, C. Double-Stranded RNA Targeting White Mold Sclerotinia Sclerotiorum Argonaute 2 for Disease Control via Spray-Induced Gene Silencing 2023. Available online: https://www.researchsquare.com/article/rs-3359704/v2 (accessed on 1 March 2024).
- Wytinck, N.; Sullivan, D.S.; Biggar, K.T.; Crisostomo, L.; Pelka, P.; Belmonte, M.F.; Whyard, S. Clathrin Mediated Endocytosis Is Involved in the Uptake of Exogenous Double-Stranded RNA in the White Mold Phytopathogen Sclerotinia Sclerotiorum. Sci. Rep. 2020, 10, 12773. [Google Scholar] [CrossRef]
- Qiao, L.; Lan, C.; Capriotti, L.; Ah-Fong, A.; Nino Sanchez, J.; Hamby, R.; Heller, J.; Zhao, H.; Glass, N.L.; Judelson, H.S.; et al. Spray-induced Gene Silencing for Disease Control Is Dependent on the Efficiency of Pathogen RNA Uptake. Plant Biotechnol. J. 2021, 19, 1756–1768. [Google Scholar] [CrossRef]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA Delivery by Extracellular Vesicles in Mammalian Cells and Its Applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Niño-Sánchez, J.; Sambasivam, P.T.; Sawyer, A.; Hamby, R.; Chen, A.; Czislowski, E.; Li, P.; Manzie, N.; Gardiner, D.M.; Ford, R.; et al. BioClayTM Prolongs RNA Interference-Mediated Crop Protection against Botrytis Cinerea. J. Integr. Plant Biol. 2022, 64, 2187–2198. [Google Scholar] [CrossRef]
- Jain, R.G. Foliar Application of Clay-Delivered RNA Interference for Whitefly Control. Nat. Plants 2022, 8, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Peng, Y.; Chen, J.; Peng, Y.; Wang, X.; Shen, Z.; Han, Z. Comparison of Efficacy of RNAi Mediated by Various Nanoparticles in the Rice Striped Stem Borer (Chilo Suppressalis). Pestic. Biochem. Physiol. 2020, 165, 104467. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Yin, M.-Z.; Shen, J. Nanoparticle-Based Nontransformative RNA Insecticides for Sustainable Pest Control: Mechanisms, Current Status and Challenges. Entomol. Gen. 2022, 43, 21–30. [Google Scholar] [CrossRef]
- De Paula, R.G.; Antoniêto, A.C.C.; Nogueira, K.M.V.; Ribeiro, L.F.C.; Rocha, M.C.; Malavazi, I.; Almeida, F.; Silva, R.N. Extracellular Vesicles Carry Cellulases in the Industrial Fungus Trichoderma Reesei. Biotechnol. Biofuels 2019, 12, 146. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, N.L.; Smagghe, G.; Sharma, R.; Oliveira, E.E.; Christiaens, O. Liposome Encapsulation and EDTA Formulation of dsRNA Targeting Essential Genes Increase Oral RNAi-caused Mortality in the Neotropical Stink Bug Euschistus Heros. Pest. Manag. Sci. 2019, 75, 537–548. [Google Scholar] [CrossRef]
- Tayler, A.; Heschuk, D.; Giesbrecht, D.; Park, J.Y.; Whyard, S. Efficiency of RNA Interference Is Improved by Knockdown of dsRNA Nucleases in Tephritid Fruit Flies. Open Biol. 2019, 9, 190198. [Google Scholar] [CrossRef]
- Schlemmer, T. Extracellular Vesicles Isolated from dsRNA-Sprayed Barley Plants Exhibit No Growth Inhibition or Gene Silencing in Fusarium Graminearum. Fungal Biol. Biotechnol. 2022, 9, 14. [Google Scholar] [CrossRef]
- Demirer, G.S.; Zhang, H.; Goh, N.S.; González-Grandío, E.; Landry, M.P. Carbon Nanotube–Mediated DNA Delivery without Transgene Integration in Intact Plants. Nat. Protoc. 2019, 14, 2954–2971. [Google Scholar] [CrossRef]
- Ahmadi, M.; Abbasi, R.; Rezaie, J. Tumor Immune Escape: Extracellular Vesicles Roles and Therapeutics Application. Cell Commun. Signal. 2024, 22, 9. [Google Scholar] [CrossRef]
- Li, Y. Comparative Study of Extracellular Vesicles Derived from Mesenchymal Stem Cells and Brain Endothelial Cells Attenuating Blood–Brain Barrier Permeability via Regulating Caveolin-1-Dependent ZO-1 and Claudin-5 Endocytosis in Acute Ischemic Stroke. J. Nanobiotechnol. 2023, 21, 70. [Google Scholar] [CrossRef] [PubMed]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal Information for Studies of Extracellular Vesicles (MISEV2023): From Basic to Advanced Approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef] [PubMed]
| Host Species | Parasite/Mutualist Microbe | EV Cargo with Biological Activity | Reference |
|---|---|---|---|
| Arabidopsis thaliana | Botrytis cinerea | 21 nt sRNAs Bc-siR3.1 Bc-siR3.2 Bc-siR5 | [48] |
| Solanum lycopersicum | Botrytis cinerea | 21 nt sRNA Bc-siR5 | [48] |
| Arabidopsis thaliana | Botrytis cinerea | Bc-siR37 | [49] |
| Triticum aestivum | Puccinia striiformis f. sp. tritici | miRNA-like (milR1) | [50] |
| Arabidopsis thaliana | Cuscuta campestris | 22 nt miRNAs e.g., miR393 | [51] |
| Solanum lycopersicum | Fusarium oxysporum f. sp. lycopersici | 23 nt miRNA-like Fol-milR1 | [52] |
| Triticum aestivum | Puccinia striiformis f.sp. tritici | 17 20–21 nt sRNAs | [53] |
| Malus × domestica | Valsa mali | miRNA-like Vm-milR1 | [54] |
| Oryza sativa | Xanthomonas oryzae pv. oryzicola | Xosr001 | [55] |
| Gossypium hirsutum | Verticillium dahliae | miR166/miR159 | [56] |
| Hordeum vulgare | Blumeria hordei | [9] | |
| Arabidopsis thaliana | Verticillium dahliae | miR166/miR159 | [57] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Lu, Y.; Xi, K.; Li, Y.; Chen, X.; Wang, P.; Huang, X. Interkingdom Communication via Extracellular Vesicles: Unraveling Plant and Pathogen Interactions and Its Potential for Next-Generation Crop Protection. Microorganisms 2024, 12, 2392. https://doi.org/10.3390/microorganisms12122392
Li F, Lu Y, Xi K, Li Y, Chen X, Wang P, Huang X. Interkingdom Communication via Extracellular Vesicles: Unraveling Plant and Pathogen Interactions and Its Potential for Next-Generation Crop Protection. Microorganisms. 2024; 12(12):2392. https://doi.org/10.3390/microorganisms12122392
Chicago/Turabian StyleLi, Fei, Yuntong Lu, Kuanling Xi, Yuke Li, Xiaoyan Chen, Puchang Wang, and Xiaolong Huang. 2024. "Interkingdom Communication via Extracellular Vesicles: Unraveling Plant and Pathogen Interactions and Its Potential for Next-Generation Crop Protection" Microorganisms 12, no. 12: 2392. https://doi.org/10.3390/microorganisms12122392
APA StyleLi, F., Lu, Y., Xi, K., Li, Y., Chen, X., Wang, P., & Huang, X. (2024). Interkingdom Communication via Extracellular Vesicles: Unraveling Plant and Pathogen Interactions and Its Potential for Next-Generation Crop Protection. Microorganisms, 12(12), 2392. https://doi.org/10.3390/microorganisms12122392






